You're using an outdated browser. Please upgrade to a modern browser for the best experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Fabian Erdel | -- | 1633 | 2023-08-08 12:11:39 | | | |
| 2 | Conner Chen | Meta information modification | 1633 | 2023-08-10 04:31:11 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Hertzog, M.; Erdel, F. The Cell Nucleus as a Multiscale Porous Medium. Encyclopedia. Available online: https://encyclopedia.pub/entry/47782 (accessed on 26 December 2025).
Hertzog M, Erdel F. The Cell Nucleus as a Multiscale Porous Medium. Encyclopedia. Available at: https://encyclopedia.pub/entry/47782. Accessed December 26, 2025.
Hertzog, Maud, Fabian Erdel. "The Cell Nucleus as a Multiscale Porous Medium" Encyclopedia, https://encyclopedia.pub/entry/47782 (accessed December 26, 2025).
Hertzog, M., & Erdel, F. (2023, August 08). The Cell Nucleus as a Multiscale Porous Medium. In Encyclopedia. https://encyclopedia.pub/entry/47782
Hertzog, Maud and Fabian Erdel. "The Cell Nucleus as a Multiscale Porous Medium." Encyclopedia. Web. 08 August, 2023.
Copy Citation
Chromatin regulatory processes physically take place in the environment of the cell nucleus, which is filled with the chromosomes and a plethora of smaller biomolecules. The nucleus contains macromolecular assemblies of different sizes, from nanometer-sized protein complexes to micrometer-sized biomolecular condensates, chromosome territories, and nuclear bodies. This multiscale organization impacts the transport processes within the nuclear interior, the global mechanical properties of the nucleus, and the way the nucleus senses and reacts to mechanical stimuli.
cell nucleus
multiscale organization
scale-dependent maerial properties
rheology
viscoelasticity
1. Introduction
The mammalian cell nucleus is the largest cellular organelle that harbors our genetic information in the form of chromosomes. It plays important roles in safeguarding genetic information and controlling its interpretation by the transcriptional machinery. To accomplish these activities, the structural organization of the genome is regulated on different levels. Inactive parts of the chromosomes are packaged into more compact, condensed domains, while active parts are less compact [1][2]. A plethora of proteins and RNA molecules are responsible for regulating this structural and functional partitioning. On the one hand, multivalent proteins can establish bridging interactions among different parts of the chromosomes and thereby package them into compact domains. On the other hand, proteins and/or RNA molecules can form condensates via liquid–liquid phase separation (LLPS), which encloses certain parts of the chromosomes and excludes others [3][4]. Active processes, including transcription or loop extrusion by proteins of the structural maintenance of chromosomes (SMC) family, might further shape the folding of chromosomes [5][6]. These mechanisms do not only drive the structural partitioning of chromosomes but also have consequences for their material properties and the mechanical features of the cell nucleus as a whole [7]. In particular, bridging interactions can make the respective chromosomal region stiffer [8][9], as such interactions have to be broken when the chromosomal conformation is altered by an external force. Bridging interactions can also modulate the permeability and accessibility of the respective chromatin region by sterically restricting access to the so-called interchromatin space between the chromosomes. This effect is conceptually similar to the regulation of the permeability of a gel in electrophoresis applications by the cross-links within the gel [10]. Furthermore, condensates formed by LLPS can also regulate the stiffness and permeability of the nucleus, among their numerous other functions in the cell. They can do so by exerting mechanical forces on chromosomal regions [3][11] and by allowing access for certain molecules while excluding others [12]. Accordingly, there is a link between the three-dimensional organization of the nuclear interior and its rheological as well as mechanical properties, i.e., the way in which the nuclear content flows and deforms under force. This link is likely important in the context of nuclear mechanosensing and mechanotransduction.
From a materials science perspective, the cell nucleus can be considered a multiscale medium because it contains objects of vastly different sizes (Figure 1A); see Table 1 for an incomplete list. At the lower end, there are small nuclear proteins and RNAs of a typical size of a few nanometers. At the upper end, there are the nucleoli, micron-sized nuclear subcompartments containing hundreds of different types of proteins and RNAs at high copy numbers [13], as well as the chromosomes, long molecules that occupy micron-sized territories [14][15]. Within their territories, chromosomes form a porous network that encloses the interchromatin space [3][14][16][17]. Due to this organization, the structural, rheological, and mechanical properties of the nucleus are scale-dependent (Figure 1B). While small displacements or mechanical deformations on the nanoscale, corresponding to the pore size of the chromatin network, will be governed by the properties of the nucleoplasmic fluid that fills the pores of the interchromatin space, large displacements at or beyond the micron scale, corresponding to the size of a nucleolus or chromosome territory, will be affected by the properties of these large nuclear subcomponents. Furthermore, if tracer particles are used to study the rheological or mechanical properties of the nuclear interior, their size will strongly influence the outcome: Small tracers can exchange between pores, whereas large tracers will not be able to do so but will rather stay in a pore and follow the motion of the chromatin network [18]. As discussed below, it is therefore useful to consider the material properties of the nucleus and the associated biological functions in the context of their respective time and length scales.
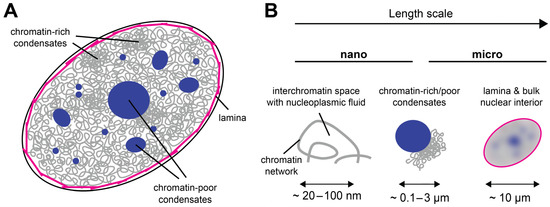
Figure 1. The cell nucleus as a multiscale medium. (A) The cell nucleus is composed of the nuclear lamina (magenta), which resides under the nuclear envelope (black). The nuclear interior contains the chromatin network (gray) and biomolecular condensates that can be chromatin-rich or chromatin-poor (dark blue). (B) The appearance of the cell nucleus is scale-dependent: On the nanoscale, the nuclear interior is partitioned into chromatin and the porous interchromatin space. On the mesoscale, chromatin domains, biomolecular condensates, and nuclear bodies can be distinguished. On the scale of a few microns, the nucleus can be considered a single organelle whose properties depend on the lamina at the periphery and all components in its interior.
Table 1. Typical sizes of nuclear subcomponents. Note that some sizes vary across cell types and species.
| Subcomponent | Size | System | References |
|---|---|---|---|
| Typical protein | 3–10 nm | mammalian cells | [19] |
| Typical RNA 1 | 2–8 nm | human cells | [20] |
| Nucleosome | 10 nm | mammalian cells | [21] |
| Transcriptional condensate | 50–500 nm | human/mouse cells | [22] |
| Compact chromatin domain | 160 nm | human cells (HeLa) | [2] |
| Polycomb body | 0.5–1 μm | mouse embryonic stem cells | [23] |
| Chromocenter | 0.5–1 μm | mouse fibroblasts | [24] |
| Chromosome territory | 1–2 μm | human/mouse cells | [15] |
| Nucleolus | 0.1–5.5 μm | human cells (HeLa) | [25] |
| Nuclear speckle | 1 to a few μm | mammalian cells | [26] |
| PML body 2 | 0.1–1 μm | mammalian cells | [27] |
| Cajal body | 0.5–1 μm | cells of higher eukaryotes | [28] |
1 assuming RNA lengths of 25–2787 nts, the latter being the median mRNA length in human cells [29]. 2 PML, promyelocytic leukemia.
2. The Cell Nucleus as a Multiscale Porous Medium
The nuclear interior can be considered as a porous medium, which is composed of the chromatin network that is soaked in nucleoplasmic fluid [30]. This arrangement is due to the large size of the chromosomes, which sets them apart from the other molecules in the nucleus, e.g., RNAs and proteins, which are much smaller (Table 1) and can therefore move faster. Even small molecules localizing to large nuclear condensates such as the nucleolus typically turn over on a timescale of seconds to minutes [31][32]. In contrast, chromosomes undergo dynamic conformational changes on this timescale but globally stay within their territories over hours [33][34][35]. The organization of the nucleus as a porous medium has consequences for its rheological and mechanical properties. In particular, both transport and stiffness coefficients are scale-dependent. The nanoscale motion of small tracer particles in a pore of the chromatin network will be determined by the properties of the fluid in this pore, while motion on larger scales will involve interactions with the chromatin network and will therefore also depend on the properties of chromatin (Figure 2A), which represents an obstacle that slows down the diffusion of tracers [16][18][36][37]. Similarly, if external forces are very transiently applied on tracers or if an oscillatory force is applied at high frequency so that it acts only briefly in one direction, nanoscale displacements in the pore are obtained, which are governed by the properties of the fluid in this pore (Figure 2B). For longer applications of a unidirectional force or lower frequencies in the case of an oscillatory force, larger displacements involving interactions between tracers and the chromatin network will be triggered, which are also influenced by the properties of chromatin. The rheological and mechanical properties of the nucleus can also be assessed by applying a force from outside, which deforms the nuclear periphery (Figure 2C). Such experiments contain a prominent contribution from the nuclear lamina, which resides at the nuclear periphery and confers mechanical stiffness to the nucleus. For these types of experiments, the outcome will depend on the indentation depth, i.e., the length scale of the induced deformation, which scales with the duration and strength of the applied force.
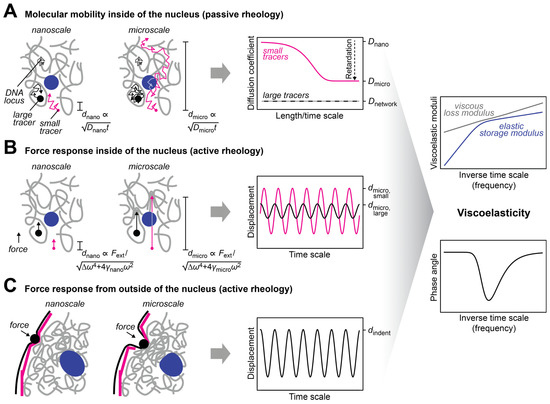
Figure 2. Approaches to measure the mechanical properties of the nucleus. (A) The diffusion coefficient of tracers in the nucleus is scale-dependent. This scale dependence can be used to infer the viscous and elastic properties of the nuclear interior [38] (right, discussed in more detail below). The dependence of the characteristic length scale d of the experiment on the diffusion coefficient D and the observation time t is indicated. Small tracers (magenta) can be used to probe the nucleoplasmic fluid that fills the pores of the chromatin network, while large tracers (black) that are trapped in pores can be used to probe the chromatin network. The latter can also be done with tracers that are attached to chromatin (gray). (B) The scale-dependent force response of tracers in the nucleus can also be used to determine the viscous and elastic properties of the nuclear interior. To this end, an external force has to be selectively applied to the tracers, which can be achieved with magnetic tracers in a magnetic field. The force response will also depend on the size of the tracers and the length of the displacement, which is regulated by the applied force. The dependence of the characteristic length scale d of the experiment on the strength of the external force Fext and the respective frequency ω is indicated. (C) To assess the bulk mechanical properties of the nucleus or its periphery, an external force can be applied from outside of the nucleus by using, for example, aspiration with a micropipette or indentation with a bead/tip. The scale dependence of the response contains information about the viscous and elastic properties of the nuclear lamina (magenta) and the nuclear interior (gray, blue).
Taken together, diffusion coefficients, viscosities, and stiffness coefficients determined on different scales with different experimental protocols represent different types of information about the cell nucleus, as different nuclear subcomponents are probed. This behavior is typical for multiscale porous media that have scale-dependent viscoelastic properties. Further details about the material properties of individual nuclear subcomponents can be found in the associated paper 10.3390/cells12151958.
References
- Boettiger, A.N.; Bintu, B.; Moffitt, J.R.; Wang, S.; Beliveau, B.J.; Fudenberg, G.; Imakaev, M.; Mirny, L.A.; Wu, C.; Zhuang, X. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 2016, 529, 418–422.
- Nozaki, T.; Imai, R.; Tanbo, M.; Nagashima, R.; Tamura, S.; Tani, T.; Joti, Y.; Tomita, M.; Hibino, K.; Kanemaki, M.T.; et al. Dynamic Organization of Chromatin Domains Revealed by Super-Resolution Live-Cell Imaging. Mol. Cell 2017, 67, 282–293.e7.
- Shin, Y.; Chang, Y.-C.; Lee, D.S.W.; Berry, J.; Sanders, D.W.; Ronceray, P.; Wingreen, N.S.; Haataja, M.; Brangwynne, C.P. Liquid nuclear condensates mechanically sense and restructure the genome. Cell 2018, 175, 1481–1491.e13.
- Erdel, F.; Rippe, K. Formation of Chromatin Subcompartments by Phase Separation. Biophys. J. 2018, 114, 2262–2270.
- Mirny, L.A.; Imakaev, M.; Abdennur, N. Two major mechanisms of chromosome organization. Curr. Opin. Cell Biol. 2019, 58, 142–152.
- Quinodoz, S.A.; Guttman, M. Essential roles for RNA in shaping nuclear organization. Cold Spring Harb. Perspect. Biol. 2022, 14, a039719.
- Lee, D.S.W.; Strom, A.R.; Brangwynne, C.P. The mechanobiology of nuclear phase separation. APL Bioeng. 2022, 6, 021503.
- Broedersz, C.P.; MacKintosh, F.C. Modeling semiflexible polymer networks. Rev. Mod. Phys. 2014, 86, 995–1036.
- Strom, A.R.; Biggs, R.J.; Banigan, E.J.; Wang, X.; Chiu, K.; Herman, C.; Collado, J.; Yue, F.; Ritland Politz, J.C.; Tait, L.J.; et al. HP1α is a chromatin crosslinker that controls nuclear and mitotic chromosome mechanics. Elife 2021, 10, e63972.
- Milster, S.; Kim, W.K.; Kanduč, M.; Dzubiella, J. Tuning the permeability of regular polymeric networks by the cross-link ratio. J. Chem. Phys. 2021, 154, 154902.
- Quail, T.; Golfier, S.; Elsner, M.; Ishihara, K.; Murugesan, V.; Renger, R.; Jülicher, F.; Brugués, J. Force generation by protein–DNA co-condensation. Nat. Phys. 2021, 17, 1007–1012.
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298.
- Andersen, J.S.; Lyon, C.E.; Fox, A.H.; Leung, A.K.L.; Lam, Y.W.; Steen, H.; Mann, M.; Lamond, A.I. Directed proteomic analysis of the human nucleolus. Curr. Biol. 2002, 12, 1–11.
- Cremer, T.; Cremer, C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001, 2, 292–301.
- Meaburn, K.J.; Misteli, T. Cell biology: Chromosome territories. Nature 2007, 445, 379–781.
- Fritsch, C.C.; Langowski, J. Anomalous diffusion in the interphase cell nucleus: The effect of spatial correlations of chromatin. J. Chem. Phys. 2010, 133, 025101.
- Ou, H.D.; Phan, S.; Deerinck, T.J.; Thor, A.; Ellisman, M.H.; O’Shea, C.C. ChromEMT: Visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 2017, 357, eaag0025.
- Valet, M.; Rosa, A. Viscoelasticity of model interphase chromosomes. J. Chem. Phys. 2014, 141, 245101.
- Erickson, H.P. Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol. Proced. Online 2009, 11, 32–51.
- Hyeon, C.; Dima, R.I.; Thirumalai, D. Size, shape, and flexibility of RNA structures. J. Chem. Phys. 2006, 125, 194905.
- Richmond, T.J.; Finch, J.T.; Rushton, B.; Rhodes, D.; Klug, A. Structure of the nucleosome core particle at 7 A resolution. Nature 1984, 311, 532–537.
- Forman-Kay, J.D.; Ditlev, J.A.; Nosella, M.L.; Lee, H.O. What are the distinguishing features and size requirements of biomolecular condensates and their implications for RNA-containing condensates? RNA 2022, 28, 36–47.
- Tatavosian, R.; Kent, S.; Brown, K.; Yao, T.; Duc, H.N.; Huynh, T.N.; Zhen, C.Y.; Ma, B.; Wang, H.; Ren, X. Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation. J. Biol. Chem. 2019, 294, 1451–1463.
- Erdel, F.; Rademacher, A.; Vlijm, R.; Tünnermann, J.; Frank, L.; Weinmann, R.; Schweigert, E.; Yserentant, K.; Hummert, J.; Bauer, C.; et al. Mouse Heterochromatin Adopts Digital Compaction States without Showing Hallmarks of HP1-Driven Liquid-Liquid Phase Separation. Mol. Cell 2020, 78, 236–249.e7.
- Caragine, C.M.; Haley, S.C.; Zidovska, A. Nucleolar dynamics and interactions with nucleoplasm in living cells. Elife 2019, 8, e47533.
- Spector, D.L.; Lamond, A.I. Nuclear speckles. Cold Spring Harb. Perspect. Biol. 2011, 3, a000646.
- Lallemand-Breitenbach, V.; de Thé, H. PML nuclear bodies. Cold Spring Harb. Perspect. Biol. 2010, 2, a000661.
- Machyna, M.; Heyn, P.; Neugebauer, K.M. Cajal bodies: Where form meets function. Wiley Interdiscip. Rev. RNA 2013, 4, 17–34.
- Piovesan, A.; Caracausi, M.; Antonaros, F.; Pelleri, M.C.; Vitale, L. GeneBase 1.1: A tool to summarize data from NCBI gene datasets and its application to an update of human gene statistics. Database 2016, 2016, baw153.
- Erdel, F. Biophysical mechanisms of chromatin patterning. Curr. Opin. Genet. Dev. 2020, 61, 62–68.
- Chen, D.; Huang, S. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J. Cell Biol. 2001, 153, 169–176.
- Phair, R.D.; Misteli, T. High mobility of proteins in the mammalian cell nucleus. Nature 2000, 404, 604–609.
- Strickfaden, H.; Zunhammer, A.; van Koningsbruggen, S.; Köhler, D.; Cremer, T. 4D chromatin dynamics in cycling cells: Theodor Boveri’s hypotheses revisited. Nucleus 2010, 1, 284–297.
- Nozaki, T.; Shinkai, S.; Ide, S.; Higashi, K.; Tamura, S.; Shimazoe, M.A.; Nakagawa, M.; Suzuki, Y.; Okada, Y.; Sasai, M.; et al. Condensed but liquid-like domain organization of active chromatin regions in living human cells. Sci. Adv. 2023, 9, eadf1488.
- Wiesmeijer, K.; Krouwels, I.M.; Tanke, H.J.; Dirks, R.W. Chromatin movement visualized with photoactivable GFP-labeled histone H4. Differentiation 2008, 76, 83–90.
- Baum, M.; Erdel, F.; Wachsmuth, M.; Rippe, K. Retrieving the intracellular topology from multi-scale protein mobility mapping in living cells. Nat. Commun. 2014, 5, 4494.
- Görisch, S.M.; Wachsmuth, M.; Tóth, K.F.; Lichter, P.; Rippe, K. Histone acetylation increases chromatin accessibility. J. Cell Sci. 2005, 118 Pt 24, 5825–5834.
- Mason, T.G.; Weitz, D.A. Optical measurements of frequency-dependent linear viscoelastic moduli of complex fluids. Phys. Rev. Lett. 1995, 74, 1250–1253.
More
Information
Subjects:
Biophysics
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
815
Revisions:
2 times
(View History)
Update Date:
10 Aug 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




