Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Muhammad Uzair Javed | -- | 2199 | 2023-08-02 02:00:32 | | | |
| 2 | Camila Xu | Meta information modification | 2199 | 2023-08-02 02:23:57 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Javed, M.U.; Hayat, M.T.; Mukhtar, H.; Imre, K. Applications of CRISPR-Cas9 System. Encyclopedia. Available online: https://encyclopedia.pub/entry/47521 (accessed on 02 March 2026).
Javed MU, Hayat MT, Mukhtar H, Imre K. Applications of CRISPR-Cas9 System. Encyclopedia. Available at: https://encyclopedia.pub/entry/47521. Accessed March 02, 2026.
Javed, Muhammad Uzair, Muhammad Tahir Hayat, Hamid Mukhtar, Kalman Imre. "Applications of CRISPR-Cas9 System" Encyclopedia, https://encyclopedia.pub/entry/47521 (accessed March 02, 2026).
Javed, M.U., Hayat, M.T., Mukhtar, H., & Imre, K. (2023, August 02). Applications of CRISPR-Cas9 System. In Encyclopedia. https://encyclopedia.pub/entry/47521
Javed, Muhammad Uzair, et al. "Applications of CRISPR-Cas9 System." Encyclopedia. Web. 02 August, 2023.
Copy Citation
The CRISPR system was initially identified in the DNA sequences of Escherichia coli bacteria and was described in Osaka University (Japan) in 1987. However, it was not until 2007 that the potential of the CRISPR system for gene editing was realized when the first experimental information about the mechanism of action of the CRISPR system was obtained by two French food scientists named Rodolphe Barrangou and Philippe Horvath working with yogurt cultures of bacteria Streptococcus thermophilus for the Danish company Danisco.
antibiotics resistance genes
target sequence
genome editing
guide RNA
1. Overview of CRISPR Cas9
The CRISPR system was initially identified in the DNA sequences of Escherichia coli bacteria and was described by Ishino et al. [1] of Osaka University (Japan) in 1987. However, it was not until 2007 that the potential of the CRISPR system for gene editing was realized when the first experimental information about the mechanism of action of the CRISPR system was obtained by two French food scientists named Rodolphe Barrangou and Philippe Horvath working with yogurt cultures of bacteria Streptococcus thermophilus for the Danish company Danisco [2]. This system functions by utilizing RNA molecules to direct the protein Cas9 to a specific site in a genome, where it can then cleave the DNA at that location, thereby enabling researchers to precisely add, delete, or modify specific genes [3].
The CRISPR system is primarily acquired through horizontal gene transfer including transformation, conjugation, and transduction. Bacteria and archaea can acquire CRISPR sequences from other bacteria and archaea via this process [4]. Upon acquisition, these sequences become integrated into the genome of the bacterium or archaeon and can be utilized to defend against foreign genetic material, such as viruses. CRISPR-Cas represents an adaptive immune system that is present in the majority of archaeal and bacterial species. This system confers protection against infections caused by viruses, phages, and other foreign genetic elements [5]. Notably, this fascinating system is present in nearly 87% of archaeal genomes and 50% of bacterial genomes [6].
The CRISPR-Cas operons are genetic elements that occur in bacteria and archaea and serve as an adaptive immune system. This system comprises a collection of short, repeated DNA sequences (known as CRISPRs) separated by spacer regions that correspond to viral or plasmid DNA sequences. Upon subsequent infections, the CRISPR-Cas system can identify and break down these exogenous genetic elements [7].
This genome editing system consists of a guide RNA molecule that directs the nuclease to a particular genomic target site of the genome and a Cas9 nuclease [8]. To make the mature gRNA, the endogenous bacterial machinery processes a single chimeric guide RNA (sgRNA) that is comprised of a combination of a CRISPR RNA (crRNA) and a fixed trans-activating crRNA (tracrRNA) [9]. The genetic loci of CRISPR-Cas systems have the CRISPR array that consists of similarly flanking sequences (spacers) and short repeated sequences (repeats). Protospacers are used as spacers in CRISPR arrays. These protospacers are in turn derived from DNA sequences from infecting plasmid or phage. The most important functional elements in CRISPR systems are Cas proteins encoded upstream of the CRISPR array and control system operation [10][11].
Class 1 and Class 2 are the two main classes in which the CRISPR-Cas system is categorized. There are six types (I to VI), and multiple subtypes of the CRISPR-Cas system, with Class 1 systems (Type I, III, and IV) having multi-Cas protein effector complexes and Class 2 systems (Type II, V, and VI) having a single effector protein [12][13]. Table 1 summarizes the classification, standard features, and representative members of each CRISPR-Cas system.
Table 1. Classification, Standard features, and Characteristics of each CRISPR-Cas system.
| Class | Type | Subtype | Signature Genes | Target | Effector | Families of Encoded Proteins | CRISPR Protein | Associated Type-Subtype | Function | References |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 (A multi-Cas protein) |
I | A, B, C, D, E, F, U | I-A; Cas8a2, Cas5 I-B; Cas8b I-C; Cas8c I-D; Cas10d I-E; Cse1, Cse2 I-F: Csy1, Csy2, Csy3, Cas6f |
dsDNA | Cascade | COG1518 | Cas 1 | I, II, III-A, III-B, IV, possibly VI |
DNA Nuclease | [5] |
| 1 | III | A, B, C, D | III-A: Csm2, Csx10 all1473 III-B Cmr5, MTH326 (Cas10 or Csx11) |
ssRNA | Cascade | COG1203 | Cas 2 | I, II, III-A, III-B, V, some VI |
RNA Nuclease | [5] |
| 1 | IV | A, B | DinG (Csf4) | dsDNA | Cascade | COG1468 | Cas 3 | I | DNA nuclease and helicase | [5][12] |
| 1 | II | A | Csn-2 | dsDNA | SpCas9 | COG1343 | Cas 4 | Mostly type I, II & V | DNA nuclease | [5][14] |
| 2 | II | A | dsDNA | SpCas9 | COG3512 | Cas 5 | Type I, I | Ribonuclease responsible for converting pre-crRNA to mature crRNA. | [14] | |
| 2 | II | B | Cas9 (Csx12 subfamily) | dsDNA/ssRNA | FnCas9 | COG1343 and COG3512 | Cas 6 | Most type IIIB and type I |
Pre-crRNA is converted to mature crRNA by ribonuclease. | [14] |
| 2 | II | C | N/A | dsDNA | NmCas9 | COG1343 and COG3512 | Cas 7 | I, III, IV | It binds with crRNA and comprises of an RNA recognition motif. | [15] |
| 2 | V | A | Csm4, Csx10, Cmr3 | dsDNA | Cas12a (Cpf1) |
COG1688 (RAMP) | Cas 8 | Most type I | It forms effector complex large subunit in type I | [16] |
| 2 | V | B | Cas5, Csy2 | dsDNA | Cas12b (C2c1) | COG1688 (RAMP) | Cas 9 | II only | DNA nuclease | [17] |
| 2 | V | C | Csc1, Csf3 | dSDNA | Cas12c (C2c3) | COG1688 (RAMP) | Cas 10 | Some type I, most type III |
It forms effector complex large subunit in type III. | [16] |
| 2 | VI | A | - | ssRNA | Cas13a (C2c2) | COG1583 and COG5551 (RAMP) | Cas 12 (cpf1) |
V | crRNA sorting, DNA nuclease | [16][18] |
| 2 | VI | B | Cmr6 | ssRNA | Cas13b (C2c4) | (RAMP) | Cas 13 (C2c2) | VI | crRNA sorting, RNA nuclease | [5][19] |
| 2 | VI | C | - | ssRNA | Cas13c (C2c7) | (RAMP) | Csm, Cmr | III | Nucleases for single-stranded DNA and RNA | [5][19] |
| 2 | VI | D | - | ssRNA | Cas13d | (RAMP) | RNase III | II | tracrRNA is processed, and crRNA maturation is aided by this system. | [12] |
2. Applications of CRISPR-Cas9 System
The CRISPR-based system has a lot of applications in curing diseases, correcting mutations, and improving crop quality and production. The detail is given below (Figure 1, Table 2).
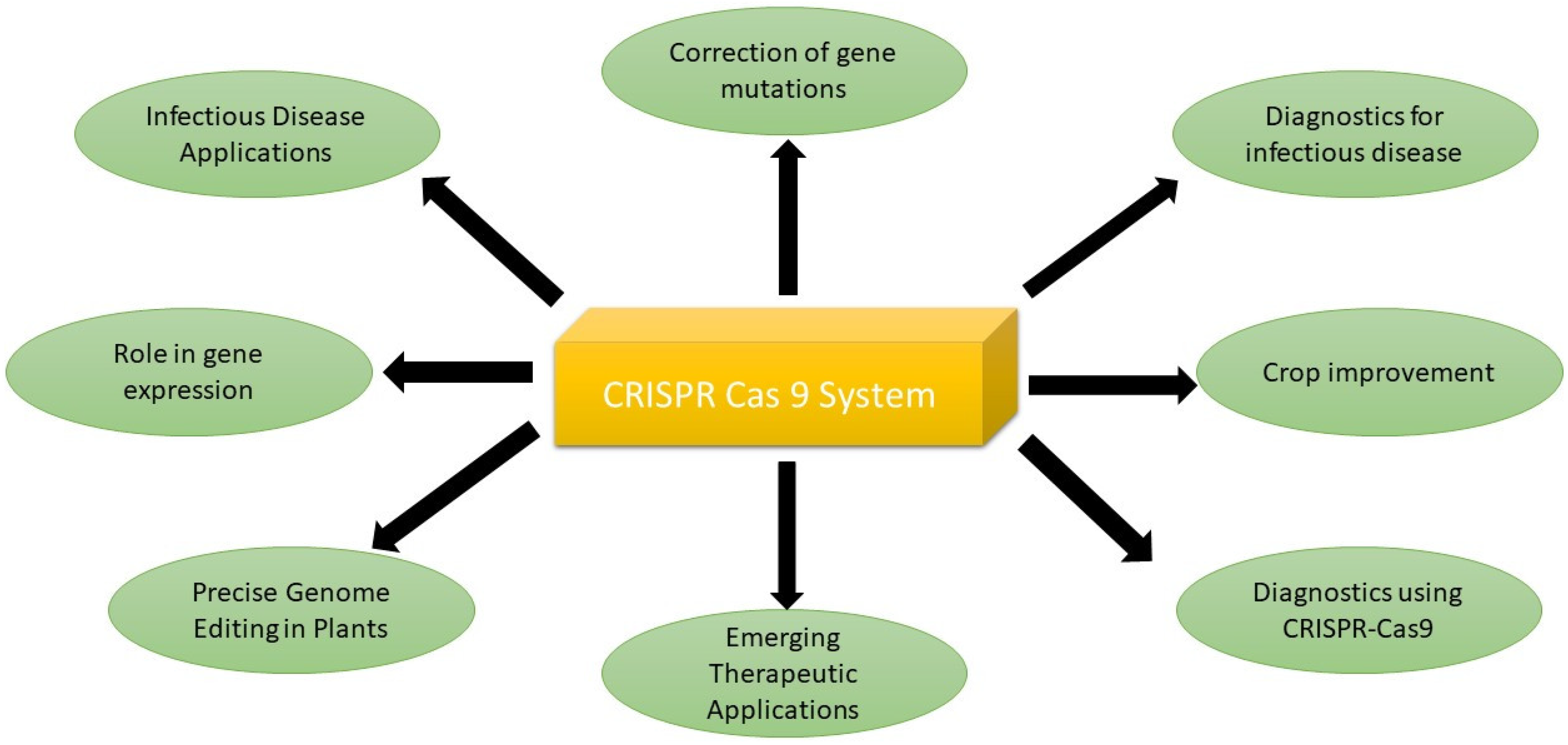
Figure 1. Applications of CRISPR-Cas9 system.
Table 2. Application of different CRISPR-Cas systems.
| CRISPR-Cas | Mechanism/Function | Delivery Vehicle |
|---|---|---|
| CRISPR-SpCas9 | single-RNA-mediated DNA endonuclease |
|
| CRISPRi | single-RNA-mediated inhibition of mRNA transcription |
|
| CRISPRa | single-RNA-mediated activation of mRNA transcription |
|
| CRISPR-SaCas9 | single-RNA-mediated DNA endonuclease |
|
| FnCas9 | single-RNA-mediated PAM-independent inhibiting of translation of target RNA |
|
| C2c1/3 | dual-RNA-guided DNA endonuclease | No mammalian expression vector |
2.1. Correction of Gene Mutations
Correcting recessive dystrophic epidermolysis bullosa (RDEB) through iPS (induced pluripotent stem) cells involves applying CRISPR/Cas9-based targeted. It is incredibly efficient and safe for gene correction. The hi-fi Cas9 (SpyFiCas9) nuclease has evident genome-wide off-target effects [20]. Targeted gene modification through CRISPR/Cas9 is an efficient way to evaluate gene function and accurately employ cellular behavior and process. Investigators can use GMOs (genetically modified organisms) to comprehend further the etiology of various disorders and elaborate the biochemical pathway utilized for an improved therapeutic strategy. These genome editing methods have helped to eradicate lethal diseases. CRISPR/Cas9 technology is used to produce chimeric antigen receptor T cells to damage malignant cells [21]. The CRISPR/Cas9 method has been used to successfully correct genetic disorders in mice or cystic fibrosis patients’ intestinal stem cell organoids. In an adult mouse model of human hereditary tyrosinemia disease, the Fah mutation has been corrected through CRISPR/Cas9 method. In this way, the symptoms of the disease have been eradicated [22].
2.2. Infectious Disease Applications
Infectious disease applications have been expanded through the CRISPR-Cas system. This technology promises to explain basic host-microbe relationships, help in the advancement of fast and precise diagnostics, and improve infectious disease prevention and care.
Knowing how bacteria, viruses, fungi, and parasites cause disease in humans is critical for providing the best health treatment and rationally designing tailored treatments and vaccinations. CRISPR Cas9-based genome editing is utilized to understand gene and protein connections to the molecular pathogenesis of a variety of pathogens.
Early detection, as well as prevention of infectious diseases, is facilitated by rapid and reliable diagnostic testing, which allows better clinical care and the prompt application of infection management and various other public health interventions to reduce disease transmission. A perfect fast diagnostic test will be responsive and specific, simple to administer and translate, compact, and inexpensive, allowing it to be used in a variety of clinical environments, including those with minimal resources. The CRISPR-Cas has aided in the advancement of fast and precise diagnostics for infectious diseases.
Many researchers are using CRISPR-Cas9 to improve diagnostics for infectious diseases. A combined nucleic acid sequence-based augmentation system known as NASBA is an example of isothermal amplification. This method is used in combination with CRISPR-Cas9 to differentiate between Zika virus strains that are closely related [22]. After applying a synthetic stimulation sequence to NASBA-amplified viral RNA, the researchers used a Cas9 and sgRNA complex to slice the resulting dsDNA. The presence or absence of a strain-specific PAM stemmed in Cas9-cleaved DNA fragments that were either abridged or full-length strands. The triggered turn was activated by full-length strands but not by truncated strands, resulting in a color shift on a paper disc and stable strain distinction [23].
2.3. Revolutionizing Fungal Disease Control with CRISPR-Cas9
Fungal infections are a serious global health issue because they may cause various diseases in plants, animals, and humans. Fungicides have been used to treat fungal diseases in the past, but they have the potential to damage the environment and breed resistant fungi. CRISPR-Cas9 technology is used in this case as a last resort. A gene-editing technique called CRISPR-Cas9 enables the precise modification of certain genes [24]. With the use of this technology, scientists may target particular genes in fungi and prevent them from being able to spread infection. This method is extremely specialized and can only target the genes that are accountable for the fungal pathogen’s virulence, leaving untargeted genes unaffected [25].
The use of CRISPR-Cas9 technologies to manage fungus infections has shown considerable potential. One use is the direct targeting and editing of fungal genes required for growth, pathogenicity, or drug resistance [26]. Researchers have successfully killed or inhibited the growth of harmful fungi by turning off these genes, perhaps improving treatment outcomes. Recently, the genomes of Candida albicans [27][28], Aspergillus [29], and Cryptococcus [30] were edited using the CRISPR/Cas9 system.
The study of Vyas et al. (2015) employed CRISPR-Cas9 to deactivate a gene required for virulence in the fungus Candida albicans. The fungus’s EFG1 gene plays a role in controlling the expression of other virulence genes. In a mouse infection model, the researchers’ disruption of EFG1 greatly decreased the virulence of C. albicans [27].
In addition to being more precise than older approaches to fungus control, CRISPR-Cas9 technology offers additional benefits. Contrary to fungicides, which need to be administered repeatedly, CRISPR-Cas9 technology may permanently change the fungus’s DNA, preventing it from spreading illness. In the long term, this strategy may also be more cost-effective because it eliminates the need for fungicides and other conventional means of preventing the growth of mold [31].
Another potential use of CRISPR-Cas9 is in developing new antifungal agents. The approach may be used to find drugs that kill fungus by either targeting certain genes or by interfering with essential cellular functions. In addition, CRISPR-Cas9 can be utilized to create fungus strains that are less aggressive or resistant to antifungal medications, which may lower the risk of infections and enhance treatment results [26][32].
Conclusively, CRISPR-Cas9 technology offers enormous potential for preventing and treating fungal infections in agricultural contexts and scenarios involving human health. With the potential for enduring impacts, this technique provides a highly specialized and possibly economical way to control fungi. While more research is needed to fully realize the potential of this technology, the use of CRISPR-Cas9 to control fungal infections represents an exciting and promising development in the field of disease control.
2.4. Emerging Therapeutic Applications
Although all bacteria do not use CRISPR-Cas systems, growing evidence supports their role in blocking the gaining of the genomic elements which impart antibiotic resistance, improving the likelihood that bacteria’s defenses could be used therapeutically against them Figure 2 depicts the CRISPR approaches towards microbiome therapies. Researchers have proved that the I-F CRISPR system in E. coli is present in E. coli which is linked with antibiotic sensitivity. CRISPR technology has been suggested as a way to grow specifically titratable antimicrobials to eradicate pathogens. This concept was used in vitro to kill single strains of E. coli as well as Salmonella enterica in pure and mixed culture experiments by utilizing a subtype I-E CRISPR-Cas system. RNA-guided Cas9 system was used by Bikard et al. that was transmitted by phagemid killed virulent. However, it did not kill the avirulent strains of S. aureus; without destroying the host bacteria, it eliminated plasmids containing the mecA methicillin resistance gene [33].
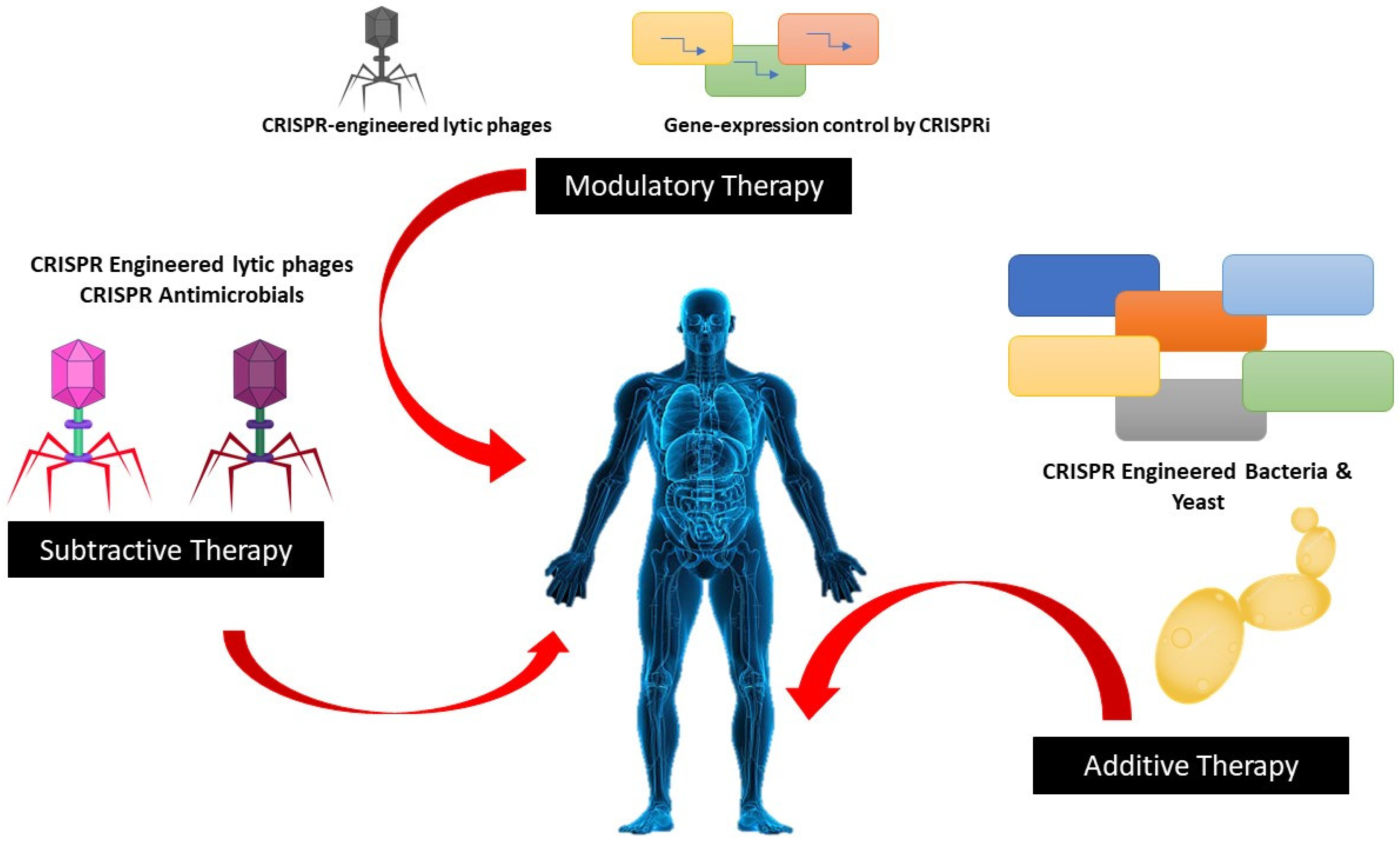
Figure 2. CRISPR approaches microbiome therapies.
Oral vaccines are being developed through Saccharomyces boulardii which is engineered with the help of the CRISPR-Cas system [34].
2.5. Role in Gene Expression
CRISPR/Cas9 is a useful tool in genetic engineering. It has been used in epigenetic studies to elicit gene expression. In a study, two methods were introduced to selectively regulate DNA methylation at the selective CpG site via utilizing CRISPR/Cas9 method. In this way, the gene expression was induced successfully [34]. The property of CRISPR/Cas9 to edit the gene has revolutionized cell therapy. The gene editing property of the CRISPR/Cas9 system is being improved and optimized through the use of artificial nucleic acid molecules (ANAMs) in cancerous cells. It is successfully proved that ANAMs improve transgene expression by inhibiting innate immune response (IIR) in the cells [34].
References
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide Sequence of the Iap Gene, Responsible for Alkaline Phosphatase Isozyme Conversion in Escherichia Coli, and Identification of the Gene Product. J. Bacteriol. 1987, 169, 5429–5433.
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR Provides Acquired Resistance against Viruses in Prokaryotes. Science 2007, 315, 1709–1712.
- Asmamaw, M.; Zawdie, B. Mechanism and Applications of CRISPR/Cas-9-Mediated Genome Editing. Biol. Targets Ther. 2021, 15, 353–361.
- Watson, B.N.J.; Staals, R.H.J.; Fineran, P.C. CRISPR-Cas-Mediated Phage Resistance Enhances Horizontal Gene Transfer by Transduction. MBio 2018, 9, e02406-17.
- Xu, Y.; Li, Z. CRISPR-Cas Systems: Overview, Innovations and Applications in Human Disease Research and Gene Therapy. Comput. Struct. Biotechnol. J. 2020, 18, 2401–2415.
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Haft, D.H.; et al. An Updated Evolutionary Classification of CRISPR-Cas Systems. Nat. Rev. Microbiol. 2015, 13, 722–736.
- Medina-Aparicio, L.; Rebollar-Flores, J.E.; Gallego-Hernández, A.L.; Vázquez, A.; Olvera, L.; Gutiérrez-Ríos, R.M.; Calva, E.; Hernandez-Lucas, I. The CRISPR/Cas Immune System Is an Operon Regulated by LeuO, H-NS, and Leucine-Responsive Regulatory Protein in Salmonella Enterica Serovar Typhi. J. Bacteriol. 2011, 193, 2396–2407.
- Jiang, C.; Lv, G.; Tu, Y.; Cheng, X.; Duan, Y.; Zeng, B.; He, B. Applications of CRISPR/Cas9 in the Synthesis of Secondary Metabolites in Filamentous Fungi. Front. Microbiol. 2021, 12, 638096.
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Eckert, M.R.; Vogel, J.; Charpentier, E. CRISPR RNA Maturation by Trans-Encoded Small RNA and Host Factor RNase III. Nature 2011, 471, 602–607.
- Heler, R.; Samai, P.; Modell, J.W.; Weiner, C.; Goldberg, G.W.; Bikard, D.; Marraffini, L.A. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature 2015, 519, 199–202.
- Horvath, P.; Barrangou, R. CRISPR/Cas, the Immune System of Bacteria and Archaea. Science 2010, 327, 167–170.
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, Classification and Evolution of CRISPR-Cas Systems. Curr. Opin. Microbiol. 2017, 37, 67–78.
- Jiang, F.; Doudna, J.A. CRISPR–Cas9 Structures and Mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529.
- Lundgren, M.; Charpentier, E.; Fineran, P.C. CRISPR: Methods and Protocols; Humana Press: New York, NY, USA, 2015; Volume 1311, pp. 1–366.
- Santajit, S.; Indrawattana, N. Mechanisms of Antimicrobial Resistance in Pasteurellaceae. BioMed Res. Int. 2016, 2016, 2475067.
- Liu, Z.; Dong, H.; Cui, Y.; Cong, L.; Zhang, D. Application of Different Types of CRISPR/Cas-Based Systems in Bacteria. Microb. Cell Fact. 2020, 19, 172.
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.M.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and Classification of the CRISPR-Cas Systems. Nat. Rev. Microbiol. 2011, 9, 467–477.
- Tang, Y.; Fu, Y. Class 2 CRISPR/Cas: An Expanding Biotechnology Toolbox for and beyond Genome Editing 06 Biological Sciences 0604 Genetics. Cell Biosci. 2018, 8, 59.
- Shmakov, S.; Smargon, A.; Scott, D.; Cox, D.; Pyzocha, N.; Yan, W.; Abudayyeh, O.O.; Gootenberg, J.S.; Makarova, K.S.; Wolf, Y.I. Diversity and Evolution of Class 2 CRISPR–Cas Systems. Nat. Rev. Microbiol. 2017, 15, 169–182.
- Jacków, J.; Guo, Z.; Hansen, C.; Abaci, H.E.; Doucet, Y.S.; Shin, J.U.; Hayashi, R.; DeLorenzo, D.; Kabata, Y.; Shinkuma, S.; et al. CRISPR/Cas9-Based Targeted Genome Editing for Correction of Recessive Dystrophic Epidermolysis Bullosa Using IPS Cells. Proc. Natl. Acad. Sci. USA 2019, 116, 26846–26852.
- Li, H.; Yang, Y.; Hong, W.; Huang, M.; Wu, M.; Zhao, X. Applications of Genome Editing Technology in the Targeted Therapy of Human Diseases: Mechanisms, Advances and Prospects. Signal Transduct. Target. Ther. 2020, 5, 1.
- Ma, Y.; Zhang, L.; Huang, X. Genome Modification by CRISPR/Cas9. FEBS J. 2014, 281, 5186–5193.
- Guk, K.; Keem, J.O.; Hwang, S.G.; Kim, H.; Kang, T.; Lim, E.K.; Jung, J. A Facile, Rapid and Sensitive Detection of MRSA Using a CRISPR-Mediated DNA FISH Method, Antibody-like DCas9/SgRNA Complex. Biosens. Bioelectron. 2017, 95, 67–71.
- Kaboli, S.; Babazada, H. CRISPR Mediated Genome Engineering and Its Application in Industry. Curr. Issues Mol. Biol. 2018, 26, 81–92.
- Liao, B.; Chen, X.; Zhou, X.; Zhou, Y.; Shi, Y.; Ye, X.; Liao, M.; Zhou, Z.; Cheng, L.; Ren, B. Applications of CRISPR/Cas Gene-Editing Technology in Yeast and Fungi. Arch. Microbiol. 2022, 204, 79.
- Doerflinger, M.; Forsyth, W.; Ebert, G.; Pellegrini, M.; Herold, M.J. CRISPR/Cas9—The Ultimate Weapon to Battle Infectious Diseases? Cell. Microbiol. 2017, 19, e12693.
- Vyas, V.K.; Barrasa, M.I.; Fink, G.R. A Candida Albicans CRISPR System Permits Genetic Engineering of Essential Genes and Gene Families. Sci. Adv. 2015, 1, e1500248.
- Min, K.; Ichikawa, Y.; Woolford, C.A.; Mitchell, A.P. Candida Albicans Gene Deletion with a Transient CRISPR-Cas9 System. MSphere 2016, 1, e00130-16.
- Fuller, K.K.; Chen, S.; Loros, J.J.; Dunlap, J.C. Development of the CRISPR/Cas9 System for Targeted Gene Disruption in Aspergillus Fumigatus. Eukaryot. Cell 2015, 14, 1073–1080.
- Wang, Y.; Wei, D.; Zhu, X.; Pan, J.; Zhang, P.; Huo, L.; Zhu, X. A ‘Suicide’CRISPR-Cas9 System to Promote Gene Deletion and Restoration by Electroporation in Cryptococcus Neoformans. Sci. Rep. 2016, 6, 31145.
- Távora, F.T.P.K.; dos Santos Diniz, F.D.A.; de Moraes Rêgo-Machado, C.; Freitas, N.C.; Arraes, F.B.M.; de Andrade, E.C.; Furtado, L.L.; Osiro, K.O.; de Sousa, N.L.; Cardoso, T.B. CRISPR/Cas-and Topical RNAi-Based Technologies for Crop Management and Improvement: Reviewing the Risk Assessment and Challenges towards a More Sustainable Agriculture. Front. Bioeng. Biotechnol. 2022, 10, 913728.
- Shanmugam, K.; Ramalingam, S.; Venkataraman, G.; Hariharan, G.N. The CRISPR/Cas9 System for Targeted Genome Engineering in Free-Living Fungi: Advances and Opportunities for Lichenized Fungi. Front. Microbiol. 2019, 10, 62.
- Bikard, D.; Euler, C.W.; Jiang, W.; Nussenzweig, P.M.; Goldberg, G.W.; Duportet, X.; Fischetti, V.A.; Marraffini, L.A. Exploiting CRISPR-Cas Nucleases to Produce Sequence-Specific Antimicrobials. Nat. Biotechnol. 2014, 32, 1146–1150.
- Bagherpour, G.; Ghasemi, H.; Zand, B.; Zarei, N.; Roohvand, F.; Ardakani, E.M.; Azizi, M.; Khalaj, V. Oral Administration of Recombinant Saccharomyces boulardii Expressing Ovalbumin-CPE Fusion Protein Induces Antibody Response in Mice. Front. Microbiol. 2018, 9, 723.
More
Information
Subjects:
Biotechnology & Applied Microbiology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.7K
Revisions:
2 times
(View History)
Update Date:
02 Aug 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





