Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Poonam Vinayamohan | -- | 2932 | 2023-07-28 15:10:02 | | | |
| 2 | Sirius Huang | Meta information modification | 2932 | 2023-07-31 02:27:53 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Vinayamohan, P.G.; Viju, L.S.; Joseph, D.; Venkitanarayanan, K. Antimicrobial-Resistant Bacteria and Genes in Fermented Foods. Encyclopedia. Available online: https://encyclopedia.pub/entry/47391 (accessed on 08 February 2026).
Vinayamohan PG, Viju LS, Joseph D, Venkitanarayanan K. Antimicrobial-Resistant Bacteria and Genes in Fermented Foods. Encyclopedia. Available at: https://encyclopedia.pub/entry/47391. Accessed February 08, 2026.
Vinayamohan, Poonam Gopika, Leya Susan Viju, Divya Joseph, Kumar Venkitanarayanan. "Antimicrobial-Resistant Bacteria and Genes in Fermented Foods" Encyclopedia, https://encyclopedia.pub/entry/47391 (accessed February 08, 2026).
Vinayamohan, P.G., Viju, L.S., Joseph, D., & Venkitanarayanan, K. (2023, July 28). Antimicrobial-Resistant Bacteria and Genes in Fermented Foods. In Encyclopedia. https://encyclopedia.pub/entry/47391
Vinayamohan, Poonam Gopika, et al. "Antimicrobial-Resistant Bacteria and Genes in Fermented Foods." Encyclopedia. Web. 28 July, 2023.
Copy Citation
Fermented food products are widely consumed for their nutritional and health-promoting properties, earning them a central place in diets around the globe. However, these foods can present a paradox, as they have the potential to harbor not only beneficial probiotics but also antibiotic-resistant (AR) microbes and genes. The impact of AR microbes and genes in fermented foods has far-reaching implications, such as potential effects on human health, repercussions in the food industry, and environmental consequences.
fermented foods
antibiotic-resistant bacteria
antibiotic-resistant genes
1. Introduction
Fermented foods have been an integral part of the global diet for centuries and are characterized by their production through controlled microbial growth and enzymatic action [1]. They encompass a wide range of food categories, including meat, dairy, vegetables, soybeans, legumes, cereals, and fruits. Fermentation serves multiple purposes, such as preservation and enhancement of organoleptic properties, offering not only unique flavors and textures but also potential health benefits. The fermentation process involves microorganisms, nutritional ingredients, and environmental conditions, resulting in numerous variations of fermented foods [2]. While fermented foods offer beneficial properties, the presence of antibiotic-resistant (AR) bacteria and genes within these food products raises concerns regarding their impact on human health. Understanding the relationship between fermented foods and AR is crucial for assessing the potential risks associated with their consumption.
Traditionally, fermentation served as a preservation method, utilizing antimicrobial metabolites to reduce the risk of contamination with pathogenic microorganisms. However, recent studies have revealed the presence of AR bacteria and genes in various types of fermented foods [3][4]. Spontaneously fermented foods pose a unique concern for AR, as the presence of naturally occurring fermentative bacteria in the food matrix makes them less amenable to safety assurance measures [5]. Additionally, lactic acid bacteria (LAB), commonly found in fermented foods, have been associated with the dissemination of AR [6][7], especially when these products are consumed without heat treatment. The consumption of such foods may facilitate the exchange of functional AR genes (ARGs) between resident microorganisms and potential pathogens through horizontal gene transfer (HGT), as represented in Figure 1 [8].
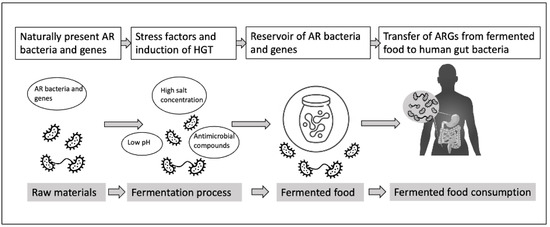
Figure 1. Potential events in fermented food production leading to the transfer of AR bacteria and genes to human gut microbiota.
2. AR Bacteria and Genes in Fermented Foods
AR in fermented foods is a concerning phenomenon closely linked to the misuse of antibiotics in the food chain [9]. The fermentation process itself might contribute to the proliferation of bacteria, leading to an overall increase in the abundance of ARGs within fermented foods [3]. As reservoirs of AR bacteria and ARGs, there is a potential for these elements to be transferred to microorganisms within the host, including pathogens. This transfer can have lasting effects on the microbial communities within the human body. Notably, fermented foods harbor a significant population of microorganisms, and the specific conditions within the fermentation environment, such as confined spaces and close cell-to-cell contact, provide an ideal setting for gene transfer. LAB and coagulase-negative Staphylococcus (CNS) species, commonly found in fermented foods, have been identified as carriers of ARGs, particularly against tetracyclines, penicillins, chloramphenicol, and macrolides [10]. Table 1 represents the major AR bacteria identified in fermented foods, along with their phenotypic resistance profiles.
In various fermented food products, metagenomic research has revealed the presence of ARGs belonging to several AR classes. Fermented dairy products exhibited an average count per million (CPM) of 3686 ARGs per sample, while brine and sugar products had lower CPM values of 426 and 261, respectively [11]. Furthermore, using starter cultures in fermentations resulted in a fivefold higher abundance of ARGs than in spontaneous fermentations [11]. This might seem contradictory, given the European Food Safety Authority’s Qualified Presumption of Safety (QPS) status and the United States Generally Recognized as Safe (GRAS) system, which both ensure a lack of AR features in microorganisms. However, this apparent discrepancy can be attributed to the enhanced characterization of starter culture microbes, their associated genomes, and associated ARG profiles, which lead to more accurate ARG assignments, rather than an actual increase in AR. Among 58 analyzed food samples, nine showed the presence of ARGs. These included kombucha, kefir, kimchi, pickled carrots, pickled vegetables, and apple cider vinegar samples. The presence of ARGs in fermented foods not only increases the risk of HGT but also amplifies the potential for ARG dissemination due to their mobility [11]. Therefore, understanding the prevalence and transferability of AR bacteria and genes in fermented foods is crucial for addressing the potential risks associated with their consumption.
Table 1. Antibiotic-resistant bacteria in fermented foods and their corresponding phenotypic resistance profile.
| Food Type | Bacteria Present | Phenotypic Resistance Profile | Reference |
|---|---|---|---|
| Cheese | LAB, Staphylococcus (CNS) | Tetracycline, chloramphenicol, erythromycin | [12][13] |
| Fermented meat/sausage | LAB, Staphylococcus (CNS) | Tetracycline, chloramphenicol, erythromycin, penicillins | [10] |
| Salmonella | Amoxicillin, gentamycin, streptomycin, tetracycline | [13] | |
| Listeria monocytogenes | Amoxicillin, benzyl penicillin, tetracycline, ciprofloxacin | [13] | |
| Fermented vegetables | LAB | Tetracycline, chloramphenicol, erythromycin, clindamycin | [10][12][13] |
| Yogurt | Lactobacillus | Mycostatin, nalidixic acid, neomycin, polymyxin b, trimethoprim, colimycin, sulfamethoxazole, and sulfonamides | [14] |
| Streptococcus | Colimycin, gentamicin, kanamycin, Mycostatin, nalidixic acid, neomycin, polymyxin b, trimethoprim–sulfamethoxazole, streptomycin, sulfonamides | [14] | |
| Vinegar | Acetic acid bacteria | Trimethoprim, ciprofloxacin, erythromycin | [15] |
| Turkish cheese | Enterococcus | Streptomycin, erythromycin, oxacillin, vancomycin | [16] |
| Fermented fish | Staphylococcus (CNS) | Tetracyclines, penicillins, chloramphenicol, macrolides | [17] |
| Ntoba Mbodi (alkaline fermented food) | Staphylococcus | Chloramphenicol, erythromycin, penicillin, sulfamethoxazole, trimethoprim | [18] |
| Nono (African traditionally fermented milk product) | Enterococcus thailandicus Streptococcus infantarius |
Tetracyclines, streptomycin Tetracyclines |
[19] |
2.1. ARGs and Associated Bacteria in Fermented Foods
2.1.1. Lactic Acid Bacteria
ARGs have been consistently observed in LAB associated with various fermented foods. Specifically, resistance to tetracyclines, penicillins, chloramphenicol, and macrolides, such as erythromycin, is frequently detected among LAB strains, particularly in certain types of cheeses, fermented meats and spontaneously fermented vegetables [12][13]. In studies analyzing LAB isolates from Indonesian traditional fermented foods (dadih, tape ketan, bekasam, and tempoyak), a significant number of strains were found to be resistant to chloramphenicol and erythromycin [20]. Lactobacilli, commonly used as starter cultures in fermented milk products, exhibit widespread resistance to penicillins. Studies have identified acquired resistance of LAB to tetracycline, erythromycin, clindamycin, and chloramphenicol in Lactobacilli isolated from fermented foods. The resistance is mediated by specific genes such as tetM and tetS (associated with tetracycline resistance) and erm(B) (associated with erythromycin resistance). Furthermore, the cat gene, encoding chloramphenicol acetyltransferase and carried on plasmids, has been identified as a resistance determinant in those isolates [21][22].
In a study examining 16 fermented Chinese foods and eight sausages and pickles, LAB isolates displayed varying levels of AR. LABs in pickle samples showed susceptibility to tetracycline, ampicillin, and clindamycin, but showed resistance to other antibiotics. LABs in sausage samples from different provinces also exhibited differing levels of resistance, with some strains showing resistance to multiple antibiotics [23]. PCR analysis of antibiotic-resistant LAB strains revealed the presence of frequently detected ARGs, such as ermB, and tetM, found on both plasmid and chromosomal DNA. The tetM gene was consistently identified in all tetracycline-resistant LAB strains, with some strains carrying the gene on both plasmids and chromosomes, indicating multiple mechanisms and locations for ARG occurrence [23]. Additionally, certain strains of Lactobacillus delbrueckii subsp. bulgaricus, commonly used in yogurt cultures, demonstrated intrinsic resistance towards Mycostatin, nalidixic acid, neomycin, polymyxin B, trimethoprim, colimycin, sulfamethoxazole, and sulfonamides [14].
2.1.2. Staphylococcus spp.
Coagulase-negative Staphylococcus (CNS) species, frequently found in fermented animal products such as certain meats, cheeses, and fermented fish products, have been associated with the presence of ARGs. The common ARGs include resistance to tetracyclines, penicillins, chloramphenicol, and macrolides [17].
In a Swiss study, tetracycline resistance was detected in Staphylococcus isolates used as starter cultures in meat, as well as in Bifidobacterium lactis and Lactobacillus reuteri [24]. Another study conducted in Nigeria examined 255 CNS isolates obtained from six different Nigerian fermented foods (kindirmo, nono, iru, wara, ogi, and kunu) and found that a substantial proportion of the isolates exhibited resistance to different antibiotics. Specifically, a high percentage of isolates demonstrated resistance to ampicillin, trimethoprim-sulfamethoxazole, amoxicillin–clavulanic acid, and oxacillin. Staphylococcus caprae isolated from the Nigerian fermented foods showed particularly high resistance levels to multiple antibiotics, including ciprofloxacin, ofloxacin, gentamicin, erythromycin, and cefotaxime [25]. In yet another study, the genetic diversity and AR profiles of Staphylococcus saprophyticus isolates from fermented foods and clinical samples were assessed, revealing notable resistance rates to lincomycin, erythromycin, and tetracycline [26].
2.1.3. Enterococcus spp.
Enterococcus species, commonly found in Turkish white cheese, have exhibited resistance to streptomycin, erythromycin, oxacillin, and vancomycin. In fact, a high prevalence of vancomycin resistance was observed in Enterococcus faecalis (96.8%) and Enterococcus faecium (76%) isolates [16]. The presence of specific resistance genes in Enterococcus faecium strain SZ109 was also investigated, with the ermB gene associated with erythromycin resistance detected on both the plasmid and chromosome. Additionally, the aphA3 gene, conferring resistance to antibiotics such as kanamycin and neomycin, was exclusively found on the plasmid, while the mefA gene, responsible for macrolide resistance, was exclusively located on the chromosome [23]. A comprehensive study assessed the resistance patterns of 55 Enterococci strains, including 41 Enterococcus faecium and 14 Enterococcus faecalis strains, isolated from various traditional fermented foods of animal and vegetable origins, as well as water samples. The analysis revealed that certain strains exhibited resistance to vancomycin, while a high incidence of AR was detected among Enterococcus faecalis isolates to rifampicin, ciprofloxacin, and quinupristin–dalfopristin. Intermediate resistance to erythromycin and tetracycline was also observed. Among Enterococcus faecium isolates, resistance was found against rifampicin, ciprofloxacin, erythromycin, levofloxacin, and nitrofurantoin [27]. These findings underscore the presence of AR in Enterococcus species isolated from fermented foods and emphasize the importance of understanding and monitoring their resistance profiles for food safety and public health considerations.
2.1.4. AR Genes in Other Bacteria
Research has focused on AR in Acetobacter and Komagataeibacter species, predominantly derived from vinegar, revealing resistance to trimethoprim, erythromycin, ciprofloxacin, and chloramphenicols [15]. Another study investigating AR microorganisms in meat and minced meat used for fermented sausage production found Salmonella strains exhibiting resistance to amoxicillin (44.5%), gentamycin (38%), streptomycin (44.5%), and tetracycline (55.5%). Similarly, Listeria monocytogenes displayed resistance to amoxicillin (38.5%), benzylpenicillin (30.8%), tetracycline (53.8%), and ciprofloxacin (38.5%). Escherichia coli strains showed resistance to amoxicillin (33.3%), neomycin (30%), streptomycin (40%), and tetracycline (40%) [13]. Moreover, 15 strains of Streptococcus thermophilus isolated from yogurt cultures exhibited varying levels of resistance to colimycin, gentamicin, kanamycin, Mycostatin, nalidixic acid, neomycin, polymyxin B, trimethoprim–sulfamethoxazole, streptomycin, and sulfonamides [14]. Furthermore, high tetracycline and erythromycin resistance have been observed in Lactococcus and Streptococcus thermophilus strains isolated from dairy products [24]. These findings highlight the prevalence of AR in diverse bacterial species associated with fermented foods and accentuate the need for continued surveillance and management strategies to mitigate the risks posed by AR strains.
3. Role of Bacterial Adaptation and Gene Transfer in Fermented Foods
Fermented foods comprise a complex microenvironment characterized by diverse microbial communities, creating an ideal environment for horizontal gene transfer [28]. HGT in fermented foods plays a crucial role in the adaptation of bacteria to evolving nutritional needs and environments [29]. For example, during the fermentation process of foods such as cheese, wine, yogurt, and vinegar, the acquisition of the horizontal transfer region occurs, facilitating the transfer of genetic material among microbial populations [30]. This can potentially improve the quality and shelf life of fermented foods. Research shows that the occurrence of HGT in a specific environment is beneficial for starter culture domestication, a process that involves selectively cultivating and adapting specific microorganisms to exhibit desired traits and behaviors during the fermentation process, leading to improved fermentation efficiency, digestibility, palatability, and longevity [30][31]. Microbial fermentation leading to HGT in fermented beverages, such as wine and beer, has also been reported, potentially impacting their shelf life and sensory attributes [32]. However, it is important to note that while HGT contributes to the adaptation and quality of fermented food, it can also facilitate the transfer of ARGs among microbial populations [3]. Within the fermentation environment, microbial interactions, such as conjugation or transduction, may also occur during manufacturing and storage [33].
3.1. Horizontal Gene Transfer in Fermented Food
The transfer of ARGs through HGT in fermented foods has been extensively studied, with sausages, kefir, yogurt, and cheese being key examples. These studies also highlight the potential of ARG transmission to both animal and human microbiota. Leroy et al., observed the transfer of plasmids carrying ARG during sausage fermentation involving Staphylococcus xylosus, isolated from fermented sausage [34]. Furthermore, the analysis of mobile genetic elements (MGE) domains in kefir revealed the presence of ARG lmrD (provides resistance to lincosamides). The contig containing this gene was found in close proximity to a transposase open reading frame (ORF) within 10 neighboring ORFs, suggesting potential gene mobility in the fermentation-associated microbial community [3]. Metagenomic analysis of fermented milk products such as yogurt, nunu, kefir, and Koumiss demonstrated the presence of plasmid-borne ARGs within the fermented milk microbiome [35]. Additionally, plasmid-mediated transfer of AR has been demonstrated between strains of L. curvatus during sausage fermentation [36]. The ripening of fermented sausages also facilitates a high frequency of plasmid transfer among Enterococcus faecalis strains, highlighting the close contact and genetic transfer facilitated during the ripening of sausages [37]. Cheese, known for its favorable conditions for microbial survival and growth, can harbor AR pathogens such as Listeria due to its moderately high water activity and low salt content, and serves as a potential reservoir for the acquisition of resistance genes by Enterococci. The horizontal transfer of the tetM gene from L. monocytogenes to E. faecalis was investigated in Minas Frescal cheese. The results revealed that both donor and recipient strains remained viable throughout the 21-day shelf life, with transconjugant bacteria detected from the second day onwards [38]. Commercial dairy-derived Lactobacillus strains from cheese and yogurts carried plasmid-mediated streptomycin and gentamicin-resistance genes in L. rhamnosus and L. plantarum, indicating the potential for HGT among probiotic strains [39]. During soybean fermentation, a plasmid carrying a lincosamide-resistance gene (lnuA) was transferred between coagulase-negative Staphylococci in the presence of lincomycin. Interestingly, the plasmid was efficiently transferred between these bacteria during passage through murine intestines, even in the absence of lincomycin, highlighting the enhanced transferability under in vivo conditions [40]. A similar study conducted by Li et al. [41] reported the transfer of vanA gene associated with vancomycin resistance during the early stages of fermented soybean meal (FSBM) production, with transconjugants steadily increasing throughout the fermentation process. Notably, these transconjugants were able to pass through the digestive tract of growing pigs and were detected after three days of feeding. These findings underscore the potential dissemination of ARGs carried by commercial probiotics used in animal feed and feed additives within the digestive tracts of animals. Such dissemination poses a risk of further infections under extreme conditions, highlighting fermented food as a potential source of ARGs to human gut microbiota via HGT.
3.2. Stress Response Contributing to Gene Transfer
During fermentation, bacteria encounter various stressors, such as low pH, high salt concentration, and the presence of antimicrobial compounds produced during the fermentation process. These stressors may exert selective pressure on bacterial populations, leading to the survival and proliferation of adapted strains. Some bacteria possess intrinsic mechanisms to counteract these stresses, while others may acquire adaptive traits through HGT. Stressful conditions, such as low temperatures, can favor the transfer of genetic elements, including ARGs [42][43]. LAB, predominantly found in cold dairy environments, has been frequently linked with AR. In the case of cheese, temperature and pH influenced antibiotic gene transfer. At a processing temperature of 30 °C, the highest levels of donor and recipient bacteria were observed after 48 h, while transconjugant bacteria peaked at 48 h. At 10 °C, growth rate and acidification were reduced, resulting in delayed detection of vancomycin-resistant transconjugants, which reached their highest number during ripening [37]. Tarrah et al. [44] discovered a genomic island closely associated with S. thermophilus isolated from dairy environments containing the tetS gene, along with genes related to plasmids, beta-lactamase class C-like, and the penicillin-binding protein superfamily. However, no HGT was detected from S. thermophilus to L. rhamnosus or L. delbrueckii during skim milk fermentation, refrigerated storage, and simulated gastrointestinal transit. On the other hand, the transfer frequencies of tetracycline-, ampicillin-, and chloramphenicol-resistance genes significantly increased following high-pressure processing exposure, commonly used in the preservation of fermented food, both in vitro and in the food matrix, when compared to the non-pressurized control strains [45].
3.3. Transfer of ARGs from Fermented Food to Human Microbiota
Transfer of ARGs within fermented foods presents a dual concern, as they impede microbial evolution within the food matrix while also posing a risk to human health. Bacteria that enter our gut through food intake have the potential to exchange functional ARGs with both saprophytic bacteria and nearby pathogens through HGT, leading to the emergence of resistant or multiresistant pathogenic strains, limiting treatment options for infections [3]. The consumption of livestock products facilitates the establishment of transfer connections between bacteria, as it brings bacterial populations harboring diverse sets of ARGs [3]. Lactobacilli, due to their wide environmental distribution, can act as potential vectors for the dissemination of AR, raising concerns about their use as probiotics due to the acquisition and transfer of transferable AR determinants [46]. Previous research has demonstrated the transfer of ARGs between Lactobacillus species and even to other pathogens. Transfer of vancomycin resistance from Enterococci (both of human and animal origin) to a probiotic Lactobacillus acidophilus was observed in vitro and in vivo in mice, highlighting the potential long-term consequences of accumulating ARGs in the gut [46]. Given the high frequency of transfer of resistant determinants in mouse models, it is likely that similar events occur in the human gut, which serves as a reservoir for ARGs [47]. Consumption of fermented products in conjunction with appropriate triggers further increases the potential for HGT within the human gut [3]. Enterococci, known for causing human diseases, possess acquired antibiotic resistance along with intrinsic resistance to multiple antibiotics, low pH, high salt concentrations, and high temperatures, enabling their survival in harsh environments. AR Enterococci are commonly found in animal-derived foods and thrive during sausage fermentation. Transfer of tetracycline-resistant determinant tet(M) from E. faecium isolated from dry sausage to clinical isolates of E. faecalis and E. faecium was associated with type 1 integron [4], highlighting the significance of fermented food as reservoirs of ARGs and their potential transfer to human strains upon consumption.
References
- Marco, M.L.; Heeney, D.; Binda, S.; Cifelli, C.J.; Cotter, P.D.; Foligné, B.; Gänzle, M.; Kort, R.; Pasin, G.; Pihlanto, A.; et al. Health Benefits of Fermented Foods: Microbiota and Beyond. Curr. Opin. Biotechnol. 2017, 44, 94–102.
- Dimidi, E.; Cox, S.R.; Rossi, M.; Whelan, K. Fermented Foods: Definitions and Characteristics, Impact on the Gut Microbiota and Effects on Gastrointestinal Health and Disease. Nutrients 2019, 11, 1806.
- Tóth, A.G.; Csabai, I.; Maróti, G.; Jerzsele, Á.; Dubecz, A.; Patai, Á.V.; Judge, M.F.; Nagy, S.Á.; Makrai, L.; Bányai, K.; et al. A Glimpse of Antimicrobial Resistance Gene Diversity in Kefir and Yoghurt. Sci. Rep. 2020, 10, 22458.
- Jahan, M.; Zhanel, G.G.; Sparling, R.; Holley, R.A. Horizontal Transfer of Antibiotic Resistance from Enterococcus faecium of Fermented Meat Origin to Clinical Isolates of E. Faecium and Enterococcus faecalis. Int. J. Food Microbiol. 2015, 199, 78–85.
- Jasiak, K.; Amund, D. Are Spontaneously Fermented Plant-Based Foods Potential Sources of Transferable Antibiotic Resistance Genes? Food Front. 2022, 3, 46–55.
- Duche, R.T.; Singh, A.; Wandhare, A.G.; Sangwan, V.; Sihag, M.K.; Nwagu, T.N.T.; Panwar, H.; Ezeogu, L.I. Antibiotic resistance in potential probiotic lactic acid bacteria of fermented foods and human origin from Nigeria. BMC Microbiol. 2023, 23, 142.
- Erginkaya, Z.; Turhan, E.U.; Tatll, D. Determination of antibiotic resistance of lactic acid bacteria isolated from traditional Turkish fermented dairy products. Iran. J. Vet. Res. 2018, 19, 53.
- Hummel, A.S.; Hertel, C.; Holzapfel, W.H.; Franz, C.M.A.P. Antibiotic Resistances of Starter and Probiotic Strains of Lactic Acid Bacteria. Appl. Environ. Microbiol. 2007, 73, 730–739.
- The WHO Regional Office for Europe. Tackling Antibiotic Resistance from a Food Safety Perspective in Europe. Available online: https://apps.who.int/iris/bitstream/handle/10665/326398/9789289014212-eng.pdf (accessed on 12 June 2023).
- Wolfe, B.E. Are Fermented Foods an Overlooked Reservoir of Antimicrobial Resistance? Curr. Opin. Food Sci. 2023, 51, 101018.
- Leech, J.; Cabrera-Rubio, R.; Walsh, A.M.; Macori, G.; Walsh, C.J.; Barton, W.; Finnegan, L.; Crispie, F.; O’Sullivan, O.; Claesson, M.J.; et al. Fermented-Food Metagenomics Reveals Substrate-Associated Differences in Taxonomy and Health-Associated and Antibiotic Resistance Determinants. MSystems 2020, 5, 10–1128.
- Flórez, A.B.; Delgado, S.; Mayo, B. Antimicrobial Susceptibility of Lactic Acid Bacteria Isolated from a Cheese Environment. Can. J. Microbiol. 2005, 51, 51–58.
- Fraqueza, M.J. Antibiotic Resistance of Lactic Acid Bacteria Isolated from Dry-Fermented Sausages. Int. J. Food Microbiol. 2015, 212, 76–88.
- Mathur, S.; Singh, R. Antibiotic Resistance in Food Lactic Acid Bacteria—A Review. Int. J. Food Microbiol. 2005, 105, 281–295.
- Cepec, E.; Trček, J. Antimicrobial Resistance of Acetobacter and Komagataeibacter Species Originating from Vinegars. Int. J. Environ. Res. Public. Health 2022, 19, 463.
- Çitak, S.; Yucel, N.; Orhan, S. Antibiotic Resistance and Incidence of Enterococcus Species in Turkish White Cheese. Int. J. Dairy Technol. 2004, 57, 27–31.
- Heo, S.; Lee, J.H.; Jeong, D.W. Food-Derived Coagulase-Negative Staphylococcus as Starter Cultures for Fermented Foods. Food Sci. Biotechnol. 2020, 29, 1023–1035.
- Ouoba, L.I.I.; Mbozo, A.B.V.; Anyogu, A.; Obioha, P.I.; Lingani-Sawadogo, H.; Sutherland, J.P.; Jespersen, L.; Ghoddusi, H.B. Environmental Heterogeneity of Staphylococcus Species from Alkaline Fermented Foods and Associated Toxins and Antimicrobial Resistance Genetic Elements. Int. J. Food Microbiol. 2019, 311, 108356.
- Obioha, P.I.; Anyogu, A.; Awamaria, B.; Ghoddusi, H.B.; Ouoba, L.I.I. Antimicrobial Resistance of Lactic Acid bacteria from Nono, A Naturally Fermented Milk product. Antibiotics 2023, 12, 843.
- Sukmarini, L.; Mustopa, A.Z.; Normawati, M.; Muzdalifah, I. Identification of Antibiotic-Resistance Genes from Lactic Acid Bacteria in Indonesian Fermented Foods. Hayati 2014, 21, 144–150.
- Çataloluk, O.; Gogebakan, B. Presence of Drug Resistance in Intestinal Lactobacilli of Dairy and Human Origin in Turkey. FEMS Microbiol. Lett. 2004, 236, 7–12.
- Danielsen, M.; Wind, A. Susceptibility of Lactobacillus spp. to Antimicrobial Agents. Int. J. Food Microbiol. 2003, 82, 1–11.
- Pan, L.; Hu, X.; Wang, X. Assessment of Antibiotic Resistance of Lactic Acid Bacteria in Chinese Fermented Foods. Food Control 2011, 22, 1316–1321.
- Verraes, C.; Van Boxstael, S.; Van Meervenne, E.; Van Coillie, E.; Butaye, P.; Catry, B.; de Schaetzen, M.A.; Van Huffel, X.; Imberechts, H.; Dierick, K.; et al. Antimicrobial Resistance in the Food Chain: A Review. Int. J. Environ. Res. Public. Health 2013, 10, 2643–2669.
- Fowoyo, P.T.; Ogunbanwo, S.T. Antimicrobial Resistance in Coagulase-Negative Staphylococci from Nigerian Traditional Fermented Foods. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 4.
- Lee, B.; Jeong, D.W.; Lee, J.H. Genetic Diversity and Antibiotic Resistance of Staphylococcus saprophyticus Isolates from Fermented Foods and Clinical Samples. J. Korean Soc. Appl. Biol. Chem. 2015, 58, 659–668.
- Sánchez Valenzuela, A.; Lavilla Lerma, L.; Benomar, N.; Gálvez, A.; Pérez Pulido, R.; Abriouel, H. Phenotypic and Molecular Antibiotic Resistance Profile of Enterococcus faecalis and Enterococcus faecium Isolated from Different Traditional Fermented Foods. Foodborne Pathog. Dis. 2013, 10, 143–149.
- Giraffa, G.; Carminati, D. Molecular Techniques in Food Fermentation: Principles and Applications. In Molecular Techniques in the Microbial Ecology of Fermented Foods; Springer: Berlin/Heidelberg, Germany, 2008; pp. 1–30.
- Gibbons, J.G.; Rinker, D.C. The Genomics of Microbial Domestication in the Fermented Food Environment. Curr. Opin. Genet. Dev. 2015, 35, 1–8.
- Wang, R.; Wu, J.; Jiang, N.; Lin, H.; An, F.; Wu, C.; Yue, X.; Shi, H.; Wu, R. Recent Developments in Horizontal Gene Transfer with the Adaptive Innovation of Fermented Foods. Crit. Rev. Food Sci. Nutr. 2023, 63, 569–584.
- Steensels, J.; Gallone, B.; Voordeckers, K.; Verstrepen, K.J. Domestication of Industrial Microbes. Curr. Biol. 2019, 29, R381–R393.
- Roach, M.J.; Borneman, A.R. New Genome Assemblies Reveal Patterns of Domestication and Adaptation across Brettanomyces (Dekkera) Species. BMC Genom. 2020, 21, 194.
- Irlinger, F.; Mounier, J. Microbial Interactions in Cheese: Implications for Cheese Quality and Safety. Curr. Opin. Biotechnol. 2009, 20, 142–148.
- Leroy, S.; Christieans, S.; Talon, R. Tetracycline Gene Transfer in Staphylococcus xylosus in situ During Sausage Fermentation. Front. Microbiol. 2019, 10, 392.
- You, L.; Yang, C.; Jin, H.; Kwok, L.Y.; Sun, Z.; Zhang, H. Metagenomic Features of Traditional Fermented Milk Products. LWT 2022, 155, 112945.
- Vogel, R.F.; Becke-Schmid, M.; Entgens, P.; Gaier, W.; Hammes, W.P. Plasmid Transfer and Segregation in Lactobacillus Curvatus LTH1432 in vitro and during Sausage Fermentations. Syst. Appl. Microbiol. 1992, 15, 129–136.
- Cocconcelli, P.S.; Cattivelli, D.; Gazzola, S. Gene Transfer of Vancomycin and Tetracycline Resistances among Enterococcus faecalis during Cheese and Sausage Fermentations. Int. J. Food Microbiol. 2003, 88, 315–323.
- Haubert, L.; dos Santos Cruxen, C.E.; Fiorentini, Â.M.; da Silva, W.P. Tetracycline Resistance Transfer from Foodborne Listeria monocytogenes to Enterococcus faecalis in Minas Frescal Cheese. Int. Dairy J. 2018, 87, 11–15.
- Seifabadi, F.S.; Baserisalehi, M. Plasmid-Mediated Antibiotic-Resistant Pattern of Lactobacillus spp. Isolated From Dairy Products. Avicenna J. Clin. Microbiol. Infect. 2021, 8, 1–4.
- Heo, S.; Bae, T.; Lee, J.H.; Jeong, D.W. Transfer of a Lincomycin-Resistant Plasmid between Coagulase-Negative Staphylococci during Soybean Fermentation and Mouse Intestine Passage. FEMS Microbiol. Lett. 2019, 366, 113.
- Li, N.; Yu, H.; Liu, H.; Wang, Y.; Zhou, J.; Ma, X.; Wang, Z.; Sun, C.; Qiao, S. Horizontal Transfer of VanA between Probiotic Enterococcus faecium and Enterococcus faecalis in Fermented Soybean Meal and in Digestive Tract of Growing Pigs. J. Anim. Sci. Biotechnol. 2019, 10, 36.
- Miller, J.H.; Novak, J.T.; Knocke, W.R.; Pruden, A. Elevation of Antibiotic Resistance Genes at Cold Temperatures: Implications for Winter Storage of Sludge and Biosolids. Lett. Appl. Microbiol. 2014, 59, 587–593.
- Schjørring, S.; Krogfelt, K.A. Assessment of Bacterial Antibiotic Resistance Transfer in the Gut. Int. J. Microbiol. 2011, 2011, 312956.
- Tarrah, A.; Pakroo, S.; Corich, V.; Giacomini, A. Identification and Transferability of Tetracycline Resistance in Streptococcus thermophilus during Milk Fermentation, Storage, and Gastrointestinal Transit. Fermentation 2021, 7, 65.
- Zarzecka, U.; Zadernowska, A.; Chajęcka-Wierzchowska, W.; Adamski, P. High-Pressure Processing Effect on Conjugal Antibiotic Resistance Genes Transfer in vitro and in the Food Matrix among Strains from Starter Cultures. Int. J. Food Microbiol. 2023, 388, 110104.
- Mater, D.D.G.; Langella, P.; Corthier, G.; Flores, M.-J. A Probiotic Lactobacillus Strain Can Acquire Vancomycin Resistance during Digestive Transit in Mice. J. Mol. Microbiol. Biotechnol. 2008, 14, 123–127.
- Broaders, E.; Gahan, C.G.; Marchesi, J.R. Gut Microbes Mobile Genetic Elements of the Human Gastrointestinal Tract Potential for Spread of Antibiotic Resistance Genes. Gut Microbes 2013, 4, 271–280.
More
Information
Subjects:
Microbiology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
865
Revisions:
2 times
(View History)
Update Date:
31 Jul 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




