Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Rajeshwar Sinha | -- | 3136 | 2023-07-25 11:01:57 | | | |
| 2 | Dean Liu | Meta information modification | 3136 | 2023-07-26 02:30:02 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Singh, V.K.; Jha, S.; Rana, P.; Gupta, A.; Singh, A.P.; Kumari, N.; Mishra, S.; Singh, P.R.; Jaiswal, J.; Sinha, R.P. High-Yield Production of Mycosporine-like Amino Acids. Encyclopedia. Available online: https://encyclopedia.pub/entry/47227 (accessed on 08 February 2026).
Singh VK, Jha S, Rana P, Gupta A, Singh AP, Kumari N, et al. High-Yield Production of Mycosporine-like Amino Acids. Encyclopedia. Available at: https://encyclopedia.pub/entry/47227. Accessed February 08, 2026.
Singh, Varsha K., Sapana Jha, Palak Rana, Amit Gupta, Ashish P. Singh, Neha Kumari, Sonal Mishra, Prashant R. Singh, Jyoti Jaiswal, Rajeshwar P. Sinha. "High-Yield Production of Mycosporine-like Amino Acids" Encyclopedia, https://encyclopedia.pub/entry/47227 (accessed February 08, 2026).
Singh, V.K., Jha, S., Rana, P., Gupta, A., Singh, A.P., Kumari, N., Mishra, S., Singh, P.R., Jaiswal, J., & Sinha, R.P. (2023, July 25). High-Yield Production of Mycosporine-like Amino Acids. In Encyclopedia. https://encyclopedia.pub/entry/47227
Singh, Varsha K., et al. "High-Yield Production of Mycosporine-like Amino Acids." Encyclopedia. Web. 25 July, 2023.
Copy Citation
Ultraviolet (UV) radiation reaching the Earth’s surface is a major societal concern, and therefore, there is a significant consumer demand for cosmetics formulated to mitigate the harmful effects of UV radiation. Synthetic sunscreens being formulated to block UV penetration include inorganic metal oxide particles and organic filters. Lately, organic UV-absorbing compounds are manufactured from non-renewable petrochemicals and, as a result, there is a need to develop a sustainable manufacturing process for efficient, high-level production of a naturally occurring group of UV-absorbing compounds, namely mycosporine-like amino acids (MAAs), for use as a sunscreen additive to skincare products.
biosynthetic gene clusters
mycosporine-like amino acids
sunscreens
1. Introduction
Chemicals, including oxybenzone, ZnO, and TiO2, are frequently used in skincare products to protect against skin damage from UV rays. However, these chemicals have several negative effects on human health and the environment. Several common chemical UV sunscreen filters are absorbed by the skin and enter the bloodstream. The usage of chemical and inorganic sunscreens has increased in recent decades to counteract the harmful effects of UV radiation, but this practice has been linked to a number of skin-related diseases, including skin allergies, rashes, premature aging, dermal cancer, and other skin problems [1]. As a result, there is an increasing need to find bio-based sunscreen chemicals that are efficient, safe, sustainable, and that have the ability to prevent UV-induced damage and boost the effectiveness of natural sunscreens [2]. Mycosporines and mycosporine-like amino acids (MAAs), synthesized by both prokaryotic as well as eukaryotic organisms such as fungi, cyanobacteria, and algae, are natural UV protectants [3]. Mycosporines, which are primarily produced in fungi, consist of the nitrogen substituent of an amino acid or an imino alcohol at the C3 position, forming the cyclohexenone ring, while MAAs have an additional nitrogen substituent conjugated via imine linkage, forming the cyclohexenimine core structure [4]. MAAs are low molecular weight (<400 Da), water-soluble, and colorless UV protectants. They have a high molar extinction coefficient (ε = 28,100–50,000 M−1 cm−1), and their maximum absorption wavelength lies within 309 to 362 nm. These compounds possess a wide array of bioactivities, such as antioxidative, anti-inflammatory, anti-aging, and antitumor activities [1]. Majority of research on cyanobacterial MAAs focuses on specific areas, such as the identification of bioactive compounds, in-depth examination of their molecular mechanisms of action, and the evaluation of their bioactivities via in vitro, in silico, and in vivo analyses.
Lately, the gene-centric approach, or bottom-up approach, has been used to explain the biosynthetic abilities of cyanobacteria by combining in silico studies with functional genomics to link the genomic context, known as biosynthetic gene clusters (BGCs), to target desired metabolites [5]. These days, it is easier to discover the cyanobacterial BGCs of diverse compound families mainly through a number of in silico analysis tools available for cyanobacterial genomic data, including antiSMASH and PRISM [6][7][8]. These resources have made it easier to understand the cyanobacterial MAAs biosynthesis.
Most of the identified cyanobacterial BGCs are silent, making them a greater challenge in MAAs research. Synthetic biology approaches, such as metabolic engineering and strain mutagenesis, have been employed to activate the silent BGCs of other bacterial phyla, for instance, actinomycetes. However, the use of these approaches is less reported for the discovery of cyanobacterial products, mainly because of the slow growth of cyanobacterial strains and because these are genetically less amenable, suggesting an open area to discover and characterize the potential novel compounds from cyanobacteria [9]. The production of the encoded molecule on a large scale for usage at an industrial level has been successfully accomplished through the introduction of targeted metabolite biosynthetic genes into a suitable heterologous host. In order to produce specific metabolite analogues or to maximize the production yield, genetic contents can be redesigned with the help of heterologous expression of the targeted secondary metabolite [10][11].
2. Application of MAAs (Sunscreen) in Cosmetics
The idea of sunscreen usage has changed from being seen as only a UV-protective compound to provide broad-spectrum defense against photoaging, DNA damage, and dyspigmentation [12]. When selecting a sunscreen, one should take into account the possibility that infrared and visible light contribute to photoaging. UV rays and visible light will be shielded against by using a broad-spectrum tinted sunscreen with SPF of at least 30, which will lessen their impacts on photoaging [12]. For mending skin aging, healing, and avoiding wrinkle development, several microalgal extracts, including those from Spirulina platensis, Chlorella vulgaris, and Dunaliella salina, can be utilized [13][14]. The extracts might provide creative and potential replacements for current cosmetics, and they encourage the development of new uses for cosmetics.
2.1. Photoprotection Prospects of MAAs
MAAs, which are hydrophilic and colorless, are synthesized by marine cyanobacteria [15][16], microalgae, macroalgae, etc., that function as an antioxidant by reducing ROS-induced DNA damage and as a photoprotectant by protecting cells from UVR [16][17]. Only a small proportion of these so-called “broad-spectrum sunscreens” are truly efficient at blocking both UV-A and UV-B rays [18]. The strong ability of MAAs to absorb UVR between 309 and 362 nm defines them as a broad-spectrum sunscreen, making it essential to incorporate MAAs as a UV-filter agent in sunscreens [19]. Due to their high photostability, and resistance to heat, pH changes, and various solvents, they can be a very stable cosmetic product [20]. The first sunscreen, called Helioguard 365, was developed by the Swiss Firm Mibelle AG Biotechnology, utilizing a naturally occurring UV-blocking component known as MAA, that contains a certain amount of porphyra-334 and shinorine derived from red algae, Porphyra umbilicalis [21][22]. The MAAs derived from the algae Dunaliella, Arthrospira, and Chlorella function as sunscreens to mitigate the harm caused by UVR. Odontella aurita, a kind of microalgae, also showed potential free radical scavenging action. Coelastrin A and Coelastrin B, two new MAAs from Coelastrella rubescens, have photoprotective characteristics [23]. The recently discovered MAAs from Klebsormidium, klebsormidin A and klebsormidin B, demonstrated that UVR exposure dramatically induces their production and intracellular enrichment, indicating the role of these molecules as natural UV sunscreens [24]. However, there are still very few MAAs that are commercially available [25]. Additionally, there are two commercial products called Helioguard®365 and Helionori® that both include an extract with an enhanced or specified MAAs content [26].
Porphyra-334 and shinorine both provided concentration-dependent protection when UV-A exposure was evaluated. The recommended dosage for the best protection was 5 μg mL−1 [27]. According to Schmid et al. [26], the formulation they utilized for their study contained 5% of Helioguard®365 (final MAA concentration of 0.005%), the same base with 4% of a synthetic UV-B sunscreen and 1% of a synthetic UV-A sunscreen. It is well-known that Helioguard®365 has anti-aging and photoprotective properties. In a dose-dependent manner, Helioguard®365 concentrations of 0.125% and 0.25% increased cell viability; at 0.25% of Helioguard®365, cell viability was increased by as much as 97.8% [26]. On applying a 2% concentration of Helioguard®365 to the cell lines, the SPF value of the sunscreen increases from 7.2 to 8.2. Sunscreen containing porphyra-334 with shinorine and mycosporine-serinol in the ratio of 4.1:2.9% have the SPF value of 8.37 ± 2.12, whereas for porphyra-334 with shinorine and mycosporine-serinol when applied separately, the SPF value was observed from 4 to 6 [28]. An algal extract having a combination of palythine, asterina-330, shinorine, porphyra-334, and palythinol obtained from H. cornea and G. longissima showed a concentration-dependent increase in the SPF value. At a density of 13.9 mg DW of algae per cm−2, the SPF values of G. longissima and H. cornea, respectively, were found to be 7.5 and 4.8. Both algal extracts increased TNF-α and IL-6 production, while exhibiting no cytotoxicity toward human cells [29]. Shinorine and porphyra-334 extract, which is found in liposomes and is encapsulated in Helioguard®365, reduce the lipid peroxidation of human skin, improve skin firmness and smoothness, and play a role in the prevention of premature aging. Helionori® can prevent sunburn and preserve membrane lipids because it has mycosporine-palythine, porphyra-334, and shinorine, which make it photo- and heat-stable [30]. Biotechnologically, MAAs can be used for a variety of commercial purposes, such as in dietary supplements, medicine, functional organic devices, and others. Therefore, the commercialization of MAAs with multiple uses is an exciting prospective endeavor [31].
2.2. Anti-Aging Prospects of MAAs
It might be challenging to characterize skin aging since it is a result of systems involving heredity and environmental variables [32]. Unlike chronological aging, premature skin aging, or photoaging, is brought on by exposure to environmental stress [33]. Clinical manifestations of photoaging include dryness, hyper- and hypo-pigmentation, leathery texture, and wrinkles [33]. As already mentioned, cyanobacteria synthesize molecules that may be used in skin moisturization, UV protection (MAAs and SCY), and shielding against ROS (MAAs, SCY, PBPs, and polyphenols), making them intriguing for use in skin anti-aging treatments [16]. These effects were also mentioned for fibroblasts, which are responsible for the skin’s firmness and elasticity, as well. The extracellular matrix (ECM), which mainly consists of collagen and elastin and provides firmness and elasticity, is produced by fibroblasts within the dermis [34]. In studies employing normal human dermal fibroblasts (nHDFs) exposed to UV-B radiation, it was shown that extracts from Arthrospira platensis increased cell viability and reduced DNA damage by inhibiting thymine dimers and matrix metalloproteinases (MMP) [35]. Mycosporine-glycine, porphyra-334, palythine, and shinorine, the most prevalent MAAs, were examined for their UV-protective properties in recent research using a variety of cell models (human skin fibroblasts and HaCaT keratinocytes) to demonstrate their effectiveness as possible sunscreens [20]. Additionally, a number of anti-aging actions, particularly those that target the elastic fibers of the extracellular matrix (ECM), such as collagen and elastin, as well as their remodeling enzymes, have been identified [36][37]. Furthermore, there are two commercial products called Helioguard®365 and Helionori® that both include an extract with an enhanced or specified MAAs content [26].
The prevention of skin aging is a result of several mechanisms, including skin hydration, UV protection, stimulation of fibroblast growth, and an increase in antioxidant capacity. Cyanobacteria produce substances that have been shown to interfere with all these processes in the aforementioned areas, making them attractive for cosmetics meant to delay the onset of skin aging.
3. Biosynthesis and Genetic Regulation of MAAs
Despite having different molecular structures, mycosporines are composed of a typical cyclohexenone ring structure, which provides them the same spectral characteristics and absorption maxima [38]. Studies have demonstrated that a cyanobacterium or a cyanobacterial ancestor acted as the progenitor of the enzymes for MAAs production [39][40][41][42][43]. MAAs are synthesized in cyanobacteria using a four-enzyme pathway. It was discovered that different cyanobacterial species exhibit significant genetic variation in the mys gene cluster, which becomes involved in MAAs biosynthesis [44]. The latest studies on the biochemical processes and genetic research have contributed to developing knowledge of the fundamental steps that are involved in the biosynthesis of MAAs. Anabaena variabilis has been used to explain the initial step in the biosynthesis of MAAs. Therefore, it is believed that cyanobacteria were the first producers of MAAs, and the genes for MAA biosynthesis most likely spread to other organisms. The shikimate pathway is suggested as a potential biosynthetic route for MAAs [45]. MAAs are synthesized from phosphoenolpyruvate (PEP) and erythrose 4-phosphate (E4P) (an intermediate in the pentose phosphate pathway). PEP and E4P react to form 3-deoxy-D-arabinoheptulosonate-7-phosphate (DAHP), which is catalyzed by the enzyme DAHP synthase. DAHP is further involved in the formation of 3-dehydoquinate (3-DHQ), which is a six-membered carbon ring-like structure (Figure 1). The 3-DHQ produces 4-deoxygadusol (4-DG), which is subsequently followed by gadusol [42]. Through a separate route, sedoheptulose 7-phosphate (S7P), another intermediate product of the PPP, is converted into 4-DG in the presence of the enzyme dimethyl 4-degadusol synthase (DDGS; MysA) and an O-methyltransferase (O-MT; MysB) [46].
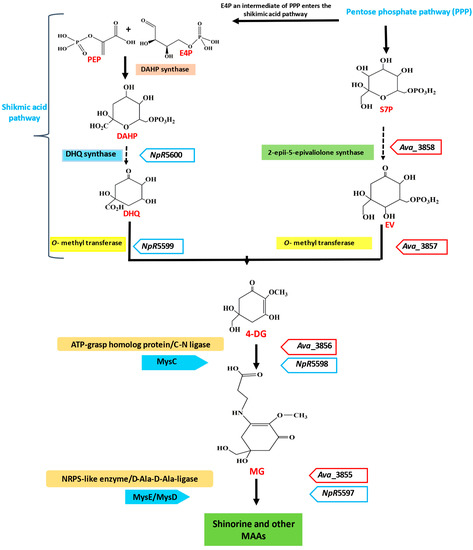
Figure 1. Schematic representation of the biosynthetic pathway of MAAs in cyanobacteria. Synthesis from the shikimic acid pathway intermediate and the pentose phosphate intermediate is shown. Enzymes and genes (in italics) involved in the bioprocess are mentioned on both sides of the arrows. PEP, phosphoenolpyruvate; E4P, erythrose 4-phosphate; DAHP, 3-deoxy-D-arabino-heptulosonate; S7P, sedoheptulose-7-phosphate; 3-DHQ, 3-dehydroquinate; EV, 2-epi-5-epi-valiolone.
The common precursor for all MAAs in both pathways is 4-DG, and the conjugation of this precursor with a glycine molecule results in a simple mono-substituted cyclohexenone-type MAA, called mycosporine-glycine (MG). This further acts as a common intermediate in the production of di-substituted (aminocyclohexene imine-type) MAAs, such as porphyra-334 (P-334) and shinorine (SH). In some species, MAAs cannot be synthesized simply by substituting an amino acid for 4-DG, but they are produced through shifting the side chains of amino acids via condensation (for the esterification process and amidation), dehydration (for the formation of double bonds), decarboxylation (for chain shortening), and oxidation and reduction (for hydroxylation) [45]. The shikimic acid pathway has been scientifically investigated in order to further comprehend how it affects the production of 4-DG, which is the precursor molecule for MAAs biosynthesis [15]. A shikimate intermediate, [Ue14C] 3-dehydroquinic acid, was selectively taken up by Trichothecium roseum, which resulted in structurally similar fungal mycosporines’ production [47]. When [14C] pyruvate was studied for MAAs biosynthesis via phosphoenolpyruvate, radiolabeled MAAs having more specificity were formed, while [14C] acetate (polyketide pathway) was unable to produce MAAs [48]. Each species requires a different set of biosynthetic pathways for producing MAAs, and these pathways are affected by many abiotic factors, such as salinity, temperature, humidity, moisture loss, and other abiotic factors [49]. Cyanobacteria were not able to produce MAAs in the presence of tyrosine, which inhibits the shikimic acid pathway. A similar result was found in the case of Nostoc commune when it was exposed to glyphosate, which inhibits the shikimic acid pathway. Therefore, the shikimic acid pathway can be suppressed or inhibited by exogenous tyrosine or glyphosate [42].
Solar radiation intensity and spectrum are additional factors that influence the biosynthesis of MAAs [50]. MG and serine are condensed by an NRPS-like enzyme with the gene ava_3855, producing shinorine as a result. O-MT is encoded by the gene ava_3857, while the enzyme S7P cyclase, known as 2-epi-5-epivaliolone synthase (EVS), is encoded by the gene ava_3858. The enzymes along with their genes that are involved in biosynthesis of MAAs in different organisms are listed in Table 1. Together, these gene products catalyze the production of 4-DG, the original precursor of mycosporine [51]. Demethyl-4-deoxygadusol synthase and O-MT enzyme are synthesized by genes such as ava_3858 and ava_3857, respectively, to produce 4-DG. The product ava_3856 promotes the addition of glycine to 4-DG to produce mycosporine-glycine (MG) [31]. The 4-DG acts as the prominent precursor utilized in both pathways to produce all MAAs [52].
Table 1. The enzymes along with their genes that are involved in the biosynthesis of MAAs in different organisms.
| Organism | Genes (Protein ID) | Accession No. | ||||
|---|---|---|---|---|---|---|
| DDG Synthase | O-MT | ATP-Grasp | NRPS-like | D-Ala D-Ala Ligase Homolog | ||
| Anaebena variabilis ATCC29413 | ava_3858 (ABA23463.1) |
ava_3857 (ABA23462.1) |
ava_3856 (ABA23461.1) |
ava_3855 (ABA23460.1) |
- | CP000117.1 |
| Nostoc punctiforme ATCC29133 | npun_R5600 (ACC83905.1) |
npun_R5599 (ACC83904.1) |
npun_R5598 (ACC83903.1) |
- | npun_F5597 (ACC83902.1) |
CP001037.1 |
| Aspergillus nidulans FGSC A4 | an6403.4 (CBF69538.1) |
an6402.4 (CBF69540.1) |
an6402.4 (CBF69540.1) |
- | - | BN001301.1 |
| Actinosynnema mirum DSM 43827 | amir_4259 (ACU38114.1) |
amir_4258 (ACU38113.1) |
amir_4257 (ACU38112.1) |
- | amir_4256 (ACU38111.1) |
CP001630.1 |
In majority of cyanobacteria, an operon usually carries the genes for a demethyl-4-deoxygadusol synthase and an O-MT, both of which are necessary for the biosynthesis of 4-DG [53]. The biosynthetic genes npR5600, npR5599, npR5598, and npR5597 are found in Nostoc punctiforme ATCC 29133, while ava_3858, ava_3857, ava_3856, and ava_3855 are found in A. variablilis. The exogenous supply of S7P in Escherichia coli resulted in heterologous expression of the genes npR5600–npR5598, thereby producing MG [54]. The biosynthesis of MAAs starts with the intermediate S7P of the PPP in N. punctiforme ATCC 29133 and A. variabilis ATCC 29413 [15]. The O-MT gene (ava-3857) deletion in A. variabilis ATCC 29413 revealed that both the shikimate and PPP depend on this gene product for the biosynthesis of MAAs [46]. When the production of MAAs was studied in four cyanobacteria, such as Anabaena sp. PCC 7120, A. variabilis PCC 7937, Synechococcus sp. PCC 6301, and Synechocystis sp. PCC 6803, MAAs were only produced in A. variabilis PCC 7937. It was reported by genome mining that two sets of the 3-dehydroquinate synthase (DHQS) genes, YP_324358 and YP_324879, were present in A. variabilis PCC 7937, and it was revealed by genomic region analysis that YP_324358 contains an O-MT gene, YP_324357, downstream to it, while the rest of the cyanobacteria lack these. Deoxygadusol, which makes up the common core of all MAAs, is synthesized in the presence of YP_324358 and YP_324357 genes. In N. punctiforme ATCC 29133, when DHQ was combined with the DHQ synthase and O-MT homologues (npR5600 and npR5599, respectively) in the presence of the cofactors: nicotinamide adeninedinucliotide (NAD+), S-adenosylmethionine (SAM), and Co2+, 6-deoxygadusol (6-DG) was not synthesized. The notion that a shikimate pathway intermediate is involved in MAA biosynthesis was disproved. The 6-DG was synthesized in the presence of SAM, NAD+, and Co2+. Therefore, it can be concluded that 6-DG and glycine are converted into MG by the A. variabilis gene ava_3856, with the involvement of ATP and Mg2+ cofactors [55]. The pathway for the production of MAA is the same in N. punctiforme ATCC 29133 and A. variabilis, and the homologous genes (npR5598–5600) are involved [56]. Figure 2 represents the MAAs biosynthetic gene clusters of different microorganisms. NpR5598 functions as an ava_3856 homologue in N. punctiforme, although its specific activity has not been identified. Homologues of ava_3855 are not found in the genome of N. punctiforme, and it was also lacking in cyanobacteria which contain MAA clusters. MG was synthesized in E. coli by heterologous expression of mysA, mysB, and mysC, represented by npR5600, npR5599, and npR5598 genes. Based on the fact that the N. punctiforme gene product NpF5597 has conserved homologues that are spatially associated with the MAA cluster in numerous cyanobacteria, it shows homologies to the recognized amino acid-ligating enzymes [54].
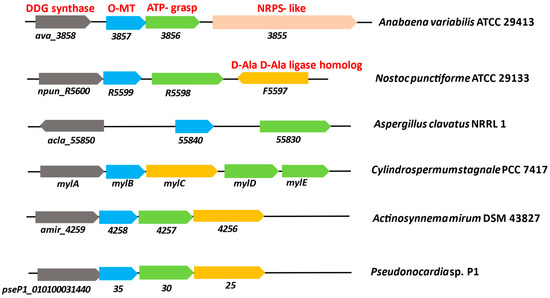
Figure 2. Genes involved in the biosynthesis of mycosporines and MAAs in cyanobacteria and other organisms. A. variabilis ATCC 29413 and N. punctiforme ATCC 29133 are known producers of MAAs. Comparison of genomic regions of Anabaena variabilis ATCC 29413 and N. punctiforme ATCC 29133 with Aspergillus clavatus NRRL 1, Cylindrospermum stagnale PCC 7417, Actinosynnema mirum DSM 43827, and Pseudonocardia sp. P1 (modified from Miyamoto et al. [57]).
In a genome-mining study, it was reported that production of MAAs takes place in cyanobacteria in the presence of homologs of the EVS gene and is absent in non-producers [58]. In A. variabilis ATCC 29413, the genetics of MAAs and key processes in MAAs production were studied [53].
4. Toolkits for Heterologous Production of MAAs
The production of MAAs from both native and heterologous hosts has been transformed by recent developments in metabolic engineering methodologies and the use of synthetic biology technologies [59]. Cloning and assembly can be the main focus for the heterologous expression of cyanobacterial MAAs, followed by BGC expression, heterologous host selection, and product optimization (Figure 3).
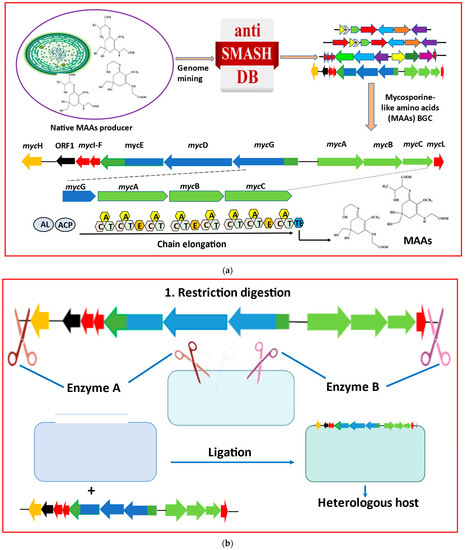
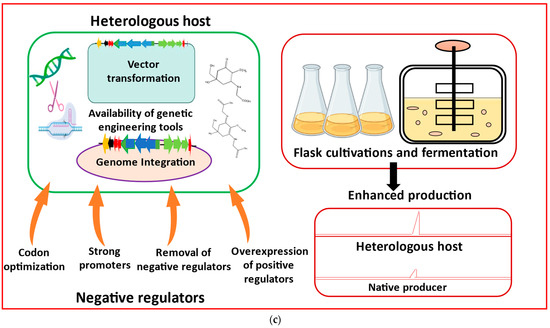
Figure 3. Major metabolic engineering and synthetic biology toolkits essential for heterologous production of mycosporine-like amino acids (MAAs) are listed as: (a) Genome mining using cyanobacterial genomic databases to identify BGCs responsible for MAAs biosynthesis. Domain ‘A’ represents adenylation, and ‘C’, ‘T’, ‘E’, and ‘TE’ represent condensation, thiolation, epimerization, and thioesterase, respectively. Acyl-CoA ligase (AL) and acyl-carrier protein (ACP) are additionally needed to initiate the secondary metabolite biosynthesis. (b) Restriction digestion and ligation, a primary approach of cloning and assembling BGCs, is most commonly utilized for heterologous expression of MAAs. (c) Considerations for heterologous expression and product analysis in heterologous hosts (modified from Sharma et al. [60]).
References
- Singh, A.; Čížková, M.; Bišová, K.; Vítová, M. Exploring mycosporine-like amino acids (MAAs) as safe and natural protective agents against UV-induced skin damage. Antioxidants 2021, 10, 683.
- Lawrence, K.P.; Long, P.F.; Young, A.R. Mycosporine-like amino acids for skin photoprotection. Curr. Med. Chem. 2018, 25, 5512.
- Shinzato, C.; Shoguchi, E.; Kawashima, T.; Hamada, M.; Hisata, K.; Tanaka, M.; Fujie, M.; Fujiwara, M.; Koyanagi, R.; Ikuta, T.; et al. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 2011, 476, 320.
- Sinha, R.P.; Singh, S.P.; Häder, D.-P. Database on mycosporines and mycosporine-like amino acids (MAAs) in fungi, cyanobacteria, macroalgae, phytoplankton and animals. J. Photochem. Photobiol. B 2007, 89, 29–35.
- Dittmann, E.; Gugger, M.; Sivonen, K.; Fewer, D.P. Natural product biosynthetic diversity and comparative genomics of the cyanobacteria. Trends Microbiol. 2015, 23, 642–652.
- Skinnider, M.A.; Dejong, C.A.; Rees, P.N.; Johnston, C.W.; Li, H.; Webster, A.L.; Wyatt, M.A.; Magarvey, N.A. Genomes to natural products prediction informatics for secondary metabolomes (PRISM). Nucleic Acids Res. 2015, 43, 9645–9662.
- Tietz, J.I.; Schwalen, C.J.; Patel, P.S.; Maxson, T.; Blair, P.M.; Tai, H.C.; Zakai, U.I.; Mitchell, D.A. A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nat. Chem. Biol. 2017, 13, 470–478.
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87.
- Berla, B.M.; Saha, R.; Immethun, C.M.; Maranas, C.D.; Moon, T.S.; Pakrasi, H. Synthetic biology of cyanobacteria: Unique challenges and opportunities. Front. Microbiol. 2013, 4, 246.
- Huo, L.; Hug, J.J.; Fu, C.; Bian, X.; Zhang, Y.; Müller, R. Heterologous expression of bacterial natural product biosynthetic pathways. Nat. Prod. Rep. 2019, 36, 1412–1436.
- Zhang, J.J.; Tang, X.; Moore, B.S. Genetic platforms for heterologous expression of microbial natural products. Nat. Prod. Rep. 2019, 36, 1313–1332.
- Guan, L.L.; Lim, H.W.; Mohammad, T.F. Sunscreens and photoaging: A review of current literature. Am. J. Clinic. Dermatol. 2021, 22, 819–828.
- Wang, H.D.; Chen, C.; Huynh, P.; Chang, J. Exploring the potential of using algae in cosmetics. Bioresour. Technol. 2015, 184, 355–362.
- Mishra, S.; Gupta, A.; Upadhye, V.; Singh, S.C.; Sinha, R.P.; Häder, D.-P. Therapeutic strategies against biofilm infections. Life 2023, 13, 172.
- Rosic, N.N. Mycosporine-like amino acids: Making the foundation for organic personalised sunscreens. Mar. Drugs 2019, 17, 638.
- Gupta, A.; Singh, A.P.; Singh, V.K.; Singh, P.R.; Jaiswal, J.; Kumari, N.; Upadhye, V.; Singh, S.C.; Sinha, R.P. Natural sun-screening compounds and DNA-repair enzymes: Photoprotection and photoaging. Catalysts 2023, 13, 745.
- Fernanda, P.M. Algae and aquatic macrophytes responses to cope to ultraviolet radiation—A Review. Emir. J. Food Agric. 2012, 24, 527–545.
- Singh, A.; Tyagi, M.B.; Kumar, A. Cyanobacteria growing on tree barks possess high amount of sunscreen compound mycosporinelike amino acids (MAAs). Plant Physiol. Biochem. 2017, 119, 110–120.
- Kumari, N.; Pandey, A.; Gupta, A.; Mishra, S.; Sinha, R.P. Characterization of UV-screening pigment scytonemin from cyanobacteria inhabiting diverse habitats of Varanasi, India. Biologia 2023, 78, 319.
- Chrapusta, E.; Kaminski, A.; Duchnik, K.; Bober, B.; Adamski, M.; Bialczyk, J. Mycosporine-like amino acids: Potential health and beauty ingredients. Mar. Drugs 2017, 15, 326.
- Bhatia, S.; Garg, A.; Sharma, K.; Kumar, S.; Sharma, A.; Purohit, A.P. Mycosporine and mycosporine-like amino acids: A paramount tool against ultra violet irradiation. Phcog. Rev. 2011, 5, 138–146.
- Siezen, R.J. Microbial sunscreens. Microb. Biotechnol. 2011, 4, 1–7.
- Zaytseva, A.; Chekanov, K.; Zaytsev, P.; Bakhareva, D.; Gorelova, O.; Kochkin, D.; Lobakova, E. Sunscreen effect exerted by secondary carotenoids and mycosporine-like amino acids in the aeroterrestrial chlorophyte Coelastrella rubescens under high light and UV-A irradiation. Plants 2021, 10, 2601.
- Hartmann, A.; Glaser, K.; Holzinger, A.; Ganzera, M.; Karsten, U. Klebsormidin A and B, two new UV-sunscreen compounds in green microalgal Interfilum and Klebsormidium species (Streptophyta) from terrestrial habitats. Front. Microbiol. 2020, 11, 499.
- Van Hal, J.W.; Huijgen, W.J.J.; López-Contreras, A.M. Opportunities and Challenges for Seaweed in the Biobased Economy. Trends Biotechnol. 2014, 32, 231–233.
- Schmid, D.; Schürch, C.; Zülli, F. Mycosporine-like amino acids from red algae protect against premature skin-ageing. Euro Cosmet. 2006, 9, 1–4.
- Schmid, D.; Schürch, C.; Zülli, F.; Nissen, H.-P.; Prieur, H. Mycosporine-like amino acids: Natural UV-screening compounds from red algae to protect the skin against photoaging. Söfw. J. 2003, 129, 38–42.
- de la Coba, F.; Aguilera, J.; Korbee, N.; de Gálvez, M.V.; Herrera-Ceballos, E.; Álvarez-Gómez, F.; Figueroa, F.L. UVA and UVB photoprotective capabilities of topical formulations containing mycosporine-like amino acids (MAAs) through different biological effective protection factors (BEPFs). Mar. Drugs 2019, 17, 55.
- Álvarez-Gómez, F.; Korbee, N.; Casas-Arrojo, V.; Abdala-Díaz, R.T.; Figueroa, F.L. UV photoprotection, cytotoxicity andimmunology capacity of red algae extracts. Molecules 2019, 24, 341.
- Soule, T.; Garcia-Pichel, F. Ultraviolet photoprotective compounds from cyanobacteria in biomedical applications. In Cyanobacteria: An Economic Perspective; Sharma, N.K., Rai, A.K., Stal, L.J., Eds.; Willey Online Library: Hoboken, NJ, USA, 2013; pp. 119–143.
- Jain, S.; Prajapat, G.; Abrar, M.; Ledwani, L.; Singh, A.; Agrawal, A. Cyanobacteria as efficient producers of mycosporine-like amino acids. J Basic Microbiol. 2017, 57, 715–727.
- Makrantonaki, E.; Brink, T.C.; Zampeli, V.; Elewa, R.M.; Mlody, B.; Hossini, A.M.; Zouboulis, C.C. Identification of biomarkers of human skin ageing in both genders. Wnt signalling—A label of skin ageing? PLoS ONE 2012, 7, e50393.
- Mukherjee, K.; Maity, N.; Nema, N.K.; Sarkar, B.K. Bioactive compounds from natural resources against skin ageing. Phytomedicine 2011, 19, 64–73.
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The extracellular matrix at a glance. J. Cell Sci. 2010, 123, 4195–4200.
- Lee, J.J.; Kim, K.B.; Heo, J.; Cho, D.H.; Kim, H.S.; Han, S.H.; Bae, S. Protective effect of Arthrospira platensis extracts against ultraviolet B-induced cellular senescence through inhibition of DNA damage and matrix metalloproteinase-1 expression in human dermal fibroblasts. J. Photochem. Photobiol. B 2017, 173, 196–203.
- Suh, S.-S.; Hwang, J.; Park, M.; Seo, H.H.; Kim, H.-S.; Lee, J.H.; Moh, S.H.; Lee, T.-K. Anti-inflammation activities of mycosporine-like amino acids (MAAs) in response to UV radiation suggest potential anti-skin ageing activity. Mar. Drugs 2014, 12, 5174–5187.
- Orfanoudaki, M.; Hartmann, A.; Alilou, M.; Gelbrich, T.; Planchenault, P.; Derbré, S.; Schinkovitz, A.; Richomme, P.; Hensel, A.; Ganzera, M. Absolute configuration of mycosporine-like amino acids, their wound healing properties and in vitro anti-ageing effects. Mar. Drugs 2019, 18, 35.
- Raj, S.; Kuniyil, A.M.; Sreenikethanam, A.; Gugulothu, P.; Jeyakumar, R.B.; Bajhaiya, A.K. Microalgae as a source of mycosporine-like amino acids (MAAs); advances and future prospects. Int. J. Environ. Res. Public Health 2021, 18, 12402.
- Rozema, J.; Björn, L.O.; Bornman, J.F.; Gaberščik, A.; Häder, D.P.; Trošt, T.; Germ, M.; Klisch, M.; Gröniger, A.; Sinha, R.P.; et al. The role of UV-B radiation in aquatic and terrestrial ecosystems—An experimental and functional analysis of the evolution of UV-absorbing compounds. J. Photochem. Photobiol. B 2002, 66, 2–12.
- Waller, R.F.; Slamovits, C.H.; Keeling, P.J. Lateral gene transfer of a multigene region from cyanobacteria to dinoagellates resulting in a novel plastid-targeted fusion protein. Mol. Biol. Evol. 2006, 23, 1437–1443.
- Starcevic, A.; Akthar, S.; Dunlap, W.C.; Shick, J.M.; Hranueli, D.; Cullum, J.; Long, P.F. Enzymes of the shikimic acid pathway encoded in the genome of a basal metazoan, Nematostella vectensis, have microbial origins. Proc. Natl. Acad. Sci. USA 2008, 105, 2533–2537.
- Singh, S.P.; Häder, D.P.; Sinha, R.P. Bioinformatics evidence for the transfer of mycosporine-like amino acid core (4-deoxygadusol) synthesizing gene from cyanobacteria to dinoflagellates and an attempt to mutate the same gene (YP_324358) in Anabaena variabilis PCC 7937. Gene 2012, 500, 155–163.
- Richa; Sinha, R.P. Biomedical applications of mycosporine-like amino acids. In Marine Microbiology: Bioactive Compounds and Biotechnological Applications; Kim, S.-K., Ed.; Wiley-VCH: Weinheim, Germany, 2016; pp. 509–534.
- Rosic, N.N. Recent advances in the discovery of novel marine natural products and mycosporine-like amino acid UV-absorbing compounds. Appl. Microbiol. Biotechnol. 2021, 105, 7053–7067.
- Wada, N.; Sakamoto, T.; Matsugo, S. Mycosporine-like amino acids and their derivatives as natural antioxidants. Antioxidants 2015, 4, 603–646.
- Sepúlveda, D.; Campusano, S.; Moliné, M.; Barahona, S.; Baeza, M.; Alcaíno, J.; Cifuentes, V. Unraveling the molecular basis of mycosporine biosynthesis in fungi. Int. J. Mol. Sci. 2023, 24, 5930.
- Favre-Bonvin, J.; Bernillon, J.; Salin, N.; Arpin, N. Biosynthesis of mycosporines: Mycosporine glutaminol in Trichothecium roseum. Phytochemistry 1987, 26, 2509–2514.
- Portwich, A.; Garcia-Pichel, F. Biosynthetic pathway of mycosporines (mycosporine-like amino acids) in the cyanobacterium Chlorogloeopsis sp. strain PCC 6912. Phycologia 2003, 42, 384–392.
- Geraldes, V.; Pinto, E. Mycosporine-like amino acids (MAAs): Biology, chemistry and identification features. Pharmaceuticals 2021, 14, 63.
- Rastogi, R.P.; Richa; Sinha, R.P.; Singh, S.P.; Häder, D.-P. Photoprotective compounds from marine organisms. J. Ind. Microbiol. Biotechnol. 2010, 37, 537–558.
- Singh, S.P.; Klisch, M.; Sinha, R.P.; Häder, D.-P. Genome mining of mycosporine-like amino acid (MAA) synthesizing and non-synthesizing cyanobacteria: A bioinformatics study. Genomics 2010, 95, 120–128.
- Rosic, N. Genome mining as an alternative way for screening the marine organisms for their potential to produce UV-absorbing mycosporine-like amino acid. Mar. Drugs 2022, 20, 478.
- Spence, E.; Dunlap, W.C.; Shick, J.M.; Long, P.F. Redundant pathways of sunscreen biosynthesis in a cyanobacterium. ChemBioChem 2012, 13, 531–533.
- Gao, Q.; Garcia-Pichel, F. An ATP-grasp ligase involved in the last biosynthetic step of the iminomycosporine shinorine in Nostoc punctiforme ATCC 29133. J. Bacteriol. 2011, 193, 5923–5928.
- Carreto, J.I.; Carignan, M.O. Mycosporine-like amino acids: Relevant secondary metabolites. Chemical and ecological aspects. Mar. Drugs 2011, 9, 387–446.
- Katoch, M.; Mazmouz, R.; Chau, R.; Pearson, L.A.; Pickford, R.; Neilan, B.A. Heterologous production of cyanobacterial mycosporine-like amino acids mycosporine-ornithine and mycosporine-lysine in Escherichia coli. Appl. Environ. Microbiol. 2016, 82, 6167–6173.
- Miyamoto, K.T.; Komatsu, M.; Ikeda, H. Discovery of gene cluster for mycosporine-like amino acid biosynthesis from Actinomycetales microorganisms and production of a novel mycosporine-like amino acid by heterologous expression. Appl. Environ. Microbiol. 2014, 80, 5028–5036.
- Rosic, N.N. Phylogenetic analysis of genes involved in mycosporine-like amino acid biosynthesis in symbiotic dinoflagellates. Appl. Microbiol. Biotechnol. 2012, 94, 29–37.
- Smanski, M.J.; Zhou, H.; Claesen, J.; Shen, B.; Fischbach, M.A.; Voigt, C.A. Synthetic biology to access and expand nature’s chemical diversity. Nat. Rev. Microbiol. 2016, 14, 135–149.
- Sharma, K.; Ghiffary, M.R.; Kim, H.U.; Lee, S.Y. Engineering heterologous hosts for the enhanced production of non-ribosomal peptides. Biotechnol. Bioprocess Eng. 2020, 25, 795–809.
More
Information
Subjects:
Biotechnology & Applied Microbiology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.8K
Revisions:
2 times
(View History)
Update Date:
26 Jul 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




