Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Seung-Hyeon Seok | -- | 1734 | 2023-07-25 09:10:50 | | | |
| 2 | Jessie Wu | + 2 word(s) | 1736 | 2023-07-25 09:33:45 | | | | |
| 3 | Jessie Wu | -1 word(s) | 1735 | 2023-07-25 09:35:14 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Seok, S. Protein Ser/Thr Phosphatase Folding. Encyclopedia. Available online: https://encyclopedia.pub/entry/47211 (accessed on 07 February 2026).
Seok S. Protein Ser/Thr Phosphatase Folding. Encyclopedia. Available at: https://encyclopedia.pub/entry/47211. Accessed February 07, 2026.
Seok, Seung-Hyeon. "Protein Ser/Thr Phosphatase Folding" Encyclopedia, https://encyclopedia.pub/entry/47211 (accessed February 07, 2026).
Seok, S. (2023, July 25). Protein Ser/Thr Phosphatase Folding. In Encyclopedia. https://encyclopedia.pub/entry/47211
Seok, Seung-Hyeon. "Protein Ser/Thr Phosphatase Folding." Encyclopedia. Web. 25 July, 2023.
Copy Citation
Post-translational modification (PTM) is a key mechanism providing the functional diversity of proteins in cellular signaling and physiology and changing the functions or stability of proteins.
Protein Kinase
Protein Phosphatase
Protein Structure
1. Introduction
Protein phosphorylation is one of the most widely observed and important PTM processes [1]. Protein phosphorylation is regulated by protein kinases, each of which covalently attaches a phosphate group to an amino acid side chain on serine (Ser), threonine (Thr), or tyrosine (Tyr), and by protein phosphatases, each of which, conversely, removes a phosphate group from a phosphoprotein (Figure 1). These reversible enzyme activities provide a regulatory mechanism, altering the changing diverse functions and stability of proteins in cellular processes and the diverse physiological functions related to musculoskeletal regulation, neurologic mechanisms and behavior, immune response, endocrine action, and so forth [2][3].
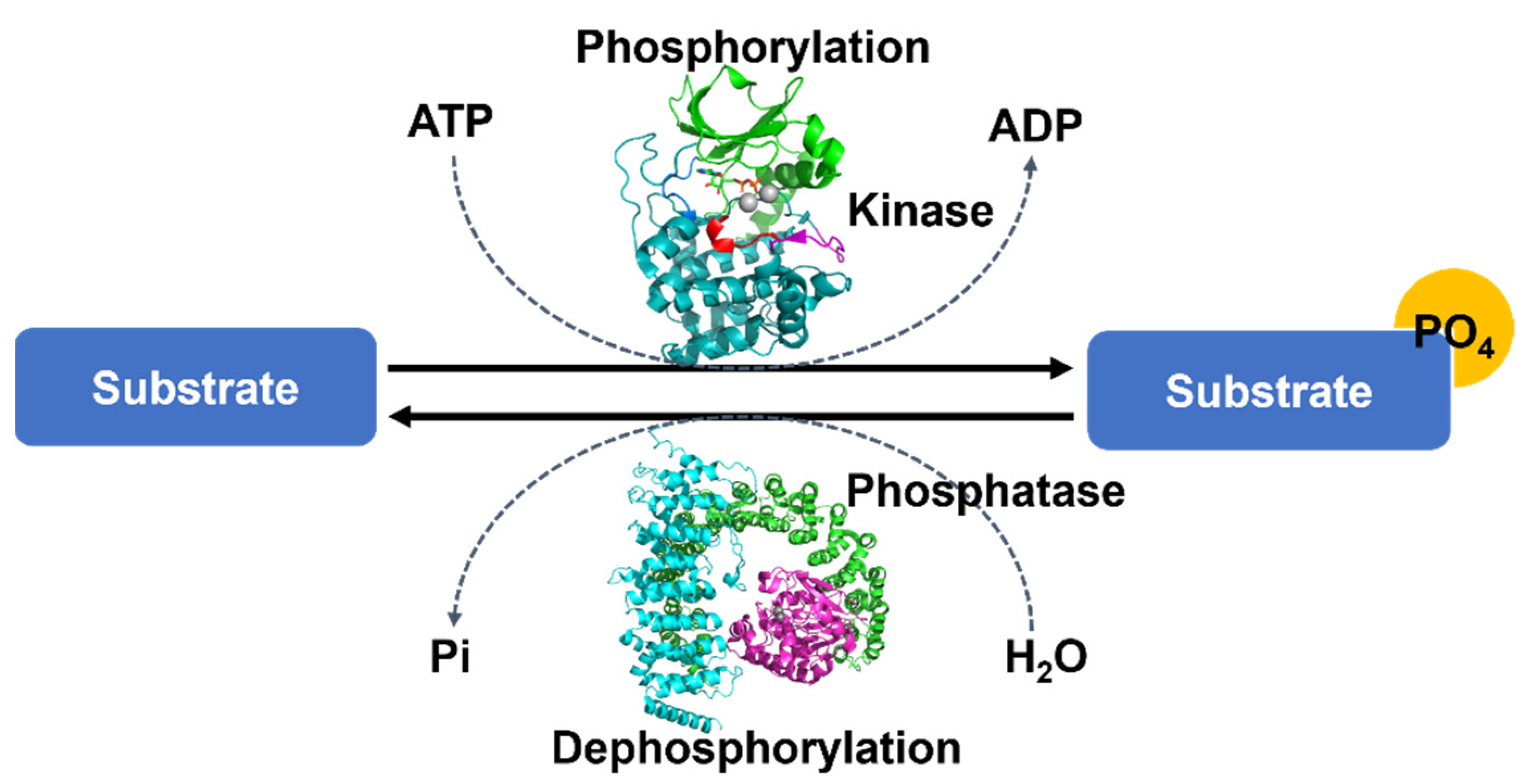
Figure 1. The overall mechanism of protein phosphorylation regulated by protein kinases and protein phosphatase. Each protein kinase covalently attaches a phosphate group from ATP to a protein substrate and each protein phosphatase removes a phosphate group from a phosphoprotein. These processes are reversible. Protein structures were drawn by the programs PyMOL (The PyMOL Molecular Graphics System, Version 2.4.1 Schrödinger, LLC., Cambridge, MA, USA).
More than two-thirds of over 20,000 proteins from human proteomes have been reported to be phosphorylated, and over 200,000 unique phosphorylation sites have been detected [4]. Reflecting the importance and abundance of protein phosphorylation, the human genome encodes more than 500 protein kinases and ~180 protein phosphatases [5]. Although the activities of protein kinases and protein phosphatases are counteracting, the ratio of the number of kinase to phosphatase species is highly unbalanced. Due to the dynamic assembly of phosphatase catalytic subunits into diverse holoenzymes that target substrates, the unbalance is, however, resolved [6]. Most phosphorylation sites in proteins are localized on their disordered or dynamic regions [7][8][9], and the reversible attachment or detachment of phosphate groups in these regions causes changes in their molecular structures to induce protein conformational shifts, interference of protein–protein or protein–nucleic acid interactions, or disorder-to-order or order-to-disorder transitions in proteins [10][11]. The structural flexibility of proteins makes it challenging to study the structural basis of kinase and phosphatase activities and the reversible phosphorylation/dephosphorylation mechanisms.
2. Protein Ser/Thr Phosphatase Folding
The extremely high stability of phosphorylated residues means that dephosphorylation by protein phosphatases is essential for the regulation of the dynamic and reversible states of proteins. As a counterreaction with protein kinases, each protein phosphatase removes a phosphate group from the phosphorylated amino acid residue of its substrate protein [12].
Most protein phosphatases can also be classified into two families, protein Ser/Thr phosphatases and protein Tyr phosphatases, depending on the dephosphorylation of the phosphorylated residues in their target proteins or substrates. In this research, some minor dephosphorylation events that occur on the phosphohistidine will be excluded and members of the protein Ser/Thr phosphatases will be described. As described above, the human genome contains more than 500 protein kinases, whereas it contains around 180 protein phosphatases [5]. Furthermore, the number of protein Ser/Thr phosphatases encoded in the human genome (~50) is 10 times lower than the number of Ser/Thr protein kinases encoded in the human genome [5][13]. This gap between protein Ser/Thr phosphatases and Ser/Thr protein kinases could be explained by the dynamic assembly and combinatorial conformation of holoenzymes with shared catalytic subunits of protein Ser/Thr phosphatases and the diverse regulatory subunits that target distinct proteins [14][15].
The protein Ser/Thr phosphatases can be divided into two families, phosphoprotein phosphatases and metal-dependent protein phosphatases, based on their sequence homology and catalytic metal dependence [16]. The phosphoprotein phosphatase family, genetically encoding approximately 15 human proteins and Zn/Fe-dependent enzymes, includes protein phosphatase 1 (PP1), PP2A, PP2B (calcineurin), PP4, PP5, PP6, and PP7 [4][17]. Most phosphoprotein phosphatases share a conserved 30 kD catalytic domain containing highly conserved sequences, GDxHG, GDxVDRG, and GNHE.
2.1. Protein Phosphatase 1
Protein phosphatase 1 (PP1), the most widely expressed protein Ser/Thr phosphatase that is responsible for more than 50% of all dephosphorylation reactions in humans, plays a key role in the regulation of a wide range of cellular processes regulated. PP1 is one of the simplest phosphatases and consists of only a highly conserved catalytic subunit, which is associated with at least one of 200 known regulatory proteins. The catalytic domain of PP1 comprises a central β-sandwich formed by two mixed β-sheets surrounded by two α-helical domains on both sides [18][19]. Two metal ions (Mn2+ or Fe2+), located in the active site of a central β-sandwich, are coordinated with six highly conserved residues, three histidines, two aspartic acids, and one asparagine. The binding and activation of a water molecule by two metal ions initiates a nucleophilic attack on the phosphorous atom (Figure 2a) [18][19].
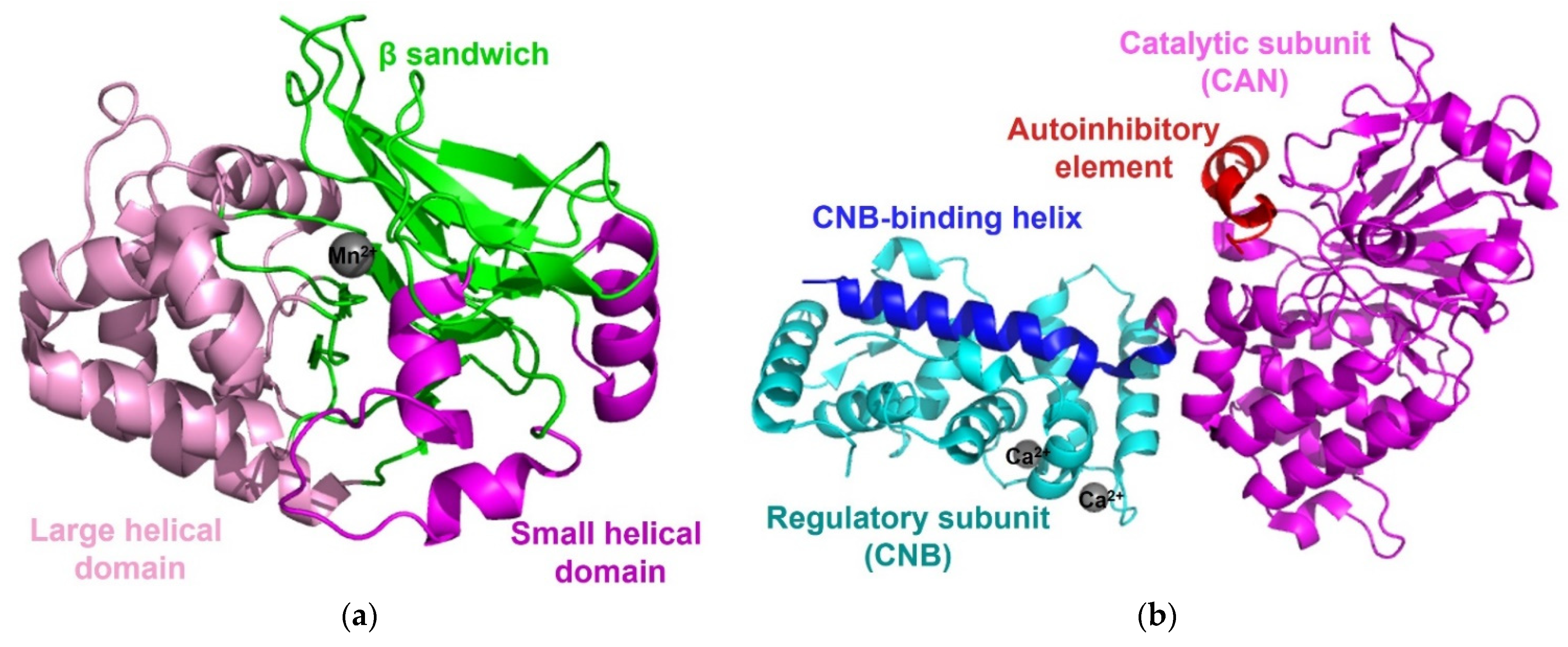
Figure 2. Structure of PP1 and calcineurin. (a) The structure of PP1 (PDB code: 6OBQ) contains a β-sandwich (green), flanked by a large helical domain (pink) and a small helical domain (magenta), and two manganese ions (grey). (b) The overall structure of calcineurin (PDB code: 4OR9) consists of a catalytic subunit (CNA, magenta), a regulatory subunit (CNB, cyan), and two calcium ions (grey). The CNB-binding helix (blue) is extended to CNB and the autoinhibitory element (red) of CNA forms an α-helix and then binds to surface residues on the phosphatase domain of the catalytic center.
2.2. Calcineurin/Protein Phosphatase 2B
Calcineurin (also known as Protein Phosphatase 2B, PP2B) regulates diverse calcium-dependent biological processes, such as neurodevelopment and memory, cardiac hypertrophy, signal transduction, muscle development, and the immune response [20]. Calcineurin consists of a ~60 kD calmodulin-binding catalytic subunit (calcineurin A or CNA) and a ~20 kD regulatory subunit (calcineurin B or CNB). The CNA subunit is highly conserved and similar to the catalytic subunit of PP1, with an identical pattern of metal ion coordination [21], and it contains an N-terminal phosphatase domain, a CNB-binding helical domain, a Ca2+-calmodulin (Ca2+-CaM) binding motif, and an autoinhibitory element. Calcineurin alone is inactive, and its phosphatase activity is activated upon interaction with Ca2+-CaM. Because the disordered autoinhibitory element of CNA forms an α-helix and then binds to surface residues on the phosphatase domain through a combination of hydrogen bonds and van der Waals interactions, access to the catalytic center can be blocked (Figure 2b) [16][21]. Although the CaM-dependent activation of calcineurin is clear, the structural information about its CaM-dependent activation mechanism remains to be clarified, because little structural information about the calcineurin-CaM complex has been reported to date. All calcineurin structures have been determined in the absence of CaM or in complex with small fragments of CaM [22][23][24][25][26][27][28]. However, combining approaches with diverse structural, biochemical, and biophysical analyses has revealed how calcineurin is activated by Ca2+-CaM. Upon binding of Ca2+-CaM to calcineurin, an autoinhibitory element becomes ordered, resulting in a stable helical structure. As a result, the displacement of the disordered autoinhibitory element from the catalytic center causes calcineurin to be activated [29][30].
2.3. Protein Phosphatase 2A
Protein phosphatase 2A (PP2A), one of the most abundant enzymes in humans, represents up to 1% of the total cellular protein in several tissues. PP2A regulates cellular processes, normal physiologies, and numerous signaling pathways [31][32]. Cellular PP2A enzymes exist in either a heterodimeric core enzyme or a heterotrimeric holoenzyme. The PP2A heterodimeric core enzyme comprises a 65 kD scaffold subunit (also known as the A, PR65A, or PPP2R1 subunit), containing 15 tandem HEAT repeats and forms a horseshoe-shaped structure, and a 35kD catalytic subunit (PP2AC and PPP2C subunit), which recognizes the conserved ridge of HEAT repeats 11-15 for association [33][34]. The PP2A core enzyme forms an active heterotrimeric holoenzyme by assembly with one of four regulatory subunits: B (B55, PR55, or PPP2R2), B′ (B56, PR61, or PPPP2R5, B′′ (PR48/PR70/PR130 or PPPP2R3), and B′′′ (Striatins or PR93/PR110) (Figure 3) [16][31]. While the sequences of the scaffold subunit and the catalytic subunit show high conservation among all eukaryotes, the regulatory subunits are more heterogeneous and play key roles in controlling the specific activity and the substrate selectivity of different holoenzymes. The structure of the PP2A holoenzyme containing the B’ subunit shows that the B’ subunit contains eight HEAT-like repeats and interacts with both the scaffold subunit and the catalytic subunit [35]. In the structure of the PP2A holoenzyme harboring the regulatory B subunit, the B subunit containing seven WD40 repeats participates in few interactions with the catalytic subunit, unlike the PP2A holoenzyme containing the B’ subunit [36]. In each PP2A holoenzyme structure, the potential substrate-binding site is on the top surface of the regulatory subunit and located close to the active site of the catalytic subunit to target substrate phosphoproteins [34][35][36].
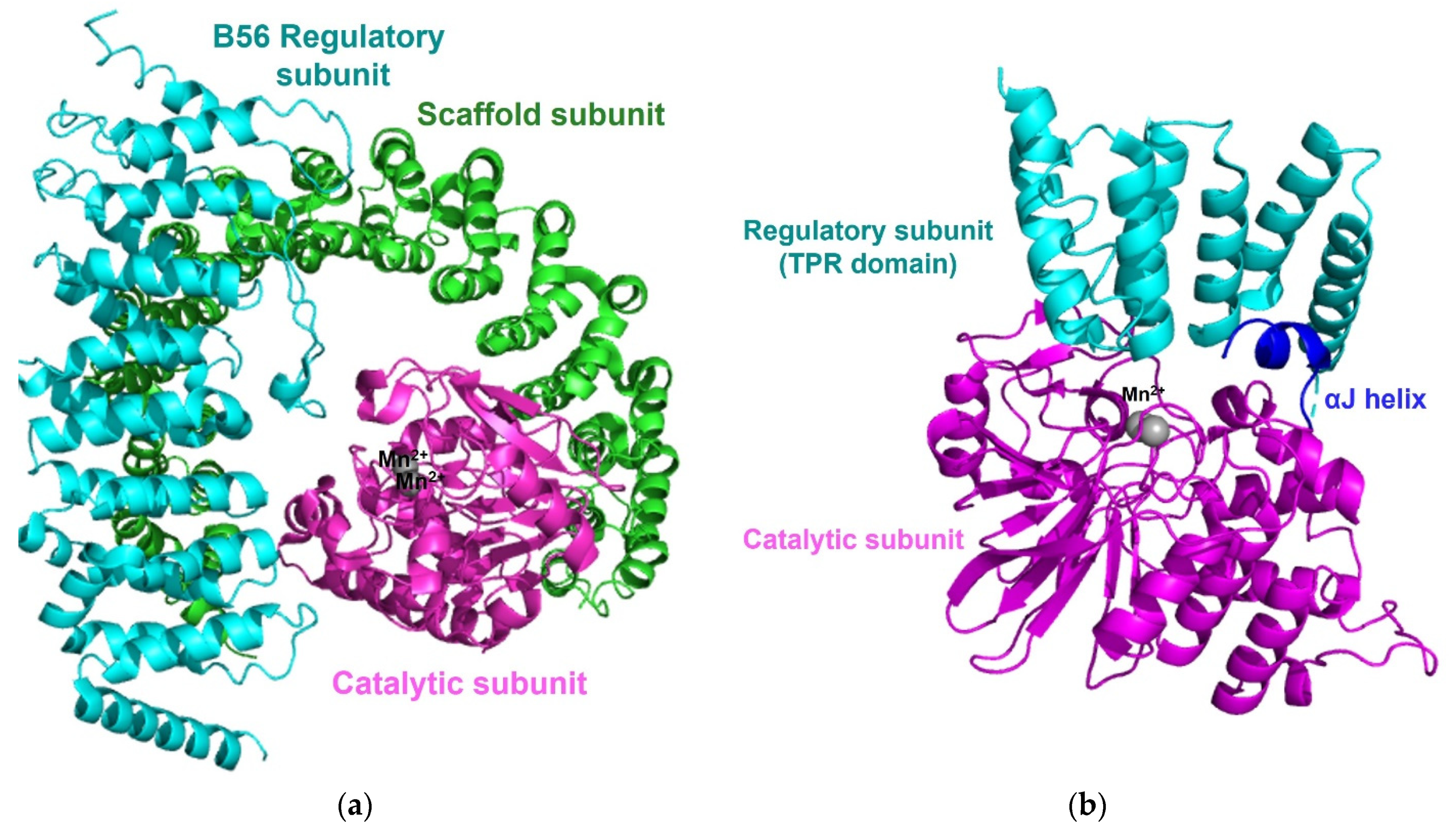
Figure 3. Overall structure of the PP2A holoenzyme and PP5. (a) The overall structure of the PP2A holoenzyme harboring a B56 regulatory subunits (PDB code: 2NYL) consists of a scaffold subunit (green), a catalytic subunit (magenta) with two manganese ions (grey), and a B56 regulatory subunit (cyan). (b) The structure of PP5 (PDB code: 1WAO) contains a catalytic domain (magenta), a regulatory domain (TPR domain, cyan), and two manganese ions (grey). The interaction between the C-terminal αJ-helix (blue) and the N-terminal TPR domain suppresses the phosphatases activity of free PP5 and maintains an autoinhibited conformation.
2.4. Other Protein Ser/Thr Phosphatases
Each protein phosphatase 4 (PP4), 5 (PP5), and 6 (PP6) also has a conserved catalytic core domain, which resembles the domain in PP1 or PP2A. The catalytic subunit of PP4 associates with its own regulatory subunits R1 or R2 [37] and the catalytic subunit of PP6 forms a heterotrimeric holoenzyme with one of three Sit4-associated protein (SAP) domain-containing subunits (PPP6R1-3 or SAPS 1-3) and one of three ankyrin repeat domain subunits (ANR28, ANR44, and ANR52) that serves as the regulatory subunit [38][39]. Unlike most phosphoprotein phosphatases, PP5 is encoded by a single gene. PP5 contains the tetratricopeptide repeat (TPR) domain, a regulatory domain, at the N-terminus, and a catalytic domain, containing an αJ-helix, at the C-terminus. The interaction between the C-terminal αJ-helix and the N-terminal TPR domain suppresses the phosphatase activity of free PP5 and maintains an autoinhibited conformation [40].
2.5. Metal-Dependent Protein Phosphatases
The metal-dependent protein phosphatase (PPM) family, genetically encoded approximately 16 human proteins and Mn2+/Mg2+-dependent enzymes, includes PP2C and pyruvate dehydrogenase phosphatase [4][17]. PP2C plays a key role in the regulation of stress signaling and other cellular signaling [41]. The conserved catalytic core domain of human PP2C shows similar domain folding to other phosphoprotein phosphatases each containing a central β-sandwich, flanked by a pair of α-helices, and coordinating the two metal ions with amino acids and water molecules.
References
- Venerando, A.; Cesaro, L.; Pinna, L.A. From phosphoproteins to phosphoproteomes: A historical account. FEBS J. 2017, 284, 1936–1951.
- Hunter, T. Signaling—2000 and beyond. Cell 2000, 100, 113–127.
- Brognard, J.; Hunter, T. Protein kinase signaling networks in cancer. Curr. Opin. Genet. Dev. 2011, 21, 4–11.
- Ardito, F.; Giuliani, M.; Perrone, D.; Troiano, G.; Lo Muzio, L. The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy. Int. J. Mol. Med. 2017, 40, 271–280.
- Chen, M.J.; Dixon, J.E.; Manning, G. Genomics and evolution of protein phosphatases. Sci. Signal. 2017, 10.
- Nilsson, J. Protein phosphatases in the regulation of mitosis. J. Cell Biol. 2019, 218, 395–409.
- Damle, N.P.; Mohanty, D. Deciphering kinase-substrate relationships by analysis of domain-specific phosphorylation network. Bioinformatics 2014, 30, 1730–1738.
- Iakoucheva, L.M.; Radivojac, P.; Brown, C.J.; O’Connor, T.R.; Sikes, J.G.; Obradovic, Z.; Dunker, A.K. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004, 32, 1037–1049.
- Turk, B.E. Understanding and exploiting substrate recognition by protein kinases. Curr. Opin. Chem. Biol. 2008, 12, 4–10.
- Collins, M.O.; Yu, L.; Campuzano, I.; Grant, S.G.; Choudhary, J.S. Phosphoproteomic analysis of the mouse brain cytosol reveals a predominance of protein phosphorylation in regions of intrinsic sequence disorder. Mol. Cell Proteom. 2008, 7, 1331–1348.
- Nishi, H.; Hashimoto, K.; Panchenko, A.R. Phosphorylation in protein-protein binding: Effect on stability and function. Structure 2011, 19, 1807–1815.
- Barford, D. Molecular mechanisms of the protein serine/threonine phosphatases. Trends Biochem. Sci. 1996, 21, 407–412.
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934.
- Moorhead, G.B.; Trinkle-Mulcahy, L.; Ulke-Lemee, A. Emerging roles of nuclear protein phosphatases. Nat. Rev. Mol. Cell Biol. 2007, 8, 234–244.
- Wlodarchak, N.; Xing, Y. PP2A as a master regulator of the cell cycle. Crit. Rev. Biochem. Mol. Biol. 2016, 51, 162–184.
- Shi, Y. Serine/threonine phosphatases: Mechanism through structure. Cell 2009, 139, 468–484.
- Almo, S.C.; Bonanno, J.B.; Sauder, J.M.; Emtage, S.; Dilorenzo, T.P.; Malashkevich, V.; Wasserman, S.R.; Swaminathan, S.; Eswaramoorthy, S.; Agarwal, R.; et al. Structural genomics of protein phosphatases. J. Struct. Funct. Genom. 2007, 8, 121–140.
- Egloff, M.P.; Cohen, P.T.; Reinemer, P.; Barford, D. Crystal structure of the catalytic subunit of human protein phosphatase 1 and its complex with tungstate. J. Mol. Biol. 1995, 254, 942–959.
- Goldberg, J.; Huang, H.B.; Kwon, Y.G.; Greengard, P.; Nairn, A.C.; Kuriyan, J. Three-dimensional structure of the catalytic subunit of protein serine/threonine phosphatase-1. Nature 1995, 376, 745–753.
- Rusnak, F.; Mertz, P. Calcineurin: Form and function. Physiol. Rev. 2000, 80, 1483–1521.
- Kissinger, C.R.; Parge, H.E.; Knighton, D.R.; Lewis, C.T.; Pelletier, L.A.; Tempczyk, A.; Kalish, V.J.; Tucker, K.D.; Showalter, R.E.; Moomaw, E.W.; et al. Crystal structures of human calcineurin and the human FKBP12-FK506-calcineurin complex. Nature 1995, 378, 641–644.
- Juvvadi, P.R.; Fox, D., 3rd; Bobay, B.G.; Hoy, M.J.; Gobeil, S.M.C.; Venters, R.A.; Chang, Z.; Lin, J.J.; Averette, A.F.; Cole, D.C.; et al. Harnessing calcineurin-FK506-FKBP12 crystal structures from invasive fungal pathogens to develop antifungal agents. Nat. Commun. 2019, 10, 4275.
- Jin, L.; Harrison, S.C. Crystal structure of human calcineurin complexed with cyclosporin A and human cyclophilin. Proc. Natl. Acad. Sci. USA 2002, 99, 13522–13526.
- Ke, H.; Huai, Q. Structures of calcineurin and its complexes with immunophilins-immunosuppressants. Biochem. Biophys. Res. Commun. 2003, 311, 1095–1102.
- Huai, Q.; Kim, H.Y.; Liu, Y.; Zhao, Y.; Mondragon, A.; Liu, J.O.; Ke, H. Crystal structure of calcineurin-cyclophilin-cyclosporin shows common but distinct recognition of immunophilin-drug complexes. Proc. Natl. Acad. Sci. USA 2002, 99, 12037–12042.
- Griffith, J.P.; Kim, J.L.; Kim, E.E.; Sintchak, M.D.; Thomson, J.A.; Fitzgibbon, M.J.; Fleming, M.A.; Caron, P.R.; Hsiao, K.; Navia, M.A. X-ray structure of calcineurin inhibited by the immunophilin-immunosuppressant FKBP12-FK506 complex. Cell 1995, 82, 507–522.
- Ye, Q.; Li, X.; Wong, A.; Wei, Q.; Jia, Z. Structure of calmodulin bound to a calcineurin peptide: A new way of making an old binding mode. Biochemistry 2006, 45, 738–745.
- Ye, Q.; Wang, H.; Zheng, J.; Wei, Q.; Jia, Z. The complex structure of calmodulin bound to a calcineurin peptide. Proteins 2008, 73, 19–27.
- Rumi-Masante, J.; Rusinga, F.I.; Lester, T.E.; Dunlap, T.B.; Williams, T.D.; Dunker, A.K.; Weis, D.D.; Creamer, T.P. Structural basis for activation of calcineurin by calmodulin. J. Mol. Biol. 2012, 415, 307–317.
- Ye, Q.; Feng, Y.; Yin, Y.; Faucher, F.; Currie, M.A.; Rahman, M.N.; Jin, J.; Li, S.; Wei, Q.; Jia, Z. Structural basis of calcineurin activation by calmodulin. Cell Signal. 2013, 25, 2661–2667.
- Janssens, V.; Goris, J. Protein phosphatase 2A: A highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 2001, 353, 417–439.
- Lechward, K.; Awotunde, O.S.; Swiatek, W.; Muszynska, G. Protein phosphatase 2A: Variety of forms and diversity of functions. Acta Biochim. Pol. 2001, 48, 921–933.
- Groves, M.R.; Hanlon, N.; Turowski, P.; Hemmings, B.A.; Barford, D. The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs. Cell 1999, 96, 99–110.
- Xing, Y.; Xu, Y.; Chen, Y.; Jeffrey, P.D.; Chao, Y.; Lin, Z.; Li, Z.; Strack, S.; Stock, J.B.; Shi, Y. Structure of protein phosphatase 2A core enzyme bound to tumor-inducing toxins. Cell 2006, 127, 341–353.
- Xu, Y.; Xing, Y.; Chen, Y.; Chao, Y.; Lin, Z.; Fan, E.; Yu, J.W.; Strack, S.; Jeffrey, P.D.; Shi, Y. Structure of the protein phosphatase 2A holoenzyme. Cell 2006, 127, 1239–1251.
- Xu, Y.; Chen, Y.; Zhang, P.; Jeffrey, P.D.; Shi, Y. Structure of a protein phosphatase 2A holoenzyme: Insights into B55-mediated Tau dephosphorylation. Mol. Cell 2008, 31, 873–885.
- Cohen, P.T.; Philp, A.; Vazquez-Martin, C. Protein phosphatase 4--from obscurity to vital functions. FEBS Lett. 2005, 579, 3278–3286.
- Guergnon, J.; Derewenda, U.; Edelson, J.R.; Brautigan, D.L. Mapping of protein phosphatase-6 association with its SAPS domain regulatory subunit using a model of helical repeats. BMC Biochem. 2009, 10, 24.
- Stefansson, B.; Ohama, T.; Daugherty, A.E.; Brautigan, D.L. Protein phosphatase 6 regulatory subunits composed of ankyrin repeat domains. Biochemistry 2008, 47, 1442–1451.
- Wang, J.; Zhu, J.; Dong, M.; Yu, H.; Dai, X.; Li, K. Inhibition of protein phosphatase 5 (PP5) suppresses survival and growth of colorectal cancer cells. Biotechnol. Appl. Biochem. 2015, 62, 621–627.
- Lu, X.; Nguyen, T.A.; Moon, S.H.; Darlington, Y.; Sommer, M.; Donehower, L.A. The type 2C phosphatase Wip1: An oncogenic regulator of tumor suppressor and DNA damage response pathways. Cancer Metastasis Rev. 2008, 27, 123–135.
More
Information
Subjects:
Biochemistry & Molecular Biology; Biophysics
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
804
Revisions:
3 times
(View History)
Update Date:
25 Jul 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




