A generative adversarial network (GAN) is essentially a two-player game composed of a generator and a discriminator. The generator’s role is to create synthetic data, while the discriminator’s task is to distinguish between real and generated data. During the training process, the generator strives to produce data that the discriminator cannot differentiate from the real data, whereas the discriminator continually improves its ability to distinguish real from generated data. This adversarial training regimen imbues GANs with the capability to model complex data distributions and produce high-quality synthetic data. Notably, their application to gene expression data systems is a fascinating and rapidly growing focus area.
1. Introduction
We are witnessing an epoch of generative artificial intelligence (GenAI), where the boundaries of creativity are being profoundly redefined. The emergence of generative deep learning models, notably the groundbreaking Generative Pretrained Transformers (GPT)
[1][2][3][4] and diffusion models
[5][6][7][8][9], has ushered in a new frontier where the very boundaries of creative expression are being reimagined. These cutting-edge advancements boldly challenge the longstanding belief that creativity is solely reserved for human intellect, unraveling a plethora of uncharted possibilities that lay before us, waiting to be explored.
In the vast array of deep learning-based GenAI, generative adversarial networks (GANs) have carved out a unique and noteworthy place
[10][11][12][13][14][15][16][17][18]. GANs, conceptualized a decade ago
[19], stand out as one of the most influential paradigms in deep-learning-based generative models. The versatility and robustness of GANs have engendered a multitude of applications across various scientific and technological fields. The defining feature of GANs, which sets them apart from other machine learning models, is their ability to generate synthetic data that closely approximates real data distributions, thus enriching the quality and diversity of available data.
Gene expression data are fundamentally constrained by the limitations imposed by the ethical and logistical challenges associated with human subject research
[20]. In stark contrast to other domains that have made substantial progress harnessing the power of big data, gene expression data are limited in size, variety, and the rate at which they can be collected. However, GANs promise a potential solution to these obstacles by enabling the synthesis of a virtually limitless supply of artificial gene expression data.
In the intersection of bioinformatics and artificial intelligence, GANs have emerged as innovative tools for the generation of gene expression data
[21]. The GAN paradigm, equipped with the capacity to create synthetic data that mirrors real-world data, offers an appealing solution to inherent challenges faced in gene expression studies, including high dimensionality, sparse data, and sample diversity.
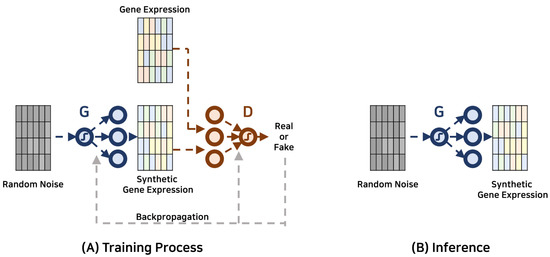
Research within this scope primarily aims to expand the breadth of existing gene expression datasets through the creation of synthetic data, offering an advanced alternative to conventional data augmentation or sampling methods. In contrast to these traditional methods that may distort the inherent data distribution, GANs excel in capturing and emulating the complex, non-linear patterns in gene expression datasets, leading to a more representative and reliable synthetic dataset. Such enriched datasets can subsequently enhance the efficacy of downstream analytical processes and predictive models. Besides expanding data quantity, the synthetic gene expression data generated by GANs can contribute significantly towards understanding and controlling data quality. GANs, with their ability to create data similar to the real-world distributions, can provide nuanced insights into the underlying biological phenomena. This knowledge can contribute to diverse areas, including disease diagnosis, pharmaceutical development, and toxicogenomics, among others. The diagram depicted in Figure 1 showcases the schematic depiction of the GAN-based framework employed for data augmentation. Table 1 offers a comprehensive compilation of recent investigations conducted within the field.
Figure 1. Gene expression data augmentation with generative adversarial networks. The generator and discriminator are represented by G and D, respectively.
Table 1. Contributions of different studies in the application of GANs in genomic data analysis.
| Author |
Contributions |
| Yu et al. [22] |
Developed MichiGAN, a network combining VAEs and GANs to generate disentangled single-cell gene expression data. |
| Yelmen et al. [23] |
Utilized GANs and RBMs to generate artificial human genomes enhancing data imputation quality for low frequency alleles. |
| Hazra et al. [24] |
Used GANs to create synthetic nucleic acid sequences of the cat genome, achieving high correlation with original data. |
| Zrimec et al. [25] |
Prototyped ExpressionGAN, generating synthetic regulatory DNA with targeted mRNA levels, exceeding natural controls in expression. |
| Ahmed et al. [26] |
Introduced omicsGAN, integrating multi-omics data, enhancing predictive signals for cancer outcomes in synthetic data. |
| Vinas et al. [27] |
Utilized a conditional GAN for generating realistic transcriptomics data preserving tissue- and cancer-specific properties. |
| Marouf et al. [28] |
Developed cscGAN, generating realistic single-cell RNA-seq data, enhancing marker gene detection and classifier reliability. |
| Chaudhari et al. [29] |
Developed MG-GAN to augment gene expression data, enhancing cancer classification accuracy significantly. |
| Kwon et al. [30] |
Used GAN for augmenting samples, enhancing prediction of cancer stages significantly. |
| Mendez-Lucio et al. [31] |
Introduced GAN model generating molecules inducing desired transcriptomic profile, a promising approach to drug discovery. |
| Chen et al. [32] |
Developed Tox-GAN, generating gene activities and expression profiles, aiding in chemical-based read-across. |
2. Recent Studies: 2019–2023
In the domain of manipulation and enhancement of gene expression data, several studies stand out. Yu et al.
[22] designed the MichiGAN model by synergistically integrating Variational Autoencoders (VAEs) and GANs. The unique advantage offered by VAEs is their ability to create a latent space of variables, which captures the underlying data distribution. This facilitates the generation of new samples by perturbing these latent variables, rendering VAEs apt for tasks requiring data augmentation. On the other hand, GANs, with their two-player adversarial framework, exhibit exceptional capabilities in generating high-quality, realistic data. In the MichiGAN model, the VAE component disentangles the representations of gene expression data, effectively mitigating the high dimensionality. The GAN component then works with these disentangled representations, generating synthetic samples that resemble real data. This seamless integration empowers the MichiGAN to generate high-quality synthetic gene expression data, thereby aiding in the prediction of cellular responses to drug treatments.
The strategy by Ahmed et al.
[26] in developing omicsGAN lies in the integration of multi-omics data, specifically mRNA and microRNA expression data, and their interaction network. mRNA and microRNA are the two key components of gene regulation processes, providing a comprehensive view of the cell’s functional elements. Integrating these data types, therefore, offers a broader and more accurate perspective of cellular states. OmicsGAN leverages this integrated data to generate synthetic data with enhanced predictive signals. The success of omicsGAN can be attributed to this integrative view of gene expression, which enables a more holistic capture of cellular phenomena, resulting in synthetic data that exhibits superior performance in predicting cancer outcomes.
Marouf et al.
[28] showcased the application of conditional GANs in the creation of their cscGAN. The strength of conditional GANs resides in their ability to guide the data generation process by conditioning on auxiliary information. This allows the model to generate data with specific attributes, contributing to the generation of more targeted and meaningful data. In the case of cscGAN, the auxiliary information was single-cell RNA-seq data, which captures gene expression at an individual cell level, providing a more nuanced understanding of cellular heterogeneity. By conditioning the GAN on these data, cscGAN was able to generate realistic synthetic gene expression data at a single-cell resolution, enhancing downstream analyses and classifier reliability.
Both Yelmen et al.
[23] and Hazra et al.
[24] utilized GANs for the generation of synthetic genetic data, albeit with different source data. In the case of Yelmen et al.
[23], artificial human genomes were generated, while Hazra et al.
[24] synthesized nucleic acid sequences of the cat genome. The performance of these models is heavily reliant on GANs’ innate ability to capture and reproduce the complex patterns inherent in genetic data. GANs, with their generator–discriminator structure, learn the data’s intricate distributions, and are thereby capable of synthesizing high-quality genetic sequences that closely mirror the authentic data. This proficiency in learning from and replicating the real-world data distributions is key to their successful application in synthetic genetic data generation.
The central theme in the works of Chaudhari et al.
[29] and Kwon et al.
[30] is the use of GANs for gene expression data augmentation to enhance cancer classification. The choice of GANs for data augmentation stems from their ability to produce novel synthetic data that not only enlarges the dataset, but also reflects the true data distribution. The Modified Generator GAN (MG-GAN) by Chaudhari et al. is particularly effective because it uses a Gaussian distribution in the generator, thereby promoting a better emulation of the real data distribution. This results in higher-quality synthetic data, leading to an improvement in the accuracy of cancer classification.
In the context of drug discovery and toxicology, the success of the models by Mendez-Lucio et al.
[31] and Chen et al.
[32] can be attributed to the effective application of GANs to gene activities and expression profiles. GANs are proficient at learning from complex multi-dimensional data distributions and reproducing them. By applying GANs to gene activities and expression profiles, these models manage to generate synthetic data that accurately represents the biological phenomena.
3. Trends, Challenges, and Future Directions in Recent Studies
Several compelling trends have emerged from recent studies exploring the application of GANs in gene expression data. A prevalent theme revolves around the manipulation and enhancement of gene expression data to improve its predictive power, as illustrated in the studies by Yu et al.
[22] and Ahmed et al.
[26]. These studies highlight the increasing trend toward harnessing the strengths of GANs, often in combination with other techniques such as VAEs, to generate synthetic data with enhanced predictive signals for biological and clinical outcomes.
The generation of synthetic genetic data, which closely mimics authentic genomic datasets, is another prominent trend. Work by Yelmen et al.
[23] and Hazra et al.
[24] exemplifies this direction, leveraging GANs and other deep learning architectures to generate realistic, artificial human and cat genomes, respectively. These synthetic datasets have proven instrumental in enhancing data imputation quality and fostering an improved understanding of complex genomic structures and functions.
Despite these advancements, a number of challenges persist. One primary concern is the need to develop more rigorous measures for evaluating the performance and reliability of GANs in gene expression analysis. Currently, there is a lack of standard evaluation metrics and validation datasets that can provide objective assessments of these models. In addition, while GANs are incredibly powerful, their complexity can make them difficult to interpret and apply in practice. Exploring ways to make these models more interpretable and user-friendly will be crucial to their broader adoption in the biological and clinical community.
Looking ahead, there are numerous intriguing prospects for future research. The application of GANs in drug discovery, as illustrated by Mendez-Lucio et al.
[31], provides a novel avenue for developing more effective therapeutics. The same applies to the work of Chen et al.
[32], where GANs were utilized to generate gene activities and expression profiles, demonstrating their potential utility in toxicogenomics. Therefore, one promising future direction may lie in further expanding the scope of GANs in translational genomics, potentially revolutionizing drug discovery, therapeutic strategies, and personalized medicine. Moreover, combining GANs with other machine learning and deep learning techniques could foster even more powerful and versatile tools for genomic data analysis. As these trends evolve, it will be exciting to see how GANs continue to reshape the landscape of genomics and bioinformatics.