Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Alexander Zhgun | -- | 1649 | 2023-07-19 14:54:57 | | | |
| 2 | Jessie Wu | + 1 word(s) | 1650 | 2023-07-20 05:15:13 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Hyvönen, M.T.; Keinänen, T.A.; Nuraeva, G.K.; Yanvarev, D.V.; Khomutov, M.; Khurs, E.N.; Kochetkov, S.N.; Vepsäläinen, J.; Zhgun, A.A.; Khomutov, A.R. Hydroxylamine Analogue of Agmatine for Arginine Decarboxylase Inhibition. Encyclopedia. Available online: https://encyclopedia.pub/entry/46985 (accessed on 07 February 2026).
Hyvönen MT, Keinänen TA, Nuraeva GK, Yanvarev DV, Khomutov M, Khurs EN, et al. Hydroxylamine Analogue of Agmatine for Arginine Decarboxylase Inhibition. Encyclopedia. Available at: https://encyclopedia.pub/entry/46985. Accessed February 07, 2026.
Hyvönen, Mervi T., Tuomo A. Keinänen, Gulgina K. Nuraeva, Dmitry V. Yanvarev, Maxim Khomutov, Elena N. Khurs, Sergey N. Kochetkov, Jouko Vepsäläinen, Alexander A. Zhgun, Alex R. Khomutov. "Hydroxylamine Analogue of Agmatine for Arginine Decarboxylase Inhibition" Encyclopedia, https://encyclopedia.pub/entry/46985 (accessed February 07, 2026).
Hyvönen, M.T., Keinänen, T.A., Nuraeva, G.K., Yanvarev, D.V., Khomutov, M., Khurs, E.N., Kochetkov, S.N., Vepsäläinen, J., Zhgun, A.A., & Khomutov, A.R. (2023, July 19). Hydroxylamine Analogue of Agmatine for Arginine Decarboxylase Inhibition. In Encyclopedia. https://encyclopedia.pub/entry/46985
Hyvönen, Mervi T., et al. "Hydroxylamine Analogue of Agmatine for Arginine Decarboxylase Inhibition." Encyclopedia. Web. 19 July, 2023.
Copy Citation
The biogenic polyamines, spermine, spermidine (Spd) and putrescine (Put) are present at micro-millimolar concentrations in eukaryotic and prokaryotic cells (many prokaryotes have no spermine), participating in the regulation of cellular proliferation and differentiation. In mammalian cells Put is formed exclusively from L-ornithine by ornithine decarboxylase (ODC) and many potent ODC inhibitors are known. In bacteria, plants, and fungi Put is synthesized also from agmatine, which is formed from L-arginine by arginine decarboxylase (ADC).
polyamines
arginine decarboxylase
ornithine decarboxylase
O-substituted hydroxylamine
nanomolar inhibitor
1. Introduction
The biogenic polyamines, spermine (Spm) and spermidine (Spd), are organic polycations present at micro-millimolar concentrations in eukaryotic and prokaryotic cells (in many of prokaryotes Spm is absent) where they participate in the regulation of vital cellular functions including proliferation and differentiation [1][2][3]. Disturbances of polyamine metabolism have been associated with the development of many diseases, including cancer (tumor cells have elevated polyamine levels) [1][4][5][6]. Therefore, it is important to investigate the cellular functions of the feasibly interconvertible and partly interchangeable Spm and Spd. Mutant and genetically modified microorganisms and cell lines, as well as transgenic animals have been widely used for these purposes during the last decades [7]. However, there is still a need for specific inhibitors and inducers of the key enzymes of polyamine metabolism, as well as functionally active mimetics of Spm and Spd [8][9][10][11][12].
In vertebrates, ornithine decarboxylase (ODC) is the key and the rate-limiting enzyme of polyamine biosynthesis, since putrescine (Put), formed from the decarboxylation of L-ornithine (L-Orn), then gives rise to Spd and Spm [2][4]. Many potent ODC inhibitors are currently known [8][12]. α-(Difluoromethyl)ornithine (DFMO) was synthesized in 1978 [13], and is the best-known and most widely used of the ODC inhibitors, serving for many years as the “gold standard inhibitor” of polyamine research [8][12]. DFMO is used as a drug to cure African sleeping sickness [14] and has also advanced in clinical trials with promising results as a cancer chemopreventive agent among populations with an elevated risk for specific epithelial cancers [15]. In addition, DFMO has fungicidal activity; this was demonstrated for the first time when it conferred protection on bean plants against infection by uredospores of the bean rust fungus Uromyces phaseoli Linnaeus in greenhouse experiments [16].
ODC is a pyridoxal-5’-phosphate (PLP) dependent enzyme, and among the carbonyl reagents typically inhibiting this class of enzymes, 1-aminooxy-3-aminopropane (APA), an isosteric hydroxylamine-containing analogue of Put, was found to inhibit mammalian ODC at nanomolar concentrations [17][18]. In some cases, APA was even more potent than DFMO [19][20][21]. Interestingly, in the case of phytopathogenic fungi Pyricularia oryzae Cav. only APA, but not DFMO bleached the mycelium, despite the fact that both exhibited fungicidal activity [22]. The biosynthesis of Put is more variable in bacteria, plants and fungi than in vertebrates [3]. In many cases, Put is synthesized from L-arginine (L-Arg), which is first decarboxylated to agmatine (the rate-limiting step in Put biosynthesis) by the biosynthetic arginine decarboxylase (ADC; also known as ArgDC, gene name SpeA for E. coli [3][23]). Agmatine is then hydrolyzed by agmatinase to yield Put and urea. ADC is a PLP-dependent enzyme, but it shares only weak sequence homology to other PLP-dependent decarboxylases, including ODC. The holoenzyme of E. coli ADC is a tetramer, having one molecule of PLP bound to each 70-kDa subunit, and the X-ray crystallographic data for E. coli ADC is available [24]. E. coli contains also an acid-inducible arginine decarboxylase (AdiA), which is an essential component in the arginine-dependent acid-resistance system [25]. The spectrum of the ADC inhibitors is not as extensive as in the case of ODC. The most well-known of these molecules is D,L-α-(difluoromethyl)arginine (DFMA) [26], which has been successfully used to inhibit ADC from different microorganisms and plants [27][28][29][30].
2. The Effects of the Arginine Decarboxylase Inhibitors on the Growth of the A. chrysogenum HY strain
ADC belongs to a large family of PLP-dependent decarboxylases, and, like the other PLP-dependent enzymes, it is sensitive to carbonyl reagents (derivatives of hydrazine, hydroxylamine, semicarbazide, etc.), which react with the coenzyme in its active center to inhibit the enzyme. However, simple carbonyl reagents, as a rule, are nonspecific and not very effective inhibitors. All the potent and specific hydroxylamine-containing inhibitors of PLP-dependent ODC [17][18], glutamate decarboxylase [31], gamma-aminobutyric (GABA) transaminase [32], aspartate aminotransferase [33], 1-aminocyclopropane-1-carboxylate synthase [34], γ-cystathionase [35], histidine decarboxylase [36] and pyruvate-dependent AdoMetDC [37][38][39], are structurally similar either to the enzyme substrate or alternatively resemble the product of the enzymatic reaction. The oximes, which are structurally similar to the external aldimines, the first intermediate of PLP-dependent transformations, are formed in the active site of the enzymes. This is the driving force behind the potent and specific inhibition. X-ray studies of the corresponding E-I complexes of aspartate aminotransferase [33], GABA transaminase [32], 1-aminocyclopropane-1-carboxylate synthase [34] and AdoMetDC [39] have confirmed these similarities. Hence, researchers postulated that AO-Agm, which is an isosteric hydroxylamine analogue of agmatine, would be an effective and specific inhibitor of ADC.
AO-Agm had an IC50 value of 30 ± 1 nM towards E. coli ADC and was a 77 times less potent inhibitor of ODC than of ADC. The guanidino group of AO-Agm anchors the inhibitor in the ADC’s active center, but not as efficiently in the active center of ODC. The properly positioned aminooxy group reacts with the internal aldimine in the enzyme’s active center, giving rise to the PLP oxime, which mimics the external aldimine (Figure 1). This is the reason for the specific and very effective inhibition of ADC. The inhibition of ADC with AO-Agm did not develop with time, as occurs in the case of the inhibition of ODC by APA [17], which is due to the much higher reactivity of Schiff base (internal aldimine) towards O-substituted hydroxylamines, if compared with that for PLP itself [40]. On the contrary, the inhibition of pyruvate-dependent S-adenosyl-L-methionine decarboxylase (AdoMetDC), which possesses a free catalytically crucial carbonyl group in its active center, with a substrate-like O-substituted hydroxylamines does develop with time [37][38]. PLP=AO-Agm binds reversibly to ADC i.e., ultrafiltration of ADC~PLP=AO-Agm complex in a PLP-containing buffer fully restored ADC activity, indicating that the inhibition was pseudo-irreversible.
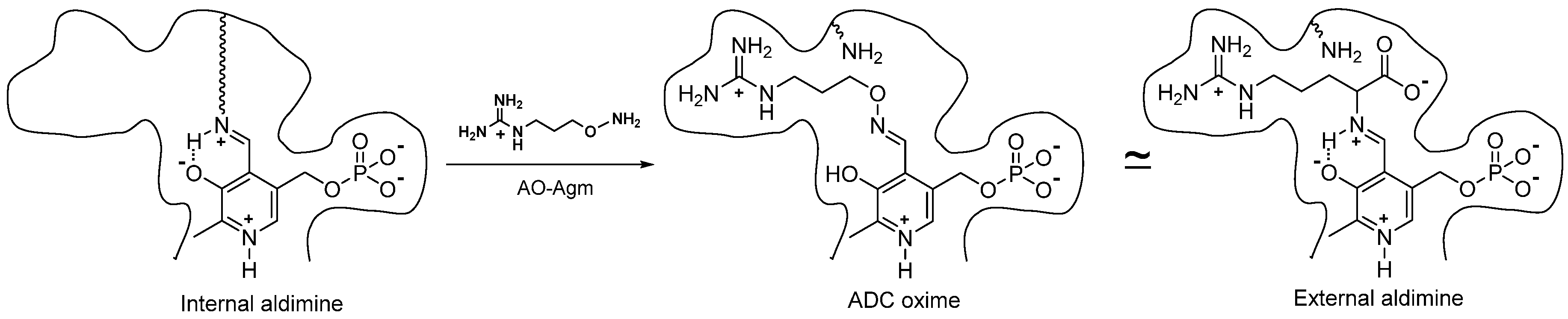
Figure 1. Structures of the internal aldimine, the oxime of ADC with AO-Agm, and the external aldimine—the first intermediate of the decarboxylation of L-Arg.
The filamentous fungi have two main biosynthetic pathways which can yield Put. The first and the most ubiquitous pathway consists of the transformation of L-Arg into L-Orn with the subsequent decarboxylation of the latter compound into Put by ODC. The second, and comparatively rare pathway, involves the transformation of L-Arg into agmatine, which is catalyzed by ADC, and the subsequent transformation of agmatine into Put by agmatinase. ODC and ADC are the key and rate-limiting enzymes in these pathways. There are no data in the literature clarifying which of these two pathways of Put biosynthesis is used by A. chrysogenum.
Here, by using an inhibitory analysis researchers attempted to elucidate which Put biosynthesis pathway is utilized in WT and HY strains of A. chrysogenum. The growth of the WT strain was completely inhibited with both of the ODC inhibitors (at 5 mM) while both ADC inhibitors were much less effective suggesting that the WT strain of A. chrysogenum uses L-Orn, but not agmatine as its major precursor of Put.
The data obtained for the HY strain of A. chrysogenum differed from those obtained for the WT strain. First, researchers found that the HY strain has an elevated polyamine content, as compared with the WT strain, i.e., the polyamine biosynthetic machinery was more active. This may be one of the spin-off results of mutagenesis and DNA damage which occurred during the development of the HY strain [41]. It is known that polyamines protect DNA from free-radical damage by reacting direct with the reactive oxygen species [42][43]. Recently it was demonstrated that polyamines can also maintain the genome integrity via homology-directed DNA repair, enhancing the DNA strand exchange activity of RAD51 recombinase. The polyamines stimulate the capture of homologous duplex DNA and promote synaptic complex formation by the RAD51-ssDNA nucleoprotein filament [44]. Therefore, the necessity to have an increased polyamine pool could have been vitally important for A. chrysogenum to survive under multi-round mutagenesis, leading to the generation of the HY strain with the elevated polyamine pool. Moreover, the elevated polyamine pool is essential not only for the survival of the fungi during the mutagenesis, but also for the production of secondary metabolites. For example, the addition of exogenous polyamines (1,3-diaminopropane, Spd) increases the production of penicillin G in Penicillium chrysogenum Wisconsin 54-1255 strain [45], and the biosynthesis of lovastatin in Aspergillus terreus [46][47].
The inhibition of the growth of the HY A. chrysogenum with APA and DFMO demonstrated the participation of ODC in the biosynthesis of Put. The difference in the activities of APA and DFMO towards the HY and WT strains might point to the existence of an additional biosynthetic pathway from L-Arg via agmatine in the HY strain. Notably, AO-Agm was more potent towards the HY strain, if compared with its activity on the WT strain. This difference is unlikely to be explained by the nonspecific inhibition of ODC observed in DU145 cells lacking ADC and also in the WT strain. The good efficacy of AO-Agm may be also considered as the preliminary indication that the HY strain might use also agmatine as a source of Put. Surprisingly, 5 mM DFMA was found to be a poor inhibitor of the growth of the HY strain. The low activity of DFMA might indicate that a 5 mM concentration was not enough to inhibit the growth of the HY strain, which has higher concentration of polyamines than the WT strain. Nothing is known about the ADC enzyme present in A. chrysogenum, and one cannot exclude the possibility that this enzyme is insensitive to DFMA, resembling the insensitive ODC from E. coli [48] and Entamoeba histolytica [49] towards DFMO (both DFMA and DFMO are enzyme-activated irreversible inhibitors, having the same mechanism of action). Therefore, the inhibitory analysis provided preliminary evidence that both biosynthetic pathways leading to Put, i.e., via L-Orn and via agmatine, might be utilized by HY A. chrysogenum.
References
- Pegg, A.E. Functions of polyamines in mammals. J. Biol. Chem. 2016, 291, 14904–14912.
- Miller-Fleming, L.; Olin-Sandoval, V.; Campbell, K.; Ralser, M. Remaining mysteries of molecular biology: The role of polyamines in the cell. J. Mol. Biol. 2015, 427, 3389–3406.
- Michael, A.J. Polyamine function in archaea and bacteria. J. Biol. Chem. 2018, 293, 18693–18701.
- Casero, R.A.; Murray Stewart, T.; Pegg, A.E. Polyamine metabolism and cancer: Treatments, challenges and opportunities. Nat. Rev. Cancer 2018, 18, 681–695.
- Ramani, D.; De Bandt, J.P.; Cynober, L. Aliphatic polyamines in physiology and diseases. Clin. Nut. 2014, 33, 14–22.
- Mounce, B.C.; Olsen, M.E.; Vignuzzi, M.; Connor, J.H. Polyamines and their role in virus infection. Microbiol. Mol. Biol. Rev. 2017, 81, e00029-17.
- Alhonen, L.; Uimari, A.; Pietilä, M.; Hyvönen, M.T.; Pirinen, E.; Keinänen, T.A. Transgenic animals modelling polyamine metabolism related diseases. Essays Biochem. 2009, 46, 125–144.
- Wallace, H.M.; Fraser, A.V. Inhibitors of polyamine metabolism: Review article. Amino Acids 2004, 26, 353–365.
- Casero, R.A.; Woster, P.M. Recent advances in the development of polyamine analogues as antitumor agents. J. Med. Chem. 2009, 52, 4551–4573.
- Khomutov, M.; Hyvönen, M.T.; Simonian, A.; Formanovsky, A.A.; Mikhura, I.V.; Chizhov, A.O.; Kochetkov, S.N.; Alhonen, L.; Vepsäläinen, J.; Keinänen, T.A.; et al. Unforeseen possibilities to investigate the regulation of polyamine metabolism revealed by novel C-methylated spermine derivatives. J. Med. Chem. 2019, 62, 11335–11347.
- Keinänen, T.A.; Hyvönen, M.T.; Alhonen, L.; Vepsäläinen, J.; Khomutov, A.R. Selective regulation of polyamine metabolism with methylated polyamine analogues. Amino Acids 2014, 46, 605–620.
- Seiler, N. Thirty years of polyamine-related approaches to cancer therapy. Retrospect and prospect. Part 1. Selective enzyme inhibitors. Curr. Drug Targets 2003, 4, 537–564.
- Metcalf, B.W.; Bey, P.; Danzin, C.; Jung, M.J.; Casara, P.; Vevert, J.P. Catalytic irreversible inhibition of mammalian ornithine decarboxylase (E.C.4.1.1.17) by substrate and product analogues. J. Am. Chem. Soc. 1978, 100, 2551–2553.
- Burri, C.C.; Brun, R. Eflornithine for the treatment of human African trypanosomiasis. Parasitol. Res. 2003, 90, S49–S52.
- Gerner, E.W.; Bruckheimer, E.; Cohen, A. Cancer pharmacoprevention: Targeting polyamine metabolism to manage risk factors for colon cancer. J. Biol. Chem. 2018, 293, 18770–18778.
- Rajam, M.V.; Weinstein, L.H.; Galston, A.W. Prevention of a plant disease by specific inhibition of fungal polyamine biosynthesis. Proc. Natl. Acad. Sci. USA. 1985, 82, 6874–6878.
- Khomutov, R.M.; Denisova, G.F.; Khomutov, A.R.; Belostotskaia, K.M.; Shlosman, R.B.; Artamonova, E.Y. Aminooxypropylamine—An effective inhibitor of ornithine decarboxylase in vitro and in vivo. Bioorg. Khim. 1985, 11, 1574–1576.
- Stanek, J.; Frei, J.; Mett, H.; Schneider, P.; Regenass, U. 2-Substituted 3-(aminooxy)propanamines as inhibitors of ornithine decarboxylase: synthesis and biological activity. J. Med. Chem. 1992, 35, 1339–1344.
- Milovica, V.; Turchanowa, L.; Khomutov, A.R.; Khomutov, R.M.; Caspary, W.F.; Stein, J. Hydroxylamine-containing inhibitors of polyamine biosynthesis and impairment of colon cancer cell growth. Biochem. Pharmacol. 2001, 61, 199–206.
- DasGupta, R.; Krause-Ihle, T.; Bergmann, B.; Muller, I.B.; Khomutov, A.R.; Muller, S.; Walter, R.D.; Luersen, K. 3-Aminooxy-1-aminopropane and derivatives have an antiproliferative effect on cultured Plasmodium falciparum by decreasing intracellular polyamine concentrations. Antimicrob. Agents Chemother. 2005, 49, 2857–2864.
- Singh, S.; Mukherjee, A.; Khomutov, A.R.; Persson, L.; Heby, O.; Chatterjee, M.; Madhubala, R. Antileishmanial effect of 3-aminooxy-1-aminopropane is due to polyamine depletion. Antimicrob. Agents Chemother. 2007, 51, 528–534.
- Khomutov, A.R.; Dzavakhia, V.G.; Voinova, T.M.; Ermolinsky, B.S.; Khomutov, R.M. Aminooxy analogue of putrescine inhibits polyketide biosynthetic pathway of natural products. Bioorg. Khim. 1989, 15, 707–709.
- Morris, S.M., Jr. Recent advances in arginine metabolism. Curr. Opin. Clin. Nutr. Metab. Care 2004, 7, 45–51.
- Forouhar, F.; Lew, S.; Seetharaman, J.; Xiao, R.; Acton, T.B.; Montelione, G.T.; Tong, L. Structures of bacterial biosynthetic arginine decarboxylases. Acta Cryst. 2010, F66, 1562–1566.
- Lin, J.; Smith, M.P.; Chapin, K.C.; Baik, H.S.; Bennett, G.N.; Foster, J.W. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 1996, 62, 3094–3100.
- Bitonti, A.J.; Casara, P.J.; McCann, P.P.; Bey, P. Catalytic irreversible inhibition of bacterial and plant arginine decarboxylase activities by novel substrate and product analogues. Biochem. J. 1987, 242, 69–74.
- Khan, A.J.; Minocha, S.C. Biosynthetic arginine decarboxylase in phytopathogenic fungi. Life Sci. 1989, 44, 1215–1222.
- Gadkari, T.V.; Cortes, N.; Madrasi, K.; Tsoukias, N.M.; Joshi, M.S. Agmatine induced NO dependent rat mesenteric artery relaxation and its impairment in salt-sensitive hypertension. Nitric Oxide 2013, 35, 65–71.
- Yarlett, N.; Waters, W.R.; Harp, J.A.; Wannemuehler, M.J.; Morada, M.; Bellcastro, J.; Upton, S.J.; Marton, L.J.; Frydman, B.J. Activities of D,L-alpha-difluoromethylarginine and polyamine analogues against Cryptosporidium parvum infection in a T-cell receptor alpha-deficient mouse model. Antimicrob. Agents Chemother. 2007, 51, 1234–1239.
- Alam, M.; Srivastava, A.; Dutta, A.; Sau, A.K. Biochemical and biophysical studies of Helicobacter pylori arginine decarboxylase, an enzyme important for acid adaptation in host. IUBMB Life 2018, 70, 658–669.
- Sashchenko, L.P.; Severin, E.S.; Khomutov, R.M. On the inhibition of L-glutamic acid decarboxylase by derivatives of hydroxylamine and related compounds. Biokhimiia (Moscow) 1968, 33, 142–147.
- Liu, W.; Peterson, P.E.; Carter, R.J.; Zhou, X.; Langston, J.A.; Fisher, A.J.; Toney, M.D. Crystal structures of unbound and aminooxyacetate-bound Escherichia coli gamma-aminobutyrate aminotransferase. Biochemistry 2004, 43, 10896–10905.
- Markovic-Housley, Z.; Schirmer, T.; Hohenester, E.; Khomutov, A.R.; Khomutov, R.M.; Karpeisky, M.Y.; Sandmeier, E.; Christen, P.; Jansonius, J.N. Crystal structure and solution studies of oxime adducts of mitochondrial aspartate aminotransferase. Eur. J. Biochem. 1996, 236, 1025–1032.
- Capitani, G.; Eliot, A.C.; Gut, H.; Khomutov, R.M.; Kirsch, J.F.; Grutter, M.G. Structure of 1-aminocyclopropane-1-carboxylate synthase in complex with an amino-oxy analogue of the substrate: Implications for substrate binding. Biochim. Biophys. Acta 2003, 1647, 55–60.
- Gabibov, A.G.; Shuster, A.M.; Khomutov, A.R.; Kostina, M.B.; Khurs, E.N.; Goriachenkova, E.V.; Khomutov, R.M. Cystathionase: Catalytic Activity of the Expression Products of cDNA Fragments, and Specific Inhibition of the Native Enzyme and the Fusion Protein by Substrate like O-substituted hydroxylamines. Dokl. Russ. Acad. Sci. 1996, 350, 405–407.
- Castro-Oropeza, R.; Pino-Ángeles, A.; Khomutov, M.A.; Urdiales, J.L.; Moya-García, A.A.; Vepsäläinen, J.; Persson, L.; Sarabia, F.; Khomutov, A.; Sánchez-Jiménez, F. Aminooxy analogue of histamine is an efficient inhibitor of mammalian L-histidine decarboxylase: combined in silico and experimental evidences. Amino Acids 2014, 46, 621–631.
- Artamonova, E.Y.; Zavalova, L.L.; Khomutov, R.M.; Khomutov, A.R. Irreversible inhibition of S-adenosylmethionine decarboxylase by hydroxylamine-containing analogues of decarboxylated S-adenosylmethionine. Bioorg. Khim. 1986, 12, 206–212.
- Pegg, A.E.; Jones, D.B.; Secrist, J.A., III. Effect of inhibitors of S-adenosylmethionine decarboxylase on polyamine content and growth of L1210 cells. Biochemistry 1988, 27, 1408–1415.
- McCloskey, D.E.; Bale, S.; Secrist, J.A., 3rd; Tiwari, A.; Moss, T.H., 3rd; Valiyaveettil, J.; Brooks, W.H.; Guida, W.C.; Pegg, A.E.; Ealick, S.E. New insights into the design of inhibitors of human S-adenosylmethionine decarboxylase: studies of adenine C8 substitution in structural analogues of S-adenosylmethionine. J. Med. Chem. 2009, 52, 1388–1407.
- Dirksen, A.; Dawson, P.E. Rapid oxime and hydrazone ligations with aromatic aldehydes for biomolecular labeling. Bioconjug. Chem. 2008, 19, 2543–2548.
- Bartoshevich, Y.; Novak, M.; Domratcheva, A.; Skryabin, K. Method of cephalosporin C biosynthesis by using new Acremonium chrysogenum strain, RNCM No F-4081D. Russian Federation Patent No 2426793, 20 August 2011.
- Murray Stewart, T.; Dunston, T.T.; Woster, P.M.; Casero, R.A., Jr. Polyamine catabolism and oxidative damage. J. Biol. Chem. 2018, 293, 18736–18745.
- Ha, H.C.; Sirisoma, N.S.; Kuppusamy, P.; Zweier, J.L.; Woster, P.A.; Casero, R.A., Jr. The natural polyamine spermine functions directly as a free radical scavenger. Proc. Natl. Acad. Sci. USA 1998, 95, 11140–11143.
- Lee, C.-Y.; Su, G.-C.; Huang, W.-Y.; Ko, M.-Y.; Yeh, H.-Y.; Chang, G.-D.; Lin, S.-J.; Chi, P. Promotion of homology-directed DNA repair by polyamines. Nat. Commun. 2019, 10, 65.
- Martín, J.; García-Estrada, C.; Kosalková, K.; Ullán, R.V.; Albillos, S.M.; Martín, J.-F. The inducers 1,3-diaminopropane and spermidine produce a drastic increase in the expression of the penicillin biosynthetic genes for prolonged time, mediated by the laeA regulator. Fungal Genet. Biol. 2012, 49, 1004–1013.
- Zhgun, A.A.; Nuraeva, G.K.; Dumina, M.V.; Voinova, T.M.; Dzhavakhiya, V.V.; Eldarov, M.A. 1,3-Diaminopropane and spermidine upregulate lovastatin production and expression of lovastatin biosynthetic genes in Aspergillus terreus via LaeA regulation. Appl. Biochem. Microbiol. 2019, 55, 244–255.
- Zhgun, A.A.; Nuraeva, G.K.; Eldarov, M.A. The role of LaeA and LovE regulators in lovastatin biosynthesis with exogenous polyamines in Aspergillus terreus. Appl. Biochem. Microbiol. 2019, 55, 542–552.
- Kallio, A.; McCann, P.P. Difluoromethylornithine irreversibly inactivates ornithine decarboxylase of Pseudomonas aeruginosa, but does not inhibit the enzymes of Escherichia coli. Biochem. J. 1981, 200, 69–75.
- Preeti; Tapas, S.; Kumar, P.; Madhubala, R.; Tomar, S. Structural insight into DFMO resistant ornithine decarboxylase from Entamoeba histolytica: An inkling to adaptive evolution. PLoS ONE 2013, 8, e53397.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
547
Revisions:
2 times
(View History)
Update Date:
20 Jul 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




