Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | VIBEKE ANDERSEN | -- | 1417 | 2023-07-18 10:14:20 | | | |
| 2 | Catherine Yang | Meta information modification | 1417 | 2023-07-18 10:17:16 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Andersen, V.; Bennike, T.B.; Bang, C.; Rioux, J.D.; Hébert-Milette, I.; Sato, T.; Hansen, A.K.; Nielsen, O.H. Essential Factors Involved in Inflammatory Bowel Disease. Encyclopedia. Available online: https://encyclopedia.pub/entry/46911 (accessed on 07 February 2026).
Andersen V, Bennike TB, Bang C, Rioux JD, Hébert-Milette I, Sato T, et al. Essential Factors Involved in Inflammatory Bowel Disease. Encyclopedia. Available at: https://encyclopedia.pub/entry/46911. Accessed February 07, 2026.
Andersen, Vibeke, Tue B. Bennike, Corinna Bang, John D. Rioux, Isabelle Hébert-Milette, Toshiro Sato, Axel K. Hansen, Ole H. Nielsen. "Essential Factors Involved in Inflammatory Bowel Disease" Encyclopedia, https://encyclopedia.pub/entry/46911 (accessed February 07, 2026).
Andersen, V., Bennike, T.B., Bang, C., Rioux, J.D., Hébert-Milette, I., Sato, T., Hansen, A.K., & Nielsen, O.H. (2023, July 18). Essential Factors Involved in Inflammatory Bowel Disease. In Encyclopedia. https://encyclopedia.pub/entry/46911
Andersen, Vibeke, et al. "Essential Factors Involved in Inflammatory Bowel Disease." Encyclopedia. Web. 18 July, 2023.
Copy Citation
Inflammatory bowel diseases (IBD) are without cure and troublesome to manage because of the considerable diversity between patients and the lack of reliable biomarkers. Diet, gut microbiota, genetics and other patient factors are essential for disease occurrence and progression. The gut epithelial barrier separates the luminal contents from the underlying tissue layers and immune cells. It controls the interactions between the patient’s immune system, the gut microbiota and environmental factors such as food components and is implicated in IBD.
gut microbiota
intestinal barrier
personalised medicine
1. Diet and IBD
Characterising the impact of diet on any disease is challenging due to the complex composition of modern diets and unreliability of self-reporting. In addition, an in-depth understanding of the functional effect of diet is lacking [1][2]. Nevertheless, strong associations between diet and IBD have been demonstrated, and recent studies have highlighted that lifestyle including diet strongly influences IBD risk [3][4][5]. In contrast, few randomised clinical trials have been performed investigating the effect of diet on patients with established IBD [6][7][8]. Prospective studies have demonstrated that a Western diet, characterised by a high intake of animal-based foods, processed foods, food additives, alcohol and sugar, is associated with IBD and increases the occurrence of flare-ups compared to a healthy diet [9][10]. High intake of ultra-processed foods has also been associated with an increased risk of IBD [11]. On the other hand, a plant-based diet such as a Mediterranean diet was reported to reduce inflammation in IBD [6]. However, other studies have failed to demonstrate an association between a specific diet and IBD, and the association remains somewhat unclear [12][13]. Consequently, evidence-based nutritional recommendations for the individual patient are scarce [1][2].
2. The Gut Microbiota and IBD
Compared with healthy control individuals, patients with IBD consistently demonstrate gut microbiota alterations. The changes include dysbiosis, characterised by lowered bacterial α-diversity (i.e., fewer defined microbial species) and altered β-diversity (i.e., significant changes in microbial species composition) [14]. The systematically described collection of microbes is known as the microbiota, while the term microbiome includes their pool of functional genes. The loss of resident microbial species, termed the “disappearing microbes”, might help to explain the rising incidence of chronic diseases in industrialised countries [15].
Many studies have found that patients with IBD have an increased abundance of Escherichia coli and Fusobacterium spp. known to promote inflammation by the adhesion and invasion of the colon epithelium. Further, a lowered abundance of the short-chain fatty acids (SCFA) producers Faecalibacterium prausnitzii and Akkermansia muciniphila has also been observed [16][17]. Reports indicate that a high abundance of the class Actinobacteria and the associated genus Bifidobacterium are protective against UC [18]. In contrast, species such as Ruminococcus gnavus and R. torques typically increase gut inflammation through their production of a TNF-α inducing polysaccharide and are abundant in patients with IBD [19].
However, apart from lower diversity, studies report inconsistent patterns of gut microbiota alterations in IBD [15]. This inconsistency is, at least in part, due to the heterogeneity of the disease [20]. Additionally, numerous factors affect human gut microbiota composition, including the sampling method, geographic location and patient factors, such as genetics, sex, age, diet, stool consistency and other lifestyle factors [15][21].
Importantly, changes observed in the gut microbiota can be a consequence or a cause of IBD. Recent data support the key role of a specific bacterium, Klebsiella pneumonia, in IBD [22]. It was found in approximately 40% of patients, the abundance correlated with disease activity, and its transfer resulted in colitis in an animal model [22]. While the exact role and mechanisms remain unclear, it is conceivable that K. pneumonia may be involved in the etiology of a subset of IBD. Complicating the aspect of causality further is that there seems to be a critical window in early life in which perturbation of the microbiome has a substantial effect on disease development [23].
Bacterial members of the microbiota are not the only microorganisms that can be altered in IBD [24][25][26]. Fungi, archaea and viruses can also significantly affect the gut immune response to IBD, although they only account for a minor proportion of the mammalian gut microbiota [24]. In particular, the faecal mycobiome differed between patients with CD and UC and between patients experiencing a flare compared to those in remission, where the mycobiome more closely resembles a healthy mycobiome [26]. In addition, viruses have been associated with IBD by activating the immune system following invasion and replication within the epithelial cells [27]. Similarly, phages can indirectly affect immune cells and other cell types through infected bacteria [22]. However, methodological biases may still complicate interpretation.
3. Gut Epithelium Barrier and Immune System in IBD
The gut epithelial barrier controls the interaction between the gut microbiota and food components on the one hand and the patient immune system on the other (Figure 1) [28]. In IBD, this barrier is compromised, giving rise to the condition commonly described as a “leaky gut”. The leaky gut is probably a key pathological factor in IBD as it has been found to precede diagnosis [29].
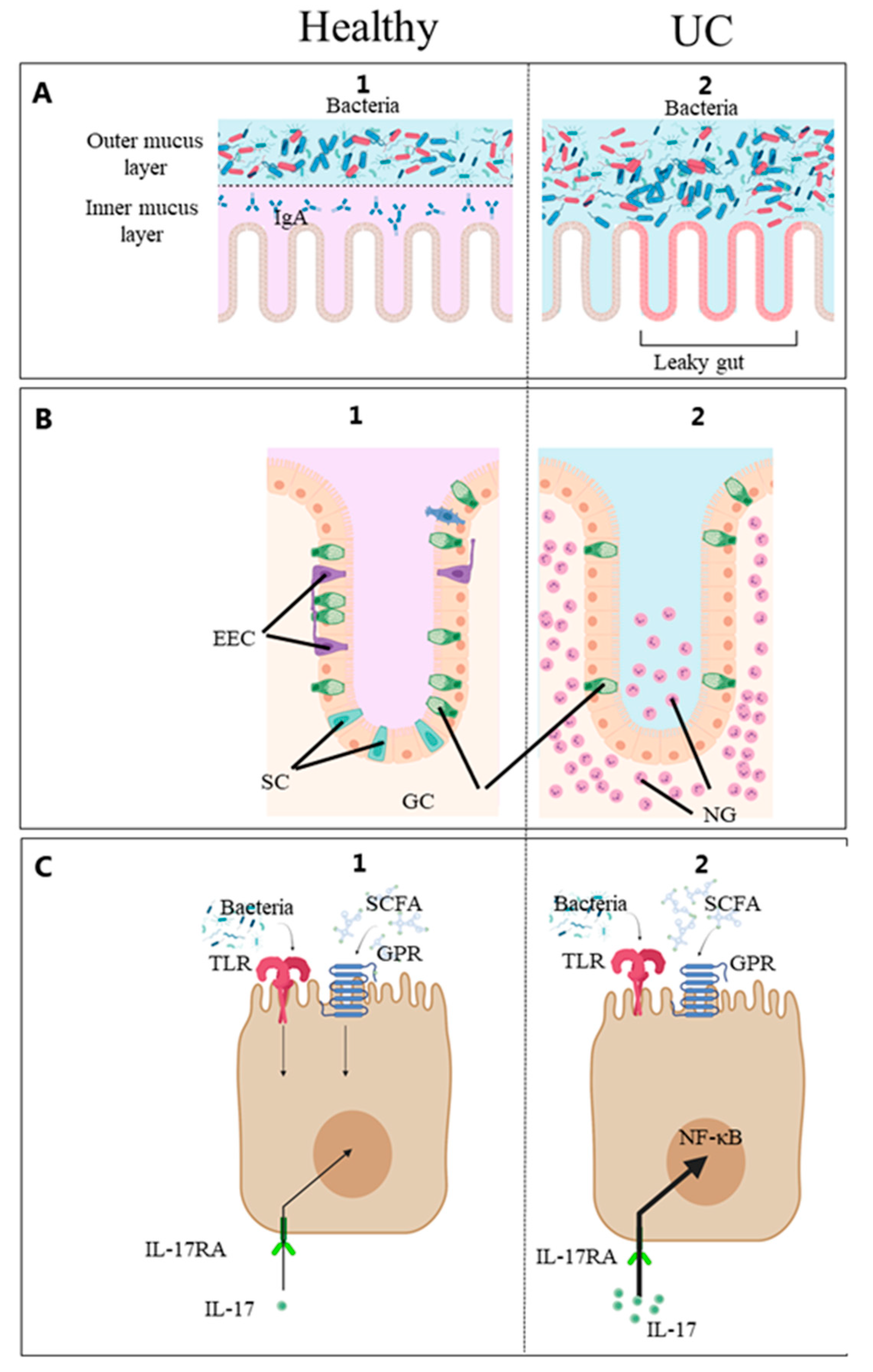
Figure 1. (A) Mucusa, (B) Epithelium, (C) Epithelial cell. Diet, gut microbes and patient factors interact at the mucosal surface. Schematic diagram of the intestinal mucosa constituting the intestinal barrier and immune system [30]. From the luminal side, it consists of the mucus and epithelial lining overlying the connective tissue. 1. (healthy) and 2. (UC). The outermost layer from the lumen side is the mucus. In the healthy gut, commensal microorganisms interact with the outer mucus layer and do not reach the inner mucus layer or epithelial cells. In IBD, the number of GCs is reduced, and this barrier is compromised, giving rise to the condition commonly described as a “leaky gut”. Certain microbial molecules activate the Toll-like receptors (TLR), and certain metabolites such as short-chain fatty acids (SCFA) activate the G protein-coupled receptors (GPR) on the intestinal epithelial cells. Enteroendocrine cells (EEC) monitor the gut microbiota and regulate inflammatory processes [28]. These processes stimulate the innate immune system resulting in gut inflammation by the pro-inflammatory IL-17 stimulating the IL-17 receptor A (IL-17RA), and neutrophilic granulocytes (NG) accumulate in the intestinal mucosa. EEC, enteroendocrine cells; goblet cells (GC), goblet cells; IL-17, interleukin-17; IL-17-RA, IL-17 receptor A; NF-ĸβ, nuclear-factor kappa beta; NG, neutrophilic granulocytes; GPR, G protein-coupled receptors; SCFA, short-chain fatty acids; TLR, Toll-like receptors; UC, ulcerative colitis. Created with Biorender.com (accessed on 2 May 2023).
The fundamental structures of the gut epithelial barrier are, from the luminal side, the mucus layer and the intestinal epithelial cells lining. The colonic mucus is a two-layered gel-like structure produced by goblet cells comprising highly glycosylated mucin proteins. In the healthy gut, commensal microorganisms interact with the outer mucus layer and cannot reach the inner mucus layer or epithelial cells [31]. Functional mucus glycosylation is essential for feeding microbes, and altered glycosylation patterns contribute to pronounced alterations in the gut microbiota [32]. In IBD patients, altered spatial patterns have been found to contribute to microbiota dysbiosis [31]. Thus, it is becoming increasingly evident that microbiota–host interactions depend highly on the microbial communities’ nature and spatial organisation [33]. Nevertheless, few studies have analysed the luminal or mucosa-associated microbiota, which are in close contact with the gut immune system and differs from the stool microbiota [34]. The gut epithelium consists of cells capable of activating the immune system when in contact with dietary materials, microbial components or metabolites [30]. For example, pattern recognition receptors and G protein-coupled receptors on intestinal epithelial cells respond to specific microbial structures and metabolites [35][36][37][38]. In recent years, new epithelial cell types such as intercrypt goblet cells [39], microfold-like (M-like) cells [40], BEST4+ cells [41] and Tuft cells have been identified. Tuft cells appear to be critical for specific immunologic responses [42][43]. M-like cells are rarely found in healthy colons but are reported to be expanded 17-fold in inflamed colons [40]. BEST4+ cells were identified as a new population of human intestinal epithelial cells by single-cell RNA-seq technology. Histologic analysis revealed their localization in the crypt top. The functional role of BEST4+ cells remains unknown, but they may be associated with bicarbonate export and a pH-sensing function based on their gene expression. Finally, the gut epithelial basement membrane is a specialized matrix that supports and separates the epithelial cells from the interstitial space and is also considered important in maintaining the epithelial barrier [44]. Understanding the host–microbial interactions at this surface will likely prove critical to gain deeper biological insights into the etiology of IBD and identifying clinically useful biomarkers.
In addition, understanding the role of the gut microbiome in the brain, joints and liver is emerging, indicating that the microbiota is a driving factor for altered cell trafficking, a crucial step for the onset and progression of extraintestinal conditions in IBD [45].
References
- Fitzpatrick, J.A.; Melton, S.L.; Yao, C.K.; Gibson, P.R.; Halmos, E.P. Dietary management of adults with IBD—The emerging role of dietary therapy. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 652–669.
- Sudhakar, P.; Wellens, J.; Verstockt, B.; Ferrante, M.; Sabino, J.; Vermeire, S. Holistic healthcare in inflammatory bowel disease: Time for patient-centric approaches? Gut 2023, 72, 192–204.
- Adolph, T.E.; Meyer, M.; Schwarzler, J.; Mayr, L.; Grabherr, F.; Tilg, H. The metabolic nature of inflammatory bowel diseases. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 753–767.
- Lopes, E.W.; Chan, S.S.M.; Song, M.; Ludvigsson, J.F.; Hakansson, N.; Lochhead, P.; Clark, A.; Burke, K.E.; Ananthakrishnan, A.N.; Cross, A.J.; et al. Lifestyle factors for the prevention of inflammatory bowel disease. Gut 2022.
- Sun, Y.; Yuan, S.; Chen, X.; Sun, J.; Kalla, R.; Yu, L.; Wang, L.; Zhou, X.; Kong, X.; Hesketh, T.; et al. The Contribution of Genetic Risk and Lifestyle Factors in the Development of Adult-Onset Inflammatory Bowel Disease: A Prospective Cohort Study. Am. J. Gastroenterol. 2023, 118, 511–522.
- Fritsch, J.; Garces, L.; Quintero, M.A.; Pignac-Kobinger, J.; Santander, A.M.; Fernandez, I.; Ban, Y.J.; Kwon, D.; Phillips, M.C.; Knight, K.; et al. Low-Fat, High-Fiber Diet Reduces Markers of Inflammation and Dysbiosis and Improves Quality of Life in Patients With Ulcerative Colitis. Clin. Gastroenterol. Hepatol. 2021, 19, 1189–1199.e30.
- Turpin, W.; Dong, M.; Sasson, G.; Raygoza Garay, J.A.; Espin-Garcia, O.; Lee, S.H.; Neustaeter, A.; Smith, M.I.; Leibovitzh, H.; Guttman, D.S.; et al. Mediterranean-Like Dietary Pattern Associations With Gut Microbiome Composition and Subclinical Gastrointestinal Inflammation. Gastroenterology 2022, 163, 685–698.
- Peters, V.; Dijkstra, G.; Campmans-Kuijpers, M.J.E. Are all dietary fibers equal for patients with inflammatory bowel disease? A systematic review of randomized controlled trials. Nutr. Rev. 2022, 80, 1179–1193.
- Peters, V.; Spooren, C.; Pierik, M.J.; Weersma, R.K.; van Dullemen, H.M.; Festen, E.A.M.; Visschedijk, M.C.; Masclee, A.A.M.; Hendrix, E.M.B.; Almeida, R.J.; et al. Dietary Intake Pattern is Associated with Occurrence of Flares in IBD Patients. J. Crohn’s Colitis 2021, 15, 1305–1315.
- Dong, C.; Chan, S.S.M.; Jantchou, P.; Racine, A.; Oldenburg, B.; Weiderpass, E.; Heath, A.K.; Tong, T.Y.N.; Tjønneland, A.; Kyrø, C.; et al. Meat Intake Is Associated with a Higher Risk of Ulcerative Colitis in a Large European Prospective Cohort Studyø. J. Crohn’s Colitis 2022, 16, 1187–1196.
- Narula, N.; Wong, E.C.L.; Dehghan, M.; Mente, A.; Rangarajan, S.; Lanas, F.; Lopez-Jaramillo, P.; Rohatgi, P.; Lakshmi, P.V.M.; Varma, R.P.; et al. Association of ultra-processed food intake with risk of inflammatory bowel disease: Prospective cohort study. BMJ 2021, 374, n1554.
- Narula, N.; Wong, E.C.L.; Dehghan, M.; Marshall, J.K.; Moayyedi, P.; Yusuf, S. Does a High-inflammatory Diet Increase the Risk of Inflammatory Bowel Disease? Results From the Prospective Urban Rural Epidemiology (PURE) Study: A Prospective Cohort Study. Gastroenterology 2021, 161, 1333–1335.
- Gubatan, J.; Kulkarni, C.V.; Talamantes, S.M.; Temby, M.; Fardeen, T.; Sinha, S.R. Dietary Exposures and Interventions in Inflammatory Bowel Disease: Current Evidence and Emerging Concepts. Nutrients 2023, 15, 579.
- Schirmer, M.; Garner, A.; Vlamakis, H.; Xavier, R.J. Microbial genes and pathways in inflammatory bowel disease. Nat. Rev. Microbiol. 2019, 17, 497–511.
- Metwaly, A.; Reitmeier, S.; Haller, D. Microbiome risk profiles as biomarkers for inflammatory and metabolic disorders. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 383–397.
- Pittayanon, R.; Lau, J.T.; Leontiadis, G.I.; Tse, F.; Yuan, Y.; Surette, M.; Moayyedi, P. Differences in Gut Microbiota in Patients With vs Without Inflammatory Bowel Diseases: A Systematic Review. Gastroenterology 2020, 158, 930–946.e931.
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019, 569, 655–662.
- Kurilshikov, A.; Medina-Gomez, C.; Bacigalupe, R.; Radjabzadeh, D.; Wang, J.; Demirkan, A.; Le Roy, C.I.; Raygoza Garay, J.A.; Finnicum, C.T.; Liu, X.; et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 2021, 53, 156–165.
- Yu, S.; Balasubramanian, I.; Laubitz, D.; Tong, K.; Bandyopadhyay, S.; Lin, X.; Flores, J.; Singh, R.; Liu, Y.; Macazana, C.; et al. Paneth Cell-Derived Lysozyme Defines the Composition of Mucolytic Microbiota and the Inflammatory Tone of the Intestine. Immunity 2020, 53, 398–416.
- Caenepeel, C.; Sadat Seyed Tabib, N.; Vieira-Silva, S.; Vermeire, S. Review article: How the intestinal microbiota may reflect disease activity and influence therapeutic outcome in inflammatory bowel disease. Aliment. Pharmacol. Ther. 2020, 52, 1453–1468.
- Vujkovic-Cvijin, I.; Sklar, J.; Jiang, L.; Natarajan, L.; Knight, R.; Belkaid, Y. Host variables confound gut microbiota studies of human disease. Nature 2020, 587, 448–454.
- Federici, S.; Kredo-Russo, S.; Valdés-Mas, R.; Kviatcovsky, D.; Weinstock, E.; Matiuhin, Y.; Silberberg, Y.; Atarashi, K.; Furuichi, M.; Oka, A.; et al. Targeted suppression of human IBD-associated gut microbiota commensals by phage consortia for treatment of intestinal inflammation. Cell 2022, 185, 2879–2898.e2824.
- Fenneman, A.C.; Weidner, M.; Chen, L.A.; Nieuwdorp, M.; Blaser, M.J. Antibiotics in the pathogenesis of diabetes and inflammatory diseases of the gastrointestinal tract. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 81–100.
- Iliev, I.D. Mycobiota-host immune interactions in IBD: Coming out of the shadows. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 91–92.
- Guzzo, G.L.; Andrews, J.M.; Weyrich, L.S. The Neglected Gut Microbiome: Fungi, Protozoa, and Bacteriophages in Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2022, 28, 1112–1122.
- Li, X.V.; Leonardi, I.; Putzel, G.G.; Semon, A.; Fiers, W.D.; Kusakabe, T.; Lin, W.Y.; Gao, I.H.; Doron, I.; Gutierrez-Guerrero, A.; et al. Immune regulation by fungal strain diversity in inflammatory bowel disease. Nature 2022, 603, 672–678.
- Shih, T.; Yusung, S.; Gonsky, R.; Dutra-Clarke, R.; Ziring, D.; Rabizadeh, S.; Kugathasan, S.; Denson, L.A.; Li, D.; Braun, J. Environmental Interaction of Resolved Human Cytomegalovirus Infection With Crohn’s Disease Location. Inflamm. Bowel Dis. 2023, 29, 328–331.
- Yu, Y.; Yang, W.; Li, Y.; Cong, Y. Enteroendocrine Cells: Sensing Gut Microbiota and Regulating Inflammatory Bowel Diseases. Inflamm. Bowel Dis. 2020, 26, 11–20.
- Turpin, W.; Lee, S.H.; Raygoza Garay, J.A.; Madsen, K.L.; Meddings, J.B.; Bedrani, L.; Power, N.; Espin-Garcia, O.; Xu, W.; Smith, M.I.; et al. Increased Intestinal Permeability Is Associated With Later Development of Crohn’s Disease. Gastroenterology 2020, 159, 2092–2100.e2095.
- Chang, J.T. Pathophysiology of Inflammatory Bowel Diseases. N. Engl. J. Med. 2020, 383, 2652–2664.
- Hansson, G.C. Mucins and the Microbiome. Annu. Rev. Biochem. 2020, 89, 769–793.
- Yao, D.; Dai, W.; Dong, M.; Dai, C.; Wu, S. MUC2 and related bacterial factors: Therapeutic targets for ulcerative colitis. eBioMedicine 2021, 74, 103751.
- Motta, J.P.; Wallace, J.L.; Buret, A.G.; Deraison, C.; Vergnolle, N. Gastrointestinal biofilms in health and disease. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 314–334.
- Buisson, A.; Sokol, H.; Hammoudi, N.; Nancey, S.; Treton, X.; Nachury, M.; Fumery, M.; Hébuterne, X.; Rodrigues, M.; Hugot, J.P.; et al. Role of adherent and invasive Escherichia coli in Crohn’s disease: Lessons from the postoperative recurrence model. Gut 2023, 72, 39–48.
- Iliev, I.D.; Cadwell, K. Effects of Intestinal Fungi and Viruses on Immune Responses and Inflammatory Bowel Diseases. Gastroenterology 2021, 160, 1050–1066.
- Abraham, C.; Abreu, M.T.; Turner, J.R. Pattern Recognition Receptor Signaling and Cytokine Networks in Microbial Defenses and Regulation of Intestinal Barriers: Implications for Inflammatory Bowel Disease. Gastroenterology 2022, 162, 1602–1616.
- Kimura, I.; Ichimura, A.; Ohue-Kitano, R.; Igarashi, M. Free Fatty Acid Receptors in Health and Disease. Physiol. Rev. 2020, 100, 171–210.
- Melhem, H.; Kaya, B.; Ayata, C.K.; Hruz, P.; Niess, J.H. Metabolite-Sensing G Protein-Coupled Receptors Connect the Diet-Microbiota-Metabolites Axis to Inflammatory Bowel Disease. Cells 2019, 8, 450–469.
- Nystrom, E.E.L.; Martinez-Abad, B.; Arike, L.; Birchenough, G.M.H.; Nonnecke, E.B.; Castillo, P.A.; Svensson, F.; Bevins, C.L.; Hansson, G.C.; Johansson, M.E.V. An intercrypt subpopulation of goblet cells is essential for colonic mucus barrier function. Science 2021, 372, eabb1590.
- Smillie, C.S.; Biton, M.; Ordovas-Montanes, J.; Sullivan, K.M.; Burgin, G.; Graham, D.B.; Herbst, R.H.; Rogel, N.; Slyper, M.; Waldman, J.; et al. Intra- and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell 2019, 178, 714–730.
- Parikh, K.; Antanaviciute, A.; Fawkner-Corbett, D.; Jagielowicz, M.; Aulicino, A.; Lagerholm, C.; Davis, S.; Kinchen, J.; Chen, H.H.; Alham, N.K.; et al. Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature 2019, 567, 49–55.
- Hendel, S.K.; Kellermann, L.; Hausmann, A.; Bindslev, N.; Jensen, K.B.; Nielsen, O.H. Tuft Cells and Their Role in Intestinal Diseases. Front. Immunol. 2022, 13, 822867.
- Howitt, M.R.; Lavoie, S.; Michaud, M.; Blum, A.M.; Tran, S.V.; Weinstock, J.V.; Gallini, C.A.; Redding, K.; Margolskee, R.F.; Osborne, L.C.; et al. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science 2016, 351, 1329–1333.
- Kobayashi, T.; Siegmund, B.; Le Berre, C.; Wei, S.C.; Ferrante, M.; Shen, B.; Bernstein, C.N.; Danese, S.; Peyrin-Biroulet, L.; Hibi, T. Ulcerative colitis. Nat. Rev. Dis. Prim. 2020, 6, 74.
- Zundler, S.; Günther, C.; Kremer, A.E.; Zaiss, M.M.; Rothhammer, V.; Neurath, M.F. Gut immune cell trafficking: Inter-organ communication and immune-mediated inflammation. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 50–64.
More
Information
Subjects:
Gastroenterology & Hepatology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
573
Revisions:
2 times
(View History)
Update Date:
18 Jul 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




