Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Xuepeng Chi | -- | 1892 | 2023-07-12 01:49:41 | | | |
| 2 | Rita Xu | -3 word(s) | 1889 | 2023-07-12 04:20:55 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Chi, X.; Wang, Z.; Wang, Y.; Liu, Z.; Wang, H.; Xu, B. Plant-Derived miRNAs in Modulating Insect Development. Encyclopedia. Available online: https://encyclopedia.pub/entry/46661 (accessed on 07 February 2026).
Chi X, Wang Z, Wang Y, Liu Z, Wang H, Xu B. Plant-Derived miRNAs in Modulating Insect Development. Encyclopedia. Available at: https://encyclopedia.pub/entry/46661. Accessed February 07, 2026.
Chi, Xuepeng, Zhe Wang, Ying Wang, Zhenguo Liu, Hongfang Wang, Baohua Xu. "Plant-Derived miRNAs in Modulating Insect Development" Encyclopedia, https://encyclopedia.pub/entry/46661 (accessed February 07, 2026).
Chi, X., Wang, Z., Wang, Y., Liu, Z., Wang, H., & Xu, B. (2023, July 12). Plant-Derived miRNAs in Modulating Insect Development. In Encyclopedia. https://encyclopedia.pub/entry/46661
Chi, Xuepeng, et al. "Plant-Derived miRNAs in Modulating Insect Development." Encyclopedia. Web. 12 July, 2023.
Copy Citation
MicroRNAs (miRNAs), a class of non-coding small RNAs, are crucial regulatory factors in plants and animals at the post-transcriptional level. These tiny molecules suppress gene expression by complementary oligonucleotide binding to sites in the target messenger.
miRNAs
cross-kingdom regulation
plants
1. Introduction
MicroRNAs (miRNAs) are a class of endogenous, highly conserved, single-stranded non-coding small RNAs (sRNAs) ranging in size from 18 to 23 nucleotides. They act as master modulators of target gene expression at the post-transcriptional level by binding to the 3′-untranslated region (3′-UTR) or open reading frame (ORF) [1]. MiRNAs have been demonstrated to play crucial roles in various biological processes, including cell growth and differentiation, cell proliferation, immune response, and cell apoptosis [2][3][4]. Since the first identified miRNA, lin-4, was discovered in Caenorhabditis elegans (Maupas, 1899) (Nematoda: Rhabditida) in 1993 [5][6], tens of thousands of miRNAs have been identified in mammals, plants, and microorganisms [3][7]. To date, the most commonly used miRNA database, miRBase version 22.1 (https://www.mirbase.org), has registered 38,589 miRNAs from 271 organisms [8].
Insects are the most abundant group of animals on earth, and miRNAs play an important role during the growth and development of insects. Studies have shown that some miRNAs play important roles in insect metamorphosis and oogenesis by regulating the hormone synthesis level or gene expression level in Kr-h1 and Notch signaling pathways [9]. MiRNAs play major and diverse roles in the biology of Drosophila melanogaster, ranging from germline development, maternal to zygotic transition, tissue growth, and physiological activity of the central nervous system [10]. There are significant differences in the miRNA and transcriptional profiles of diploid females relative to haploid drone males, and between reproductively distinct females, indicating that miRNA plays a crucial role in caste determination in the honeybee [11]. In addition, miRNAs are potential targets for insect pest control applications [12]. Studies on insect miRNAs have demonstrated their important roles in insect development, and there is a wide range of fields to explore in reproductive manipulation, caste differentiation, and so on.
Recently, the discovery of plant-derived miRNAs showing cross-kingdom abilities to regulate gene expression in mammals, insects, and even viruses has prompted exciting discussion [13][14][15]. The digestive system of animals can absorb plant miRNAs and release them into the circulatory system, which then delivers them to target cells to regulate the functions of recipient cells [16]. In 2012, Zhang et al. [17] reported that plant-derived miRNAs were present in the sera and tissues of various animals and that these exogenous miRNAs were primarily acquired through food intake. They found miR168a from rice could bind to the human/mouse low-density lipoprotein receptor adapter protein 1 (LDLRAP1) mRNA, inhibit LDLRAP1 expression in the liver, and consequently decrease LDL removal from mouse plasma. In 2014, miR172 from Brassica oleracea was also detected in the blood, spleen, liver, and kidney in mice [13]. The abundance of plant miR159 in human serum is negatively correlated with the incidence and progression of breast cancer, and oral miR159 mimics significantly inhibit the growth of mouse xenograft breast tumors [18]. Plant miR5338 was also enriched in the posterior lobes of the prostate gland of rats fed with rape bee pollen, suggesting that plant-derived miR5338 has potential utility in the treatment of rat benign prostatic hyperplasia through inhibiting Mfn1 in the prostate [19]. These findings indicate that plant-derived miRNAs can be absorbed into animals through the gastrointestinal tract and remain stable and selectively functional in target organs, target tissues, or target cells, with subsequent physiological effects. Although the findings of cross-kingdom regulation by plant-derived miRNAs are exciting, several groups of researchers have questioned these discoveries due to a lack of reproducibility [20]. Therefore, these findings must be examined dialectically.
Mutualism is a crucial outcome of the interactions between multiple species that provide the energy, nutrients, and services for ecosystems to function and persist. Over the course of their long-term evolution, plants have evolved strategies to deter herbivores whilst attracting beneficial insects. The interactions between plants and herbivorous insects are close and complex. Flowering plants depend on insects for pollination, while insects have been feeding on plants for 400 million years [21]. Studies have found that after feeding on melon phloem sap, plant-derived miRNAs can be detected in Aphis gossypii tissues [22]. This finding indicates that plant-derived miRNAs can enter insects and exhibit the potential to exert regulatory effects. In addition, the coevolution of plants and insects may depend on cross-kingdom regulation by plant-derived miRNAs. However, knowledge of the involvement of miRNAs in these reciprocal interactions is in its infancy [23]. The influence of miRNAs on coevolution between plants and insects is also unclear.
2. Comparison of miRNA Biogenesis and Mechanisms in Plants and Insects
The biogenesis and other mechanisms of animal- and plant-derived miRNAs are similar overall. The process of biogenesis begins in the nucleus. As shown in Figure 1, in both animals and plants, capped and polyadenylated primary miRNA (pri-miRNA) transcripts are first transcribed by miRNA-encoding genes with the help of RNA polymerase Ⅱ, followed by removal of the hairpin stem to form the precursor-miRNA (pre-miRNA) and then further cleavage of the hairpin loop, resulting in the miRNA duplex [23]. Mature guide miRNAs are loaded on Argonaute (Ago) proteins to form the RNA-induced silencing complex (RISC) that binds to mRNA via target sequence complementarity to repress gene expression through inhibition of translation or degradation of mRNA [24].
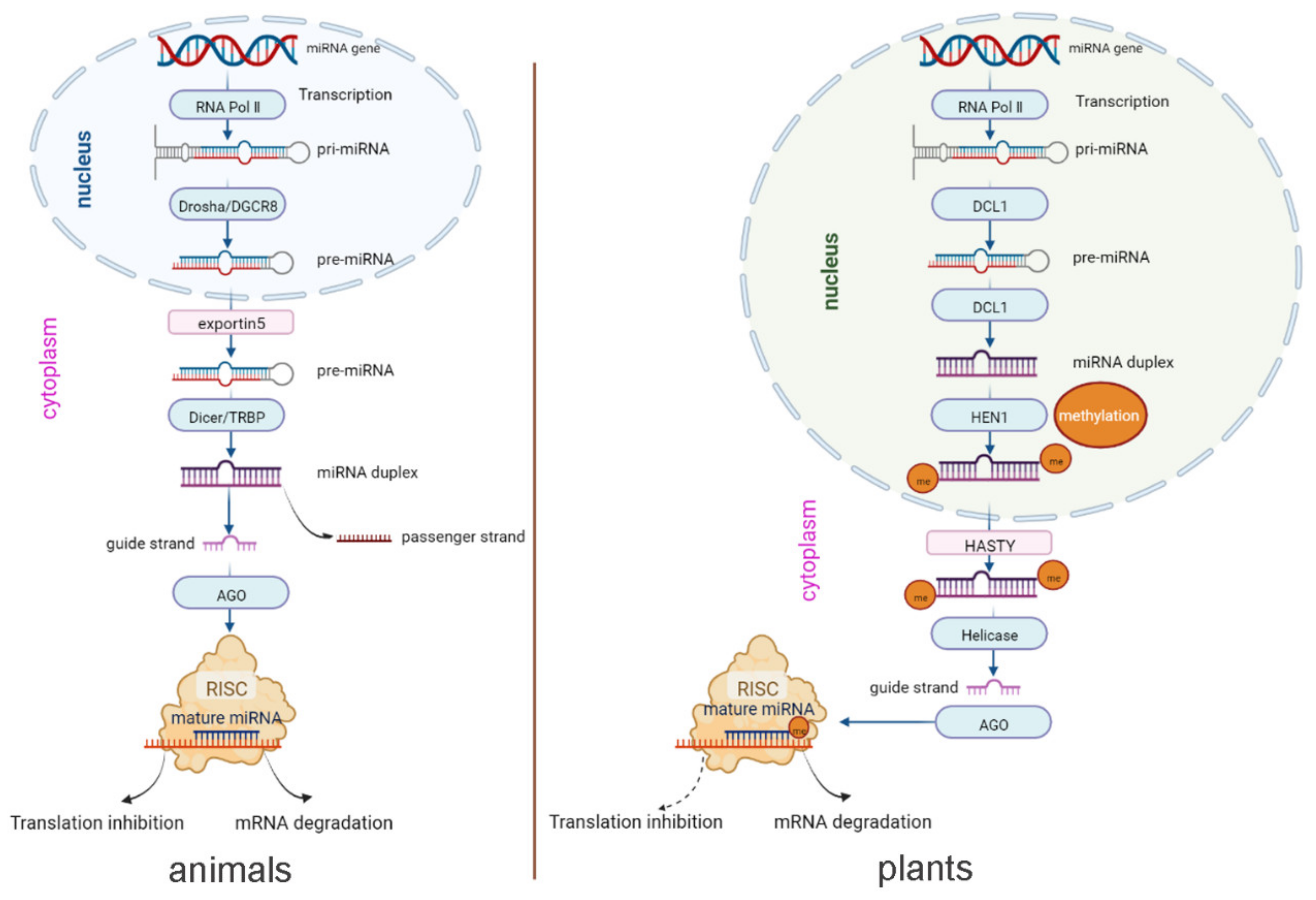
Figure 1. miRNA biogenesis pathways in animals and plants.
However, the biogenesis of animal and plant-derived miRNAs biogenesis exhibits some distinct features. In plants, pri-miRNAs are cleaved through the action of the endonuclease DCL1 until they become mature miRNA duplexes. Subsequently, methylation at the 3′-terminus of the duplex is carried out by HEN1. Then, HASTY exports the methylated miRNA duplex from the nucleus to the cytoplasm [25]. In contrast, in animals, pre-miRNAs are produced through the activity of endonuclease Drosha and exported from the nucleus to the cytoplasm by the transport protein exportin5. Then, endonuclease Dicer converts the pre-miRNAs into mature miRNA duplexes [26]. To sum up, first, the maturation of miRNA occurs in the nucleus in animals but in the cytoplasm in plants. Second, the types of enzymes are different in animals and plants, although their functions are similar. In addition, in plants, the dominant mechanism (solid arrow) occurs via mRNA cleavage, in which the miRNA binds its target with high complementarity, while the alternative mechanism (dashed arrow) occurs by translational inhibition (Figure 1) [25].
3. Coevolution between Insects and Plants
Coevolution is the process in which two or more distinct taxa radiate and speciate in association with one another. The huge number of species of plants and insects is thought to be the result of adaptive radiation driven by the coevolution between plants and their beneficial animal pollinators or foragers [27]. In 1964, Ehrlich and Raven published “Butterflies and Plants: A Study in Coevolution” [28]. The authors fostered a new way of thinking about the ecology and evolution of plant–herbivore interactions. Ehrlich and Raven theorized on the importance of coevolution to both the origin of species and the maintenance of species diversity, and suggested that coevolution between plants and their insect herbivores could drive the adaptive diversification of both groups. This theory laid the foundation for over five decades of research on anti-herbivore defense and coevolution.
In the course of developing and refining coevolutionary theory, plant secondary metabolites are considered defense compounds because they deter herbivory, reduce food digestibility, or directly interact with molecular targets in non-adapted insects [29]. In order to survive, insects must evolve to resist the plant’s secondary metabolites. The never-ending arms race drives coevolution between pathogens and hosts. In other words, insect herbivores can drive real-time ecological and evolutionary change in plant populations. In 2016, Marquis et al. [30] coupled the evolution of novel plant chemistry with reproductive isolation, thereby enabling plant speciation. According to this theory, the evolution of plant defenses could be associated with plant speciation. Meanwhile, phytophagous insects have been adapting to exploit their hosts [31]. Insects have developed sophisticated morphological, behavioral, and physiological adaptations that enable them to exploit plants as a resource [29]. The model system of Heliconiines and Passiflora (Passiflora quadrangularis L., 1758) (Magnoliatae: Malpighiales) plants was one of the first examples used to exemplify coevolution theory. In this model system, Passiflora plants evolved yellow structures mimicking heliconiine eggs and their extensive diversity of defense compounds known as cyanogenic glucosides. However, after a complex process of coevolution, Heliconiines can synthesize cyanogenic glucosides themselves, and the Heliconius (Insecta: Lepidoptera) adults have highly accurate visual and chemosensory systems. Further, the expansion of brain structures that can process such information allows Heliconius adults to memorize shapes and display elaborate pre-oviposition behavior [32]. Under some circumstances, plant defenses may impact pollinator health, foraging behavior, and reproductive success. Therefore, in order to survive and reproduce, flowering plants must balance the conflicting selective pressures of herbivore avoidance and pollinator attraction [33]. In brief, plants and animals have always been in a dynamic process of coevolution.
4. The Coevolution of miRNAs and miRNA Targets
Because plants are hosts for many species of insects, the coevolution between plants and insects is a complementary process, involving molecular pathways comprising important interactions between plants and insects. The non-coding RNAs, miRNAs, serve as a paradigm for studying functional divergence between paralogs and the possible coevolutionary processes between the duplicated miRNAs and the genomic contexts [34]. Studies of cross-kingdom regulation suggest that miRNAs play a significant role in the process of coevolution. The changes in the expression patterns of an existing miRNA may affect new sets of targets, forming novel regulatory circuits and changing pre-existing ones [35]. In other words, to maintain their regulatory functions, miRNAs have been forced to coevolve with their target genes when the targets experience functional divergence [36], which may be the molecular mechanism through which coevolution occurs.
The coevolution events for specific miRNAs and their targets in Drosophila and for miRNA-941 in primate evolution have been predicted and validated, although predictions of coevolution were limited to a few miRNAs and their target sites [37]. Barbash et al. [35] identified the coevolution of miRNAs and their targets by performing detailed genomic dissection using a combination of computational approaches. In addition, the evolutionary patterns of miRNAs and their targets during soybean domestication have been revealed through a comprehensive investigation of their genetic diversity [38]. Barik et al. [39] found that conserved miR167 sequences and their target auxin response factors (ARFs) have undergone coevolution, leading to functional diversification among diverse plant species. A family of X-linked miRNAs named spermatogenesis-related miRNAs also underwent strong coevolution with their target genes [40]. In a fungus–plant model (Botrytis cinerea and Arabidopsis thaliana) of RNA-based communication, the majority of sequences that were predicted to target host genes were shown to come from retrotransposons and intergenic regions in the fungal genome [41].
However, to date, no study has demonstrated that miRNAs play an important role in the coevolution of animals and plants. The cross-kingdom regulatory effects of plant-derived miRNAs on insects are, however, undisputed. Claycomb [42] postulated that extracellular small RNAs act as part of a pathogen’s arsenal by binding to target mRNA, thus enabling coevolution. Most studies emphasize the interaction between plant secondary metabolites and insect stress resistance. For example, in protein-related studies, the coevolution between viruses and hosts has been proven. The sequence of interactions that occur between MICA, a key natural killer (NK) cell activating receptor that recognizes a family of stress-induced ligands, and human cytomegalovirus, illustrates the dynamic and ongoing coevolution of virus and host, which enables the former to be so exquisitely tailored to the latter [43]. As knowledge in the field develops in depth, the cross-kingdom functions of miRNAs in coevolution will be further revealed.
References
- Kataria, P.; Surela, N.; Chaudhary, A.; Das, J. MiRNA: Biological Regulator in Host-Parasite Interaction during Malaria Infection. Int. J. Environ. Res. Public Health 2022, 19, 2395.
- Wang, J.; Mei, J.; Ren, G. Plant microRNAs: Biogenesis, Homeostasis, and Degradation. Front. Plant Sci. 2019, 10, 360.
- Li, D.; Yang, J.; Yang, Y.; Liu, J.; Li, H.; Li, R.; Cao, C.; Shi, L.; Wu, W.; He, K. A Timely Review of Cross-Kingdom Regulation of Plant-Derived MicroRNAs. Front. Genet. 2021, 12, 613197.
- Li, M.; Yu, B. Recent advances in the regulation of plant miRNA biogenesis. RNA Biol. 2021, 18, 2087–2096.
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854.
- Esquela-Kerscher, A. The lin-4 microRNA: The ultimate micromanager. Cell Cycle 2014, 13, 1060–1061.
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 2019, 234, 5451–5465.
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162.
- Song, J.; Zhou, S. Post-transcriptional regulation of insect metamorphosis and oogenesis. Cell. Mol. Life Sci. 2020, 77, 1893–1909.
- Carthew, R.W.; Agbu, P.; Giri, R. MicroRNA function in Drosophila melanogaster. Semin. Cell Dev. Biol. 2017, 65, 29–37.
- Ashby, R.; Forêt, S.; Searle, I.; Maleszka, R. MicroRNAs in Honey Bee Caste Determination. Sci. Rep. 2016, 6, 18794.
- Zhang, Q.; Dou, W.; Taning, C.N.T.; Smagghe, G.; Wang, J.J. Regulatory roles of microRNAs in insect pests: Prospective targets for insect pest control. Curr. Opin. Biotechnol. 2021, 70, 158–166.
- Liang, G.; Zhu, Y.; Sun, B.; Shao, Y.; Jing, A.; Wang, J.; Xiao, Z. Assessing the survival of exogenous plant microRNA in mice. Food Sci. Nutr. 2014, 2, 380–388.
- Zhu, W.J.; Liu, Y.; Cao, Y.N.; Peng, L.X.; Yan, Z.Y.; Zhao, G. Insights into Health-Promoting Effects of Plant MicroRNAs: A Review. J. Agric. Food Chem. 2021, 69, 14372–14386.
- Majumdar, R.; Galewski, P.J.; Eujayl, I.; Minocha, R.; Vincill, E.; Strausbaugh, C.A. Regulatory Roles of Small Non-coding RNAs in Sugar Beet Resistance against Beet Curly Top Virus. Front. Plant Sci. 2021, 12, 780877.
- Chen, Q.; Zhang, F.; Dong, L.; Wu, H.; Xu, J.; Li, H.; Wang, J.; Zhou, Z.; Liu, C.; Wang, Y.; et al. SIDT1-dependent absorption in the stomach mediates host uptake of dietary and orally administered microRNAs. Cell Res. 2021, 31, 247–258.
- Zhang, L.; Hou, D.; Chen, X.; Li, D.; Zhu, L.; Zhang, Y.; Li, J.; Bian, Z.; Liang, X.; Cai, X.; et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: Evidence of cross-kingdom regulation by microRNA. Cell Res. 2012, 22, 107–126.
- Chin, A.R.; Fong, M.Y.; Somlo, G.; Wu, J.; Swiderski, P.; Wu, X.; Wang, S.E. Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Res. 2016, 26, 217–228.
- Chen, X.; Wu, R.Z.; Zhu, Y.Q.; Ren, Z.M.; Tong, Y.L.; Yang, F.; Dai, G.H. Study on the inhibition of Mfn1 by plant-derived miR5338 mediating the treatment of BPH with rape bee pollen. BMC Complement. Altern. Med. 2018, 18, 38.
- Zhang, Y.; Wiggins, B.E.; Lawrence, C.; Petrick, J.; Ivashuta, S.; Heck, G. Analysis of plant-derived miRNAs in animal small RNA datasets. BMC Genom. 2012, 13, 381.
- Labandeira, C.C. A paleobiologic perspective on plant-insect interactions. Curr. Opin. Plant Biol. 2013, 16, 414–421.
- Sattar, S.; Addo-Quaye, C.; Song, Y.; Anstead, J.A.; Sunkar, R.; Thompson, G.A. Expression of small RNA in Aphis gossypii and its potential role in the resistance interaction with melon. PLoS ONE 2012, 7, e48579.
- Li, C.; Wong, A.Y.P.; Wang, S.; Jia, Q.; Chuang, W.P.; Bendena, W.G.; Tobe, S.S.; Yang, S.H.; Chung, G.; Chan, T.F.; et al. miRNA-Mediated Interactions in and between Plants and Insects. Int. J. Mol. Sci. 2018, 19, 3239.
- Moran, Y.; Agron, M.; Praher, D.; Technau, U. The evolutionary origin of plant and animal microRNAs. Nat. Ecol. Evol. 2017, 1, 27.
- Samad, A.F.A.; Kamaroddin, M.F.; Sajad, M. Cross-Kingdom Regulation by Plant microRNAs Provides Novel Insight into Gene Regulation. Adv. Nutr. 2021, 12, 197–211.
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610.
- Yuan, Y.W.; Byers, K.J.; Bradshaw, H.D., Jr. The genetic control of flower-pollinator specificity. Curr. Opin. Plant Biol. 2013, 16, 422–428.
- Ehrlich, P.R.; Raven, P.H. Butterflies and Plants: A Study in Coevolution. Evolution 1964, 18, 586–608.
- Beran, F.; Petschenka, G. Sequestration of Plant Defense Compounds by Insects: From Mechanisms to Insect-Plant Coevolution. Annu. Rev. Entomol. 2022, 67, 163–180.
- Marquis, R.J.; Salazar, D.; Baer, C.; Reinhardt, J.; Priest, G.; Barnett, K. Ode to Ehrlich and Raven or how herbivorous insects might drive plant speciation. Ecology 2016, 97, 2939–2951.
- Anderson, J.T.; Mitchell-Olds, T. Ecological genetics and genomics of plant defenses: Evidence and approaches. Funct. Ecol. 2011, 25, 312–324.
- de Castro, É.C.P.; Zagrobelny, M.; Cardoso, M.Z.; Bak, S. The arms race between heliconiine butterflies and Passiflora plants—New insights on an ancient subject. Biol. Rev. Camb. Philos. Soc. 2018, 93, 555–573.
- Jacobsen, D.J.; Raguso, R.A. Lingering Effects of Herbivory and Plant Defenses on Pollinators. Curr. Biol. 2018, 28, R1164–R1169.
- Luo, J.; Wang, Y.; Yuan, J.; Zhao, Z.; Lu, J. MicroRNA duplication accelerates the recruitment of new targets during vertebrate evolution. RNA 2018, 24, 787–802.
- Barbash, S.; Shifman, S.; Soreq, H. Global coevolution of human microRNAs and their target genes. Mol. Biol. Evol. 2014, 31, 1237–1247.
- Felippes, F.F.; Schneeberger, K.; Dezulian, T.; Huson, D.H.; Weigel, D. Evolution of Arabidopsis thaliana microRNAs from random sequences. RNA 2008, 14, 2455–2459.
- Hu, H.Y.; He, L.; Fominykh, K.; Yan, Z.; Guo, S.; Zhang, X.; Taylor, M.S.; Tang, L.; Li, J.; Liu, J.; et al. Evolution of the human-specific microRNA miR-941. Nat. Commun. 2012, 3, 1145.
- Liu, T.; Fang, C.; Ma, Y.; Shen, Y.; Li, C.; Li, Q.; Wang, M.; Liu, S.; Zhang, J.; Zhou, Z.; et al. Global investigation of the co-evolution of MIRNA genes and microRNA targets during soybean domestication. Plant J. 2016, 85, 396–409.
- Barik, S.; Kumar, A.; Sarkar Das, S.; Yadav, S.; Gautam, V.; Singh, A.; Singh, S.; Sarkar, A.K. Coevolution Pattern and Functional Conservation or Divergence of miR167s and Their Targets across Diverse Plant Species. Sci. Rep. 2015, 5, 14611.
- Zhang, F.; Zhang, Y.; Lv, X.; Xu, B.; Zhang, H.; Yan, J.; Li, H.; Wu, L. Evolution of an X-Linked miRNA Family Predominantly Expressed in Mammalian Male Germ Cells. Mol. Biol. Evol. 2019, 36, 663–678.
- Weiberg, A.; Wang, M.; Lin, F.M.; Zhao, H.; Zhang, Z.; Kaloshian, I.; Huang, H.D.; Jin, H. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 2013, 342, 118–123.
- Claycomb, J.; Abreu-Goodger, C.; Buck, A.H. RNA-mediated communication between helminths and their hosts: The missing links. RNA Biol. 2017, 14, 436–441.
- Seidel, E.; Le, V.T.K.; Bar-On, Y.; Tsukerman, P.; Enk, J.; Yamin, R.; Stein, N.; Schmiedel, D.; Oiknine Djian, E.; Weisblum, Y.; et al. Dynamic Co-Evolution of Host and Pathogen: HCMV Downregulates the Prevalent Allele MICA∗008 to Escape Elimination by NK Cells. Cell Rep. 2015, 10, 968–982.
More
Information
Subjects:
Developmental Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
641
Revisions:
2 times
(View History)
Update Date:
12 Jul 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




