Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jose Antonio Velázquez-Aragón | -- | 3782 | 2023-07-04 21:09:29 | | | |
| 2 | Rita Xu | Meta information modification | 3782 | 2023-07-05 03:35:25 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
De Sales-Millán, A.; Aguirre-Garrido, J.F.; González-Cervantes, R.M.; Velázquez-Aragón, J.A. Microbiome–Gut–Mucosal–Immune–Brain Axis and Autism Spectrum Disorder. Encyclopedia. Available online: https://encyclopedia.pub/entry/46413 (accessed on 01 March 2026).
De Sales-Millán A, Aguirre-Garrido JF, González-Cervantes RM, Velázquez-Aragón JA. Microbiome–Gut–Mucosal–Immune–Brain Axis and Autism Spectrum Disorder. Encyclopedia. Available at: https://encyclopedia.pub/entry/46413. Accessed March 01, 2026.
De Sales-Millán, Amapola, José Félix Aguirre-Garrido, Rina María González-Cervantes, José Antonio Velázquez-Aragón. "Microbiome–Gut–Mucosal–Immune–Brain Axis and Autism Spectrum Disorder" Encyclopedia, https://encyclopedia.pub/entry/46413 (accessed March 01, 2026).
De Sales-Millán, A., Aguirre-Garrido, J.F., González-Cervantes, R.M., & Velázquez-Aragón, J.A. (2023, July 04). Microbiome–Gut–Mucosal–Immune–Brain Axis and Autism Spectrum Disorder. In Encyclopedia. https://encyclopedia.pub/entry/46413
De Sales-Millán, Amapola, et al. "Microbiome–Gut–Mucosal–Immune–Brain Axis and Autism Spectrum Disorder." Encyclopedia. Web. 04 July, 2023.
Copy Citation
Autism Spectrum Disorder (ASD) is a complex neurodevelopmental disorder characterised by deficits in social interaction and communication, as well as restricted and stereotyped interests. Due of the high prevalence of gastrointestinal disorders in individuals with ASD, researchers have investigated the gut microbiota as a potential contributor to its aetiology. The relationship between the microbiome, gut, and brain (microbiome–gut–brain axis) has been acknowledged as a key factor in modulating brain function and social behaviour, but its connection to the aetiology of ASD is not well understood.
microbiome-gut-brain axis
autism spectrum disorder
dysbiosis
1. Introduction
Autism spectrum disorder (ASD) refers to a complex neurodevelopmental disorder characterised by deficits in social interaction and communication and restricted and stereotyped behaviours, interests or activities [1]. ASD has a significant financial and health impact on individuals with autism, their families and society [2]. The cause of ASD is not yet fully understood, but research suggests that a combination of genetic and environmental factors are involved [3]. Different reports propose that, in addition to the core symptoms of ASD, gastrointestinal symptoms, including constipation, abdominal pain, vomiting, diarrhoea and gas, are frequent in people with ASD, with estimates ranging from 9% to 70% [4]. The gut microbiome is a crucial component of human physiology [5], and the frequent gastrointestinal symptoms in individuals with ASD have led to the investigation of a possible connection between the gut microbiota and the symptoms of the disorder [4]. The mode of birth and feeding type during the first years of life can condition changes in the gut microbiota. Children born vaginally and fed with breastmilk present more Bifidobacteria than children born by caesarean section and fed with formula. In the complementary feeding stage, the diversity of foods can determine the conformation of different microbial groups that will be maintained in the adult stage. A long-term fibre-deficient diet can lead to changes in the composition of gut microbiota (dysbiosis). These changes cause alterations in the production of important neurotransmitters for neurodevelopment [5]. There is increasing evidence supporting the importance of gut microbiota in brain development and function, showing communication along the gut–brain axis [6]. During the first 3 years of life, the consolidation of the microbiota–gut–brain axis is of vital importance because during this stage, the central nervous system (CNS) develops rapidly [7], and gut microbiota also suffer important changes due to changes in feeding patterns during this developmental stage [8][9]. Recently, the focus has been expanded to the role of the gut microbiota [10], generating increased interest with findings suggesting specific microorganisms associated with memory, stress, mood, neurodevelopment and even ASD [11][12][13].
The gut–microbiome–brain axis is a multidisciplinary research area that has attracted the attention of different researchers from around the world. Multiple studies have found evidence that the gut–microbiome–brain axis is important for the mental and cognitive health of children with ASD [14][15][16]. Despite this, there have been relatively few clinical studies in humans that provide clear evidence of the role in the aetiology of neurodevelopmental disorders [17][18]. On the other hand, a paper published in 2021 by Yap et al. [19] establishes that the microbiota does not participate in autistic manifestations and that gut dysbiosis is a consequence of central symptoms in ASD.
2. The Microbiota–Gut–Brain Axis
The gut microbiota plays a fundamental role in the physiological functioning of the host and alterations in this microenvironment can have harmful effects on key points in the development of various organs systems, including the brain and digestive system (brain–gut–microbiota or microbiota–gut–brain axis). The brain–gut axis consists of the brain, the spinal cord, ANS, ENS and the hypothalamic–pituitary–adrenal (HPA) axis. Disturbances in the microbiota–gut–brain axis are the principal cause of the most frequent gastrointestinal motility disorders [20]. Studies in germ-free (GF) animals, those treated with antimicrobials or those exposed to environmental modifications that alter the gut microbiota from the prenatal or postnatal stage have been related to problems in brain immunity, blood–brain barrier (BBB) permeability, brain architecture and neural circuits [21].
2.1. Animal Models of Altered Gut Microbiota and Effects in the CNS
There are bacterial strains such as Escherichia coli or Lactobacillus sp. that interact directly with the host’s CNS through neurotransmitters dopamine, norepinephrine, histamine, acetylcholine, GABA or serotonin. An alteration in the composition of these strains can lead to an alteration in the metabolic state of the microbiome, resulting in metabolic disorders that may be responsible for the severity or progression of neurological disorders, such as Parkinson’s, Alzheimer’s, ASD and depression, among others [22][23].
Experimental studies have shown that the production of the bacterial metabolite 4-methylphenol (para-cresol or p-cresol) can alter the composition of the intestinal microbiota, leading to the recolonisation of Clostridium difficile. Fermenting tyrosine via the p-hydroxyphenylacetate (p-HPA) pathway is how C. difficile produces p-cresol. Since C. difficile is related to decreased growth of Proteobacteria, experimental studies showed that there were animals infected with C. difficile mutants (hpdC), since they found bacterial families belonging to Proteobacteria. In addition, the studies found that Bifidobacterium adolescentis is more sensitive to the presence of p-cresol than other Gram-positive species [24][25].
p-cresol negatively affects the homeostasis of epithelial cells, and its excess negatively affects the integrity of colonic epithelial cells [26]. As a result of the disruption in epithelial cells, a proinflammatory phenotype involving LPS may be promoted. Inflammation is closely related to the pathophysiology of mental disorders. Multiple communication pathways between the microbiota and the CNS have been identified, including immune pathways [27].
2.2. Effects of Gut Microbiota Metabolites in Immune Cells of the CNS
The gut microbiota has a complex and specific communication system with the CNS. The communication between the microbiota, the gut and the brain involves the secretion of different metabolites, including short-chain fatty acids (SCFA), the structural components of bacteria and signalling molecules [28].
Bacterial metabolites such as LPS can easily cross the intestinal barrier and cause inflammation, which affects the brain by altering cytokine levels [29]. Additionally, cytokines produced locally in the gastrointestinal mucosa travel peripherally and can cross the BBB [30]. During inflammation, the brain releases arginine vasopressin, a metabolite that affects social behaviour and is a considered biomarker in ASD [14]. There is a bidirectional relationship between the gut and the brain that includes nerve fibres [31]. Enteroendocrine cells of the intestinal epithelial barrier can detect the composition of the intestinal lumen, as well as nutrients and bacterial metabolites. These cells synapse with afferent fibres that directly connect the intestinal lumen to the brainstem [32].
The SCFA are the result of the fermentation of dietary fibre by anaerobic commensal bacteria in the colon. The host recognises SCFA (acetate, propionate and butyrate). Recently, butyrate has been identified as protective of the mucosa via goblet cells, as they regulate the response to the upregulation of MUC gene expression [16][33][34][35].
Altered concentrations of metabolites may have functional consequences in ASD. Different studies have identified various altered metabolites in the urinary profile of children with autism, some even correlate with the severity of autistic behaviour such as p-cresol [36][37]. Additionally, the metabolic pathways for tryptophan, vitamin B6, purine and phenylalanine are altered in ASD [38][39].
Combination therapy with vancomycin and Bifidobacterium improved in autistic symptoms. Additionally, the above therapy helped normalise the levels of the metabolites 3-(3-hydroxyphenyl)-3-hydroxypropionic acid, 3-hydroxyphenylacetic acid and 3-hydroxyhippuric acid in the urine of children with ASD, indicating an alteration in the production of phenylalanine in ASD [40].
3. Gut Microbiome, Immune System and Neurodevelopment Disorders
In mammals, enteric neurogenesis and gliogenesis occur primarily during the embryonic and foetal stages, but a considerable fraction of enteric neurons and glia are born in the colonising postnatal gut [41]. Functional maturation of gut neural networks is completed within the microenvironment of the postnatal gut, under the influence of gut microbiota and the mucosal immune system [31].
The gut microbiome lies at the intersection between the environment and the host, with the ability to modify host responses to disease-relevant exposures and stimuli. This is evident in the way that enteric microbes interact with the immune system, for example, by supporting immune maturation in the first years of life, affecting the efficacy of drugs through modulation of immune responses, or influencing the development of immune cell populations and their mediators [42].
3.1. Altered Gut Microbiota and Neurodevelopment Disorders
Gut microbiota alterations have been linked to various pathogenic pathways, and an increasing number of studies are linking changes in the gut microbiota to a range of neuropsychiatric diseases [43][44]. Similarly, the decrease in microbial diversity throughout life could be related to neurodegeneration [45][46].
The neuroinflammation produced by the different metabolites of a dysbiotic microbiota can be a pathogenic factor in severe neurodegenerative disorders [47]. In a severe inflammatory state, the activation of microglia releases proinflammatory cytokines such as TNF-α, IL-6 or MCP-1, or the inflammasome, as well as reactive oxygen species from microglial cells and resident macrophages, which could cause chronic neuroinflammation [17][48][49]. Therefore, the decrease or loss of the integrity of intestinal epithelial cells and chronic inflammation are highlighted as the main consequences of dysbiosis. Neuroinflammation and neurodegenerative and neuropsychiatric disorders can be the result of dysbiosis [43][50][51].
CNS inflammation is related to ASD. TNF-α is found at high levels in children with ASD, which was correlated with the severity of gastrointestinal symptoms [52]. Increases in TNF-α (increase Lachnoclostridium bolteae), IL-2, IL-4, IL-6 (increase Clostridium lituseburense), IL-8, IL-10 and IL-17 (increase Clostridium tertium) indicate a higher level of inflammation in the CNS of children with autism [18][29][53]. Additionally, children with ASD also have dysregulated T-cell production, leading to biases in the Th1 to Th2 ratio and immune cell activation associated with altered behaviour due to further neurodevelopmental impairment [54].
3.2. Altered Gut Microbiota and ASD
Microbial colonisation of the gastrointestinal tract begins prenatally, as microorganisms are detected in the placenta and meconium [55][56]. A recent study found that mothers of children with ASD harbour altered gut microbiomes [57], supporting the idea that maternal gut microbiota variation and infections during pregnancy may increase the risk of ASD in offspring [15]. In this same study, a clear relationship was found between the gut microbiome profiles of children and their mothers.
Children with ASD have unique bacterial biomarkers [57]. Previous studies have suggested that the gut microbiome of children with ASD contains harmful genera or species that contribute to the severity of autism symptoms, such as Bacteroides [58][59] or Desulfovibrio, which is related to the modulation of p-cresol production [36]. Changes in the gut microenvironment caused by the gut microbiome affect the production of signalling substances, leading to inadequate functioning of the brain and thus the prenatal and postnatal CNS [11].
The most common gut microbiota findings in children with ASD were a decreasing trend from Bacteroidetes to Firmicutes [60] and increased abundance of Clostridium [61][62][63]. Despite these findings, there are inconsistencies about the phenotypic signature of the gut microbiome of children with ASD, and a reason for these inconsistent results is that the composition of the gut microbiota is influenced by several factors, such as diet, lifestyle and medical history, among others [58]. In this regard, the use of antibiotics showed a correlation with the improvement of symptoms in ASD [64]. Although recently, it has been seen this is not always the case since the prolonged use of oral antibiotics can increase the proliferation of anaerobic bacteria in the intestine. For example, Clostridia, Bacteroidetes and Desulfovibrio are common bacteria that, in addition to modulating the intestinal immune system, can promote gastrointestinal symptoms and autistic behaviour in ASD [65].
The fungi Candida also appears to play a role in children with ASD [60]. In a dysbiotic environment, as often seen in the population with autism, Candida proliferates and produces ammonia and toxins, which increases autistic behaviour. Candida also causes malabsorption of minerals and carbohydrates that play an important role in the pathophysiology of ASD [66]. A subset of people with ASD shows gastrointestinal disturbances [67], and results from different studies indicate that eliminating some foods from the diet may help improve gastrointestinal symptoms in ASD. Associating diet is an important factor in the composition of the gut microbiota [68].
On the other hand, in a study with a large group of ASD patients, their gut metagenome showed a relationship with diet, reduced taxonomic diversity and stool consistency, but no relationship was found between the diagnosis of ASD and the gut microbiome. It was suggested that softer faecal consistency is more closely related to decreased taxonomic diversity and that there is a downstream relationship to reduced dietary diversity, which is a common feature of patients with ASD. In this work, authors proposed that this mechanism could explain the relationship between the increase in gastrointestinal problems and increase in repetitive behaviours in ASD. Maintain that sensory sensitivity could be the basis of restricted diets in ASD but found no relationship between the sensory profile and ASD severity. The study found that all psychometric characteristics had more significant correlations with dietary diversity than with taxonomic diversity. In conclusion, the results suggest that dysbiosis is a consequence of autistic manifestations and that it has no causal role in the disease. Therefore, microbiome-directed treatment is not a suitable therapeutic target for treating comorbidities in children with ASD [19].
The cause of ASD remains undetermined, complex and incompletely understood, with increasing evidence pointing to abnormal synaptic development and aberrant immune responses as possible effectors of autistic symptoms [69][70]. Microglial cells have been strongly associated with physiological processes and the development of autistic symptoms, as they are part of the main cells of the CNS that provide an innate immune response to the tissue with inflammatory and tissue repair functions [69].
Some metabolites produced by specific bacterial groups have anti-inflammatory effects on microglia, such as butyrate, an essential SCFA for the modulation of excitatory and inhibitory neuronal pathways in ASD [28][71]. Patients with ASD showed low levels of SCFA [72]. The deficiency of these metabolites could be the cause of a disruption in the intercommunication between the ENS and the mucosal immune system [31], what could be generate changes in the intestinal motility of children with ASD [73]. For example, SCFA activate G protein-coupled receptors (GPR41 and GPR43) on enteroendocrine cells of the intestinal epithelium, resulting in increased production of GLP-1 and 5-HT and changes in intestinal motility [31].
Not studies that suggest that dysbiosis has a leading role in the cause of ASD, since it has always been presented as a multifactorial disease with a very high genetic component, which seems to be the main cause of autistic symptoms [57][74][75][76][77][78]. Although, dysbiosis has been proposed as another factor in the cause of ASD, its relevance has not been well clarified since the results of various studies have not been conclusive. Some studies point to dysbiosis as a possible factor in autistic symptoms [61][79], and other studies rule out the possibility that dysbiosis is a determining factor in the aetiology of ASD [19][80].
Recently, there are increased reports of evidence regarding the possible involvement of intestinal dysbiosis as an aetiologic factor in ASD with moderate effects. The relevance of dysbiosis as a factor in the aetiology of ASD relies on the fact that it is modifiable. There are reports of interventions that have had beneficial but modest effects on autistic manifestations. For example, faecal transplant therapies and probiotic supplementation. A pioneer group in faecal transplantation in children with ASD found that after ten weeks of intervention, there was a reduction of almost 80% in gastrointestinal symptoms and improvement in behavioural symptoms, and that these improvements persisted after eight weeks of treatment. In addition, there were beneficial changes in the abundance of Bifidubacterium, Prevotella and Desulfovibrio; these changes also persisted after the suspension of the intervention and two years later [81][82]. Regarding probiotics, a successful study that supplemented with probiotics children with ASD for 6 months found positive effects on some gastrointestinal symptoms, adaptive functions and sensory profiles versus the placebo group [83]. Another study found that after supplementation with probiotics and prebiotics, there was an increase in beneficial bacteria (Bifidobacterial and B. longum) and suppression of pathogenic bacteria (Clostridium) with a significant reduction in the severity of autistic and gastrointestinal symptoms [84].
ASD can be syndromic or non-syndromic. Syndromic ASD is often associated with chromosomal abnormalities or monogenic alterations such as Rett and Fragile X syndrome among other syndromes. Non-syndromic ASD is a complex and multifactorial condition influenced by both genetic and environmental factors [55][72][74][75][76][77]. It is widely recognised that the causative traits of ASD predominantly have a genetic basis. A meta-analysis of 13 twin studies estimated the heritability of ASD to be 74%, with an environmental effect of 25% [85]. However, it is important to acknowledge that the heritability value may be prone to overestimation due to statistical artifacts or an underestimation of unaccounted shared environmental factors, which cannot be accurately estimated in twin and sibling studies. Another estimation of ASD heritability based on 192 twin pairs found a lower value of 38% for heritability and a 58% contribution from environmental factors [86].
The contribution of genetic and environmental factors to the presentation of ASD can vary, but genetic factors are considered to be the primary determinants. Approximately 1000 human loci have been associated with ASD according to SFARI database [87]. Additionally, a meta-analysis of genome-wide association studies (GWAS) involving over 16,000 individuals identified chromosomal regions 10q24.32, 3p13, 3p25 and 8p11.23 that are associated with ASD. It is worth noting that the 10q24.32 region is also linked to schizophrenia [88].
In terms of environmental factors, advanced parental age [89], prenatal exposure to antibiotics or certain drugs [90], maternal immune activation [91], imbalances in prenatal micronutrients [92], epilepsy [93][94] and maternal obesity [95] have been shown to be associated with an increased risk of ASD. Therefore, these factors are considered as potential environmental drivers in the aetiology of ASD.
While there is a wealth of evidence suggesting the potential involvement of the gut microbiota in the manifestation of core ASD symptoms, it is important to note that the precise role of gut microbiota in the pathogenesis of ASD remains unclear. Numerous studies have focused on animal models, such as germ-free mice, while others have analysed dysbiosis in ASD patients and its impact on immunoregulators, neurotransmitters and relevant metabolite production in ASD [25][34][35][37]. However, despite the available information, no study to date has definitively established the intestinal microbiota as a causative environmental factor in ASD. Nevertheless, it has been a therapeutic target in various interventions aimed at managing ASD symptoms in patients.
The relevance of dysbiosis as a factor in the aetiology of ASD relies on the fact that it is modifiable. There are reports of interventions that have had beneficial but modest effects on autistic manifestations. For example, faecal transplant therapies and probiotic supplementation. A pioneer group in faecal transplantation in children with ASD found that after ten weeks of intervention, there was a reduction of almost 80% in gastrointestinal symptoms and improvement in behavioural symptoms, and that these improvements persisted after eight weeks of treatment. In addition, there were beneficial changes in the abundance of Bifidubacterium, Prevotella and Desulfovibrio; these changes also persisted after the suspension of the intervention and two years later [81][82]. Regarding probiotics, a successful study that supplemented with probiotics children with ASD for 6 months found positive effects on some gastrointestinal symptoms, adaptive functions and sensory profiles versus the placebo group [83]. Another study found that after supplementation with probiotics and prebiotics, there was an increase in beneficial bacteria (Bifidubacterium and B. longum) and suppression of pathogenic bacteria (Clostridium) with a significant reduction in the severity of autistic and gastrointestinal symptoms [84].
Dysbiosis has been proposed as a potential contributing factor in the development of ASD. However, its relevance in ASD aetiology remains unclear due to inconclusive results from various studies. Some studies suggest a possible association between dysbiosis and autistic symptoms [62][96], while others dismiss the notion that dysbiosis plays a decisive role in the development of ASD [19][80].
Yap and colleagues proposed an alternative perspective, suggesting that gut dysbiosis observed in ASD patients could be a consequence of the restrictive and behavioural patterns inherent to ASD. They argue that alterations in eating patterns, rather than dysbiosis, might have a minor or non-causal role in ASD. This study highlights the unconvincing evidence supporting the participation of gut microbiota in ASD. Many studies lacked statistical power due to small sample sizes and failed to consider important confounders such as sex, stool consistency and diet [19].
Researchers propose a novel model that incorporates the role of gut microbiota in ASD while considering the findings presented by Yap and colleagues aims to explain the limited but evident effects of microbiota intervention on ASD core symptoms in patients (Figure 1).
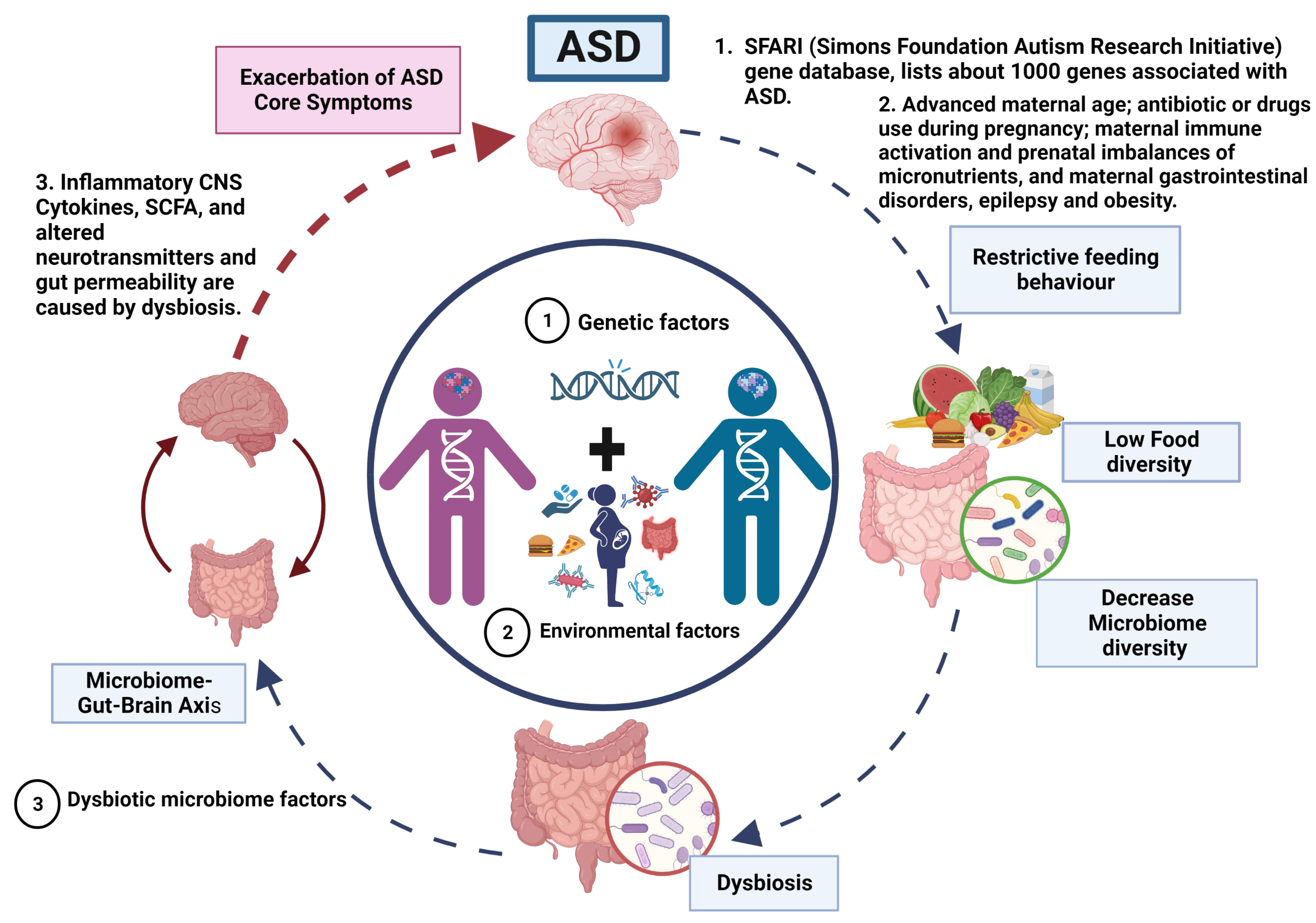
Figure 1. Vicious cycle and phenotype relationship in ASD.
ASD is a neurodevelopmental disorder of multifactorial aetiology where the main factors are genetic variations interacting with environmental factors (dysbiosis). (1) Genetics factors. The genetic predisposition considered as the main cause of ASD. Where most genes are related to synaptogenesis and many others with metabolic abnormalities and immune response. (2) Dysbiotic microbiome factors. Approximately 70% of the population with ASD present gastrointestinal problems that, together with a restricted diet (diet reduced in fruits and vegetables and high in fat and sugar), exacerbate the imbalance in the microbiome composition that may altered homeostasis and causing dysbiotic state, potentially triggering inflammatory responses and dysregulation on the production of neurotransmitters that control the microbiome–gut–brain axis. Therefore, a vicious cycle is generated until an equilibrium is reached in the severity of ASD manifestations that gives rise to the different phenotypes in ASD.
According to the model, the primary driving factors in the aetiology of ASD are genetic and environmental, with no direct involvement of gut microbiota in the manifestation of ASD. However, researchers propose that the restricted feeding patterns observed in ASD patients lead to decreased food diversity, subsequently resulting in a reduction of microbiota diversity and the development of gut dysbiosis.
The dysbiosis of the gut microbiota affects the gut–brain axis through mechanisms (Figure 2). Therefore, the severity of autistic manifestations is accentuated. Within the proposed model, gut dysbiosis acts as a secondary factor that exacerbates the core symptoms of ASD and contributes to the occurrence of gastrointestinal manifestations until a certain threshold of severity is reached.
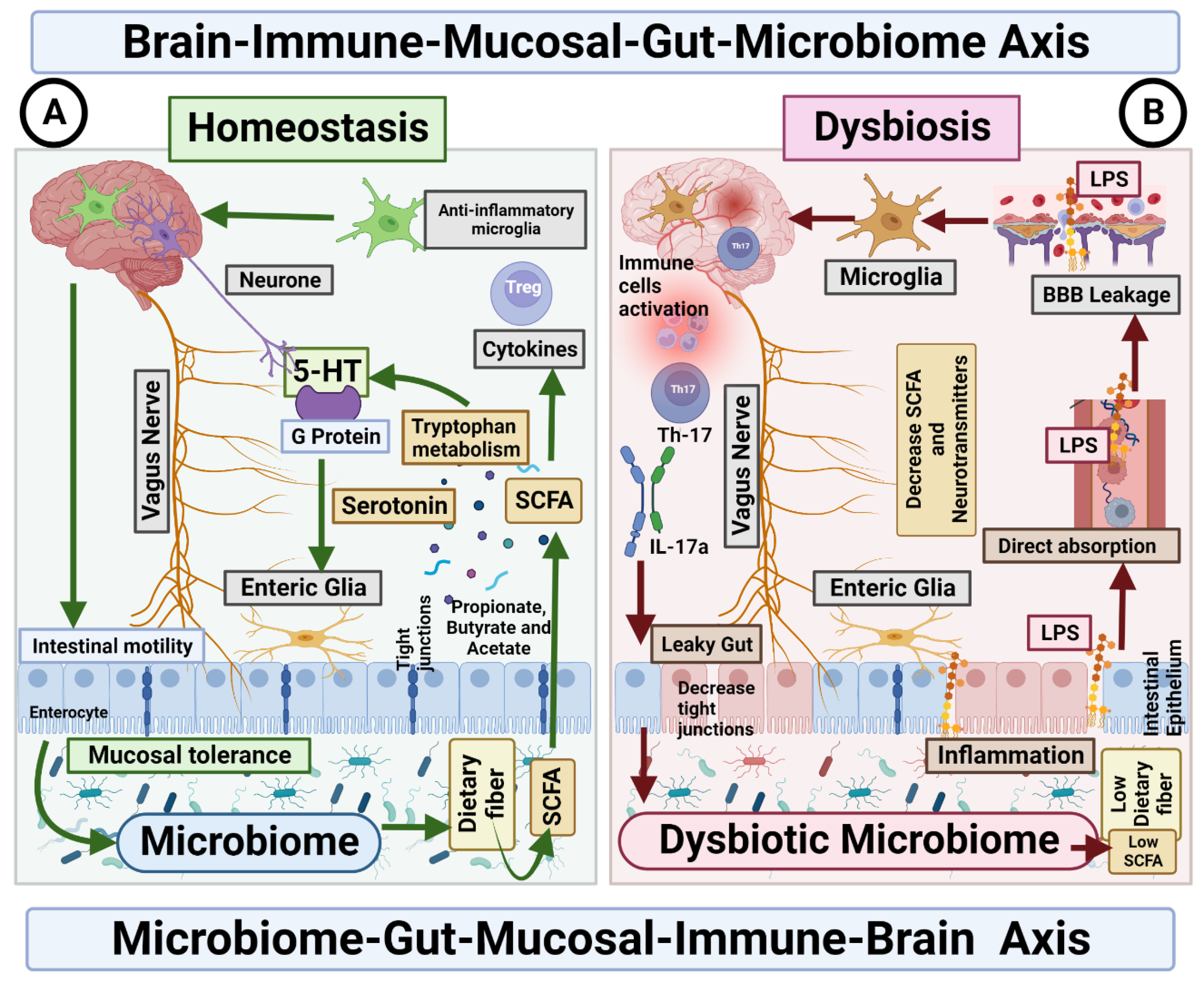
Figure 2. Microbiome–Gut–Mucosal–Immune–Brain bidirectional relationship. The vagus nerve transfers information on the state of the digestive system to the brain through sensory fibres. It sends neuronal, endocrine and immune signals. The gut microbiota metabolites are involved in permeability of the intestinal epithelium, neurotransmitter synthesis and cytokines release. (A) Homeostasis. Under homeostasis conditions, gut microbiome and diet control the activity of ENS cells through SCFA, which in turn activate the enteroendocrine cells of the intestinal epithelium, which increases the production of serotonin and therefore changes intestinal motility. In addition, the microbiome is essential for the maintenance of mucosal glial cells that express neurotrophic factor. (B) Dysbiosis. The imbalance in the homeostasis of the gut microbiome is associated with the decreased production of metabolites, such as butyrate, responsible for the modulation of inhibitory and excitatory pathways, and with anti-inflammatory effects on microglia. Deficiency of this microbial metabolite could be the cause of a disruption in the intercommunication between the ENS and the mucosal immune system.
References
- American Psychiatric Association; DSM-5 Task Force. Diagnostic and Statistical Manual of Mental Disorders: DSM-5™, 5th ed.; American Psychiatric Publishing, Inc.: Washington, DC, USA, 2013.
- Rogozin, I.B.; Gertz, E.M.; Baranov, P.; Poliakov, E.; Schaffer, A.A. Genome-Wide Changes in Protein Translation Efficiency Are Associated with Autism. Genome Biol. Evol. 2018, 10, 1902–1919.
- Sauer, M.A.K.; Stanton, M.J.; Hans, M.S.; Grabrucker, A.M. Autism Spectrum Disorders: Etiology and Pathology. In Autism Spectrum Disorders; Grabrucker, A.M., Ed.; Chapter 1; Exon Publications: Brisbane, Australia, 2021; pp. 1–16.
- Krigsman, A.; Walker, S.J. Gastrointestinal disease in children with autism spectrum disorders: Etiology or consequence? World J. Psychiatry 2021, 11, 605–618.
- Fobofou, S.A.; Savidge, T. Microbial metabolites: Cause or consequence in gastrointestinal disease? Am. J. Physiol. Liver Physiol. 2022, 322, G535–G552.
- Wu, W.-L.; Adame, M.D.; Liou, C.-W.; Barlow, J.T.; Lai, T.-T.; Sharon, G.G.; Schretter, C.E.; Needham, B.D.; Wang, M.I.; Tang, W.; et al. Microbiota regulate social behaviour via stress response neurons in the brain. Nature 2021, 595, 409–414.
- Parkin, K.; Christophersen, C.T.; Verhasselt, V.; Cooper, M.N.; Martino, D. Risk Factors for Gut Dysbiosis in Early Life. Microorganisms 2021, 9, 2066.
- Nagpal, R.; Tsuji, H.; Takahashi, T.; Nomoto, K.; Kawashima, K.; Nagata, S.; Yamashiro, Y. Ontogenesis of the Gut Microbiota Composition in Healthy, Full-Term, Vaginally Born and Breast-Fed Infants over the First 3 Years of Life: A Quantitative Bird’s-Eye View. Front. Microbiol. 2017, 8, 1388.
- Dizzell, S.; Stearns, J.C.; Li, J.; van Best, N.; Bervoets, L.; Mommers, M.; Penders, J.; Morrison, K.M.; Hutton, E.K.; on behalf of the GI-MDH Consortium Partners. Investigating colonization patterns of the infant gut microbiome during the introduction of solid food and weaning from breastmilk: A cohort study protocol. PLoS ONE 2021, 16, e0248924.
- Guo, M.; Miao, M.; Wang, Y.; Duan, M.; Yang, F.; Chen, Y.; Yuan, W.; Zheng, H. Developmental differences in the intestinal microbiota of Chinese 1-year-old infants and 4-year-old children. Sci. Rep. 2020, 10, 19470.
- Seki, D.; Mayer, M.; Hausmann, B.; Pjevac, P.; Giordano, V.; Goeral, K.; Unterasinger, L.; Klebermaß-Schrehof, K.; De Paepe, K.; Van de Wiele, T.; et al. Aberrant gut-microbiota-immune-brain axis development in premature neonates with brain damage. Cell Host Microbe 2021, 29, 1558–1572.e6.
- Garcia-Gutierrez, E.; Narbad, A.; Rodríguez, J.M. Autism Spectrum Disorder Associated with Gut Microbiota at Immune, Metabolomic, and Neuroactive Level. Front. Neurosci. 2020, 14, 578666.
- Rothenberg, S.E.; Chen, Q.; Shen, J.; Nong, Y.; Nong, H.; Trinh, E.P.; Biasini, F.J.; Liu, J.; Zeng, X.; Zou, Y.; et al. Neurodevelopment correlates with gut microbiota in a cross-sectional analysis of children at 3 years of age in rural China. Sci. Rep. 2021, 11, 7384.
- Eshraghi, R.S.; Davies, C.; Iyengar, R.; Perez, L.; Mittal, R.; Eshraghi, A.A. Gut-Induced Inflammation during Development May Compromise the Blood-Brain Barrier and Predispose to Autism Spectrum Disorder. J. Clin. Med. 2020, 10, 27.
- Jaini, R.; Wolf, M.R.; Yu, Q.; King, A.T.; Frazier, T.W.; Eng, C. Maternal genetics influences fetal neurodevelopment and postnatal autism spectrum disorder-like phenotype by modulating in-utero immunosuppression. Transl. Psychiatry 2021, 11, 348.
- Tran, S.M.-S.; Mohajeri, M.H. The Role of Gut Bacterial Metabolites in Brain Development, Aging and Disease. Nutrients 2021, 13, 732.
- Nagpal, J.; Cryan, J.F. Host genetics, the microbiome & behaviour—A ‘Holobiont’ perspective. Cell Res. 2021, 31, 832–833.
- Lungba, R.M.; Khan, S.Z.A.; Ajibawo-Aganbi, U.; Bastidas, M.V.P.; Veliginti, S.; Saleem, S.; Cancarevic, I. The Role of the Gut Microbiota and the Immune System in the Development of Autism. Cureus 2020, 12, 11226.
- Yap, C.X.; Henders, A.K.; Alvares, G.A.; Wood, D.L.; Krause, L.; Tyson, G.W.; Restuadi, R.; Wallace, L.; McLaren, T.; Hansell, N.K.; et al. Autism-related dietary preferences mediate autism-gut microbiome associations. Cell 2021, 184, 5916–5931.e17.
- Agirman, G.; Hsiao, E.Y. SnapShot: The microbiota-gut-brain axis. Cell 2021, 184, 2524–2524.e1.
- Wang, Y.; Kasper, L.H. The role of microbiome in central nervous system disorders. Brain Behav. Immun. 2014, 38, 1–12.
- Marć, M.A.; Jastrząb, R.; Mytych, J. Does the Gut Microbial Metabolome Really Matter? The Connection between GUT Metabolome and Neurological Disorders. Nutrients 2022, 14, 3967.
- Passmore, I.J.; Letertre, M.P.M.; Preston, M.D.; Bianconi, I.; Harrison, M.A.; Nasher, F.; Kaur, H.; Hong, H.A.; Baines, S.D.; Cutting, S.M.; et al. Para-cresol production by Clostridium difficile affects microbial diversity and membrane integrity of Gram-negative bacteria. PLoS Pathog. 2018, 14, e1007191.
- Harrison, M.A.; Kaur, H.; Wren, B.W.; Dawson, L.F. Production of p-cresol by Decarboxylation of p-HPA by All Five Lineages of Clostridioides difficile Provides a Growth Advantage. Front. Cell Infect. Microbiol. 2021, 11, 757599.
- Rogers, A.P.; Mileto, S.J.; Lyras, D. Impact of enteric bacterial infections at and beyond the epithelial barrier. Nat. Rev. Genet. 2023, 21, 260–274.
- Cenit, M.C.; Sanz, Y.; Codoñer-Franch, P. Influence of gut microbiota on neuropsychiatric disorders. World J. Gastroenterol. 2017, 23, 5486–5498.
- Doroszkiewicz, J.; Groblewska, M.; Mroczko, B. The Role of Gut Microbiota and Gut-Brain Interplay in Selected Diseases of the Central Nervous System. Int. J. Mol. Sci. 2021, 22, 10028.
- Srikantha, P.; Mohajeri, M.H. The Possible Role of the Microbiota-Gut-Brain-Axis in Autism Spectrum Disorder. Int. J. Mol. Sci. 2019, 20, 2115.
- Banks, W.A.; Kastin, A.J.; Broadwell, R.D. Passage of Cytokines across the Blood-Brain Barrier. Neuroimmunomodulation 1995, 2, 241–248.
- Kaelberer, M.M.; Buchanan, K.L.; Klein, M.E.; Barth, B.B.; Montoya, M.M.; Shen, X.; Bohórquez, D.V. A gut-brain neural circuit for nutrient sensory transduction. Science 2018, 361, aat5236.
- Jacobson, A.; Yang, D.; Vella, M.; Chiu, I.M. The intestinal neuro-immune axis: Crosstalk between neurons, immune cells, and microbes. Mucosal Immunol. 2021, 14, 555–565.
- Gaudier, E.; Jarry, A.; Blottière, H.M.; de Coppet, P.; Buisine, M.P.; Aubert, J.P.; Laboisse, C.; Cherbut, C.; Hoebler, C. Butyrate specifically modulates MUC gene expression in intestinal epithelial goblet cells deprived of glucose. Am. J. Physiol. Liver Physiol. 2004, 287, G1168–G1174.
- Donohoe, D.R.; Garge, N.; Zhang, X.; Sun, W.; O’Connell, T.M.; Bunger, M.K.; Bultman, S.J. The Microbiome and Butyrate Regulate Energy Metabolism and Autophagy in the Mammalian Colon. Cell Metab. 2011, 13, 517–526.
- Levy, M.; Blacher, E.; Elinav, E. Microbiome, metabolites and host immunity. Curr. Opin. Microbiol. 2017, 35, 8–15.
- Vasquez, A. Biological plausibility of the gut-brain axis in autism. Ann. N. Y. Acad. Sci. 2017, 1408, 5–6.
- Ho, L.K.H.; Tong, V.J.W.; Syn, N.; Nagarajan, N.; Tham, E.H.; Tay, S.K.; Shorey, S.; Tambyah, P.A.; Law, E.C.N. Gut microbiota changes in children with autism spectrum disorder: A systematic review. Gut Pathog. 2020, 12, 6.
- Clayton, T.A. Metabolic differences underlying two distinct rat urinary phenotypes, a suggested role for gut microbial metabolism of phenylalanine and a possible connection to autism. FEBS Lett. 2012, 586, 956–961.
- Gevi, F.; Zolla, L.; Gabriele, S.; Persico, A.M. Urinary metabolomics of young Italian autistic children supports abnormal tryptophan and purine metabolism. Mol. Autism 2016, 7, 47.
- Xiong, X.; Liu, D.; Wang, Y.; Zeng, T.; Peng, Y. Urinary 3-(3-Hydroxyphenyl)-3-hydroxypropionic Acid, 3-Hydroxyphenylacetic Acid, and 3-Hydroxyhippuric Acid Are Elevated in Children with Autism Spectrum Disorders. BioMed Res. Int. 2016, 2016, 9485412.
- Hitch, T.C.A.; Hall, L.J.; Walsh, S.K.; Leventhal, G.E.; Slack, E.; de Wouters, T.; Walter, J.; Clavel, T. Microbiome-based interventions to modulate gut ecology and the immune system. Mucosal Immunol. 2022, 15, 1095–1113.
- Obata, Y.; Pachnis, V. The Effect of Microbiota and the Immune System on the Development and Organization of the Enteric Nervous System. Gastroenterology 2016, 151, 836–844.
- Santos, J.; Barbara, G. Editorial: Human Intestinal Permeability, Mucosal Inflammation and Diet. Front. Nutr. 2022, 9, 894869.
- Toledo, A.R.L.; Monroy, G.R.; Salazar, F.E.; Lee, J.-Y.; Jain, S.; Yadav, H.; Borlongan, C.V. Gut-Brain Axis as a Pathological and Therapeutic Target for Neurodegenerative Disorders. Int. J. Mol. Sci. 2022, 23, 1184.
- Marogianni, C.; Sokratous, M.; Dardiotis, E.; Hadjigeorgiou, G.M.; Bogdanos, D.; Xiromerisiou, G. Neurodegeneration and Inflammation—An Interesting Interplay in Parkinson’s Disease. Int. J. Mol. Sci. 2020, 21, 8421.
- Xie, J.; Van Hoecke, L.; Vandenbroucke, R.E. The Impact of Systemic Inflammation on Alzheimer’s Disease Pathology. Front. Immunol. 2022, 12, 796867.
- Giri, R.; Hoedt, E.C.; Khushi, S.; Salim, A.A.; Bergot, A.-S.; Schreiber, V.; Thomas, R.; McGuckin, M.A.; Florin, T.H.; Morrison, M.; et al. Secreted NF-κB suppressive microbial metabolites modulate gut inflammation. Cell Rep. 2022, 39, 110646.
- Saresella, M.; Piancone, F.; Marventano, I.; Zoppis, M.; Hernis, A.; Zanette, M.; Trabattoni, D.; Chiappedi, M.; Ghezzo, A.; Canevini, M.P.; et al. Multiple inflammasome complexes are activated in autistic spectrum disorders. Brain Behav. Immun. 2016, 57, 125–133.
- Jyonouchi, H.; Geng, L. Associations between Monocyte and T Cell Cytokine Profiles in Autism Spectrum Disorders: Effects of Dysregulated Innate Immune Responses on Adaptive Responses to Recall Antigens in a Subset of ASD Children. Int. J. Mol. Sci. 2019, 20, 4731.
- Wanchao, S.; Chen, M.; Zhiguo, S.; Futang, X.; Mengmeng, S. Protective effect and mechanism of Lactobacillus on cerebral ischemia reperfusion injury in rats. Braz. J. Med. Biol. Res. 2018, 51, e7172.
- Dinan, T.G.; Cryan, J.F. Gut instincts: Microbiota as a key regulator of brain development, ageing and neurodegeneration. J. Physiol. 2017, 595, 489–503.
- Tomova, A.; Husarova, V.; Lakatosova, S.; Bakos, J.; Vlkova, B.; Babinska, K.; Ostatnikova, D. Gastrointestinal microbiota in children with autism in Slovakia. Physiol. Behav. 2015, 138, 179–187.
- Luna, R.A.; Oezguen, N.; Balderas, M.; Venkatachalam, A.; Runge, J.K.; Versalovic, J.; Veenstra-VanderWeele, J.; Anderson, G.M.; Savidge, T.; Williams, K.C. Distinct Microbiome-Neuroimmune Signatures Correlate with Functional Abdominal Pain in Children with Autism Spectrum Disorder. Cell Mol. Gastroenterol. Hepatol. 2016, 3, 218–230.
- Wang, Y.; Li, N.; Yang, J.-J.; Zhao, D.-M.; Chen, B.; Zhang, G.-Q.; Chen, S.; Cao, R.-F.; Yu, H.; Zhao, C.-Y.; et al. Probiotics and fructo-oligosaccharide intervention modulate the microbiota-gut brain axis to improve autism spectrum reducing also the hyper-serotonergic state and the dopamine metabolism disorder. Pharmacol. Res. 2020, 157, 104784.
- Careaga, M.; Rogers, S.; Hansen, R.L.; Amaral, D.G.; Van de Water, J.; Ashwood, P. Immune Endophenotypes in Children with Autism Spectrum Disorder. Biol. Psychiatry 2017, 81, 434–441.
- Groer, M.W.; Gregory, K.E.; Louis-Jacques, A.; Thibeau, S.; Walker, W.A. The very low birth weight infant microbiome and childhood health. Birth Defects Res. Part C Embryo Today Rev. 2015, 105, 252–264.
- Collado, M.C.; Rautava, S.; Aakko, J.; Isolauri, E.; Salminen, S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 2016, 6, 23129.
- Li, N.; Yang, J.; Zhang, J.; Liang, C.; Wang, Y.; Chen, B.; Zhao, C.; Wang, J.; Zhang, G.; Zhao, D.; et al. Correlation of Gut Microbiome Between ASD Children and Mothers and Potential Biomarkers for Risk Assessment. Genom. Proteom. Bioinform. 2019, 17, 26–38.
- Tamana, S.K.; Tun, H.M.; Konya, T.; Chari, R.S.; Field, C.J.; Guttman, D.S.; Becker, A.B.; Moraes, T.J.; Turvey, S.E.; Subbarao, P.; et al. Bacteroides-dominant gut microbiome of late infancy is associated with enhanced neurodevelopment. Gut Microbes 2021, 13, 1930875.
- Strati, F.; Cavalieri, D.; Albanese, D.; De Felice, C.; Donati, C.; Hayek, J.; Jousson, O.; Leoncini, S.; Renzi, D.; Calabrò, A.; et al. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome 2017, 5, 24.
- Suzuki, T.A.; Fitzstevens, J.L.; Schmidt, V.T.; Enav, H.; Huus, K.E.; Ngwese, M.M.; Grießhammer, A.; Pfleiderer, A.; Adegbite, B.R.; Zinsou, J.F.; et al. Codiversification of gut microbiota with humans. Science 2022, 377, 1328–1332.
- Ma, B.; Liang, J.; Dai, M.; Wang, J.; Luo, J.; Zhang, Z.; Jing, J. Altered Gut Microbiota in Chinese Children with Autism Spectrum Disorders. Front. Cell Infect. Microbiol. 2019, 9, 40.
- Ding, X.; Xu, Y.; Zhang, X.; Zhang, L.; Duan, G.; Song, C.; Li, Z.; Yang, Y.; Wang, Y.; Wang, X.; et al. Gut microbiota changes in patients with autism spectrum disorders. J. Psychiatr. Res. 2020, 129, 149–159.
- Alshammari, M.K.; AlKhulaifi, M.M.; Al Farraj, D.A.; Somily, A.; Albarrag, A.M. Incidence of Clostridium perfringens and its toxin genes in the gut of children with autism spectrum disorder. Anaerobe 2020, 61, 102114.
- Taguer, M.; Maurice, C. The complex interplay of diet, xenobiotics, and microbial metabolism in the gut: Implications for clinical outcomes. Clin. Pharmacol. Ther. 2016, 99, 588–599.
- Kovtun, A.S.; Averina, O.V.; Alekseeva, M.G.; Danilenko, V.N. Antibiotic Resistance Genes in the Gut Microbiota of Children with Autistic Spectrum Disorder as Possible Predictors of the Disease. Microb. Drug Resist. 2020, 26, 1307–1320.
- Kantarcioglu, A.S.; Kiraz, N.; Aydin, A. Microbiota-Gut-Brain Axis: Yeast Species Isolated from Stool Samples of Children with Suspected or Diagnosed Autism Spectrum Disorders and In Vitro Susceptibility against Nystatin and Fluconazole. Mycopathologia 2016, 181, 1–7.
- McElhanon, B.O.; McCracken, C.; Karpen, S.; Sharp, W.G. Gastrointestinal Symptoms in Autism Spectrum Disorder: A Meta-analysis. Pediatrics 2014, 133, 872–883.
- Ristori, M.V.; Quagliariello, A.; Reddel, S.; Ianiro, G.; Vicari, S.; Gasbarrini, A.; Putignani, L. Autism, Gastrointestinal Symptoms and Modulation of Gut Microbiota by Nutritional Interventions. Nutrients 2019, 11, 2812.
- Zhou, R.; Qian, S.; Cho, W.C.S.; Zhou, J.; Jin, C.; Zhong, Y.; Wang, J.; Zhang, X.; Xu, Z.; Tian, M.; et al. Microbiota-microglia connections in age-related cognition decline. Aging Cell 2022, 21, 13599.
- Goines, P.; Van de Water, J. The immune system’s role in the biology of autism. Curr. Opin. Neurol. 2010, 23, 111–117.
- Kim, C.H. Immune regulation by microbiome metabolites. Immunology 2018, 154, 220–229.
- Adams, J.B.; Johansen, L.J.; Powell, L.D.; Quig, D.; Rubin, R.A. Gastrointestinal flora and gastrointestinal status in children with autism–Comparisons to typical children and correlation with autism severity. BMC Gastroenterol. 2011, 11, 22.
- Ehrlich, S.D. The MetaHIT Consortium MetaHIT: The European Union Project on Metagenomics of the Human Intestinal Tract. In Metagenomics of the Human Body; Nelson, K., Ed.; Springer: New York, NY, USA, 2011.
- Chang, J.; Gilman, S.R.; Chiang, A.H.; Sanders, S.; Vitkup, D. Genotype to phenotype relationships in autism spectrum disorders. Nat. Neurosci. 2015, 18, 191–198.
- Zhang, Y.; Li, N.; Li, C.; Zhang, Z.; Teng, H.; Wang, Y.; Zhao, T.; Shi, L.; Zhang, K.; Xia, K.; et al. Genetic evidence of gender difference in autism spectrum disorder supports the female-protective effect. Transl. Psychiatry 2020, 10, 4.
- Duda, M.; Zhang, H.; Li, H.-D.; Wall, D.P.; Burmeister, M.; Guan, Y. Brain-specific functional relationship networks inform autism spectrum disorder gene prediction. Transl. Psychiatry 2018, 8, 56.
- Lefebvre, A.; Tillmann, J.; Cliquet, F.; Amsellem, F.; Maruani, A.; Leblond, C.; Beggiato, A.; Germanaud, D.; Amestoy, A.; Moal, M.L.; et al. Tackling hypo and hyper sensory processing heterogeneity in autism: From clinical stratification to genetic pathways. Autism Res. 2023, 16, 364–378.
- Trost, B.; Thiruvahindrapuram, B.; Chan, A.J.; Engchuan, W.; Higginbotham, E.J.; Howe, J.L.; Loureiro, L.O.; Reuter, M.S.; Roshandel, D.; Whitney, J.; et al. Genomic architecture of autism from comprehensive whole-genome sequence annotation. Cell 2022, 185, 4409–4427.e18.
- Liu, S.; Li, E.; Sun, Z.; Fu, D.; Duan, G.; Jiang, M.; Yu, Y.; Mei, L.; Yang, P.; Tang, Y.; et al. Altered gut microbiota and short chain fatty acids in Chinese children with autism spectrum disorder. Sci. Rep. 2019, 9, 287.
- Nova, E.; Gómez-Martinez, S.; González-Soltero, R. The Influence of Dietary Factors on the Gut Microbiota. Microorganisms 2022, 10, 1368.
- Kang, D.-W.; Adams, J.B.; Gregory, A.C.; Borody, T.; Chittick, L.; Fasano, A.; Khoruts, A.; Geis, E.; Maldonado, J.; McDonough-Means, S.; et al. Microbiota Transfer Therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: An open-label study. Microbiome 2017, 5, 10.
- Kang, D.-W.; Adams, J.B.; Coleman, D.M.; Pollard, E.L.; Maldonado, J.; McDonough-Means, S.; Caporaso, J.G.; Krajmalnik-Brown, R. Long-term benefit of Microbiota Transfer Therapy on autism symptoms and gut microbiota. Sci. Rep. 2019, 9, 5821.
- Taniya, M.A.; Chung, H.-J.; Al Mamun, A.; Alam, S.; Aziz, A.; Emon, N.U.; Islam, M.; Hong, S.-T.S.; Podder, B.R.; Mimi, A.A.; et al. Role of Gut Microbiome in Autism Spectrum Disorder and Its Therapeutic Regulation. Front. Cell Infect. Microbiol. 2022, 12, 915701.
- Zou, R.; Wang, Y.; Duan, M.; Guo, M.; Zhang, Q.; Zheng, H. Dysbiosis of Gut Fungal Microbiota in Children with Autism Spectrum Disorders. J. Autism Dev. Disord. 2021, 51, 267–275.
- Tick, B.; Bolton, P.; Happé, F.; Rutter, M.; Rijsdijk, F. Heritability of autism spectrum disorders: A meta-analysis of twin studies. J. Child Psychol. Psychiatry 2016, 57, 585–595.
- Hallmayer, J.; Cleveland, S.; Torres, A.; Phillips, J.; Cohen, B.; Torigoe, T.; Miller, J.; Fedele, A.; Collins, J.; Smith, K.; et al. Genetic Heritability and Shared Environmental Factors Among Twin Pairs with Autism. Arch. Gen. Psychiatry 2011, 68, 1095–1102.
- Abrahams, B.S.; Arking, D.E.; Campbell, D.B.; Mefford, H.C.; Morrow, E.M.; Weiss, L.A.; Menashe, I.; Wadkins, T.; Banerjee-Basu, S.; Packer, A. SFARI Gene 2.0: A community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol. Autism 2013, 4, 36.
- The Autism Spectrum Disorders Working Group of The Psychiatric Genomics Consortium. Meta-analysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol. Autism 2017, 8, 21.
- Idring, S.; Magnusson, C.; Lundberg, M.; Ek, M.; Rai, D.; Svensson, A.C.; Dalman, C.; Karlsson, H.; Lee, B.K. Parental age and the risk of autism spectrum disorders: Findings from a Swedish population-based cohort. Int. J. Epidemiol. 2014, 43, 107–115.
- Christensen, J.; Grønborg, T.K.; Sørensen, M.J.; Schendel, D.; Parner, E.T.; Pedersen, L.H.; Vestergaard, M. Prenatal Valproate Exposure and Risk of Autism Spectrum Disorders and Childhood Autism. JAMA 2013, 309, 1696–1703.
- Massarali, A.; Adhya, D.; Srivastava, D.P.; Baron-Cohen, S.; Kotter, M.R. Virus-Induced Maternal Immune Activation as an Environmental Factor in the Etiology of Autism and Schizophrenia. Front. Neurosci. 2022, 16, 834058.
- Behl, S.; Mehta, S.; Pandey, M.K. Abnormal Levels of Metal Micronutrients and Autism Spectrum Disorder: A Perspective Review. Front. Mol. Neurosci. 2020, 13, 586209.
- Veiby, G.; Daltveit, A.K.; Schjølberg, S.; Stoltenberg, C.; Øyen, A.-S.; Vollset, S.E.; Engelsen, B.A.; Gilhus, N.E. Exposure to antiepileptic drugs in utero and child development: A prospective population-based study. Epilepsia 2013, 54, 1462–1472.
- Jokiranta, E.; Sourander, A.; Suominen, A.; Timonen-Soivio, L.; Brown, A.S.; Sillanpää, M. Epilepsy Among Children and Adolescents with Autism Spectrum Disorders: A Population-Based Study. J. Autism Dev. Disord. 2014, 44, 2547–2557.
- Carter, S.A.; Lin, J.C.; Chow, T.; Yu, X.; Rahman, M.; Martinez, M.P.; Feldman, K.; Eckel, S.P.; Chen, J.-C.; Chen, Z.; et al. Maternal obesity, diabetes, preeclampsia, and asthma during pregnancy and likelihood of autism spectrum disorder with gastrointestinal disturbances in offspring. Autism 2023, 27, 916–926.
- Williams, B.L.; Hornig, M.; Buie, T.; Bauman, M.L.; Paik, M.C.; Wick, I.; Bennett, A.; Jabado, O.; Hirschberg, D.L.; Lipkin, W.I. Impaired Carbohydrate Digestion and Transport and Mucosal Dysbiosis in the Intestines of Children with Autism and Gastrointestinal Disturbances. PLoS ONE 2011, 6, e24585.
More
Information
Subjects:
Neurosciences
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.1K
Revisions:
2 times
(View History)
Update Date:
05 Jul 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





