Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jacek Z. Kubiak | -- | 2922 | 2023-07-04 11:43:34 | | | |
| 2 | Sirius Huang | Meta information modification | 2922 | 2023-07-05 02:41:31 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
El Dika, M.; Dudka, D.; Kloc, M.; Kubiak, J.Z. CDC6 as Key Inhibitory Regulator of CDK1 Activation. Encyclopedia. Available online: https://encyclopedia.pub/entry/46391 (accessed on 07 February 2026).
El Dika M, Dudka D, Kloc M, Kubiak JZ. CDC6 as Key Inhibitory Regulator of CDK1 Activation. Encyclopedia. Available at: https://encyclopedia.pub/entry/46391. Accessed February 07, 2026.
El Dika, Mohammed, Damian Dudka, Malgorzata Kloc, Jacek Z. Kubiak. "CDC6 as Key Inhibitory Regulator of CDK1 Activation" Encyclopedia, https://encyclopedia.pub/entry/46391 (accessed February 07, 2026).
El Dika, M., Dudka, D., Kloc, M., & Kubiak, J.Z. (2023, July 04). CDC6 as Key Inhibitory Regulator of CDK1 Activation. In Encyclopedia. https://encyclopedia.pub/entry/46391
El Dika, Mohammed, et al. "CDC6 as Key Inhibitory Regulator of CDK1 Activation." Encyclopedia. Web. 04 July, 2023.
Copy Citation
The kinetics of Cyclin Dependent Kinase 1 (CDK1) activation must be strictly controlled to guarantee a timely and physiological entry into mitosis. CDC6, a known S-phase regulator, has been found as a critical component in mitotic CDK1 activation cascade in early embryonic divisions. It acts due to association with Xic1 serving as a bona fide CDK1 inhibitor upstream of Aurora A and Polo-Like Kinase 1 (PLK1), both of which are CDK1 activators.
time of mitosis
cell cycle
CDC6
CDK1
cyclins
CDC25
Xic1
mitotic entry
1. Introduction
The entry into mitosis is controlled by CDK1/Cyclin B, also known as MPF (maturation promoting factor). Previous research has shown that cyclin B synthesis is the key for driving the embryonic cell cycle and determining the timing of mitosis in Xenopus leavis [1][2][3]. Apparently, it is a key, but it is not the only controlling factor. Cyclin B accumulation during interphase is necessary for CDK1 activation because without cyclin, CDK1 cannot be active as a protein kinase. On the other hand, cyclin degradation through the ubiquitin pathway is necessary for CDK1 inactivation and mitotic exit [1][2][3]. Cyclin B is encoded by several genes and creates a family of B-type cyclins required for mitosis (e.g., cyclins B1, B2, and B3 in human; or cyclins B1-B5 in Xenopus leavis [4][5]). De novo B-type cyclin synthesis is required between meiosis I and II during Xenopus oocyte maturation [4]; however, despite a steady increase in cyclin B levels in G2 [6], CDK1 activation is biphasic, characterized by a slow phase, followed by a rapid phase attributed to the positive feedback between newly activated CDK1 and its major activating phosphatase CDC25 [6]. How the stable increase in cyclin B level may lead to this biphasic CDK1 activation before the CDK1/CDC25 positive feedback acceleration triggering the final peak of CDK1 activity has been intriguing for the long time. This particularity suggested that the control of CDK1 activation at the initial stages of its mitotic activation might not solely depend on cyclin B accumulation but might also involve an unidentified inhibitor that could counterbalance CDK1 activation, assuring the slow and biphasic mode of this process. A research group performed a proteomic screen to find novel CDK1 partners in M-phase-arrested Xenopus leavis eggs vs. freshly activated ones (5 min post-activation [7]). The researchers showed a rapid 5 min, change in the composition of the CDK1 complex during the period between the MII arrest of oocytes and their activation for development; however, researchers did not find any classical CDK1 inhibitors, such as INK4 or Cip/Kip, in this screen, which potentially could slow down CDK1 activation [8] in a CDK1 complex. Surprisingly, researchers found a CDC6 protein associated with the active mitotic CDK. CDC6 inhibits CDK1 throughout the whole period of the M-phase (including the highest peak of CDK1 activity just prior its inactivation) [9][10][11][12][13][14]. CDC6 is an evolutionarily conserved member of the AAA + ATPase family and was only known as the S-phase activator [6]. It initiates DNA replication at each origin once per cell cycle to maintain genomic stability and is regulated during the S-phase by another member of the CDK family, namely CDK2. The CDK2/cyclin A-mediated phosphorylation of CDC6 causes its translocation from the nucleus to the cytoplasm, blocking DNA replication [15]. CDK1 can additionally phosphorylate CDC6 to maintain it in the cytoplasm (ibid.). Furthermore, researchers showed that two proteins, namely CDC6 and Xic1 (the last one is a bona fide CDK1 inhibitor identified for its role in later stages of Xenopus embryo development), interact within the mitotic CDK1/CDC6 complex. Upon arriving at the peak of the CDK1 activity (measured by histone H1 kinase activity), the Xic1 momentarily dissociates from the mitotic CDK1 allowing for the maximum activation of CDK1 during the M-phase [9]. It is critical to understand how CDC6 and Xic1 receive signals, how they interact, and how they are interdependent; however, the identification of this new inhibitory mechanism towards CDK1 was surprising because it has been well known already for years that CDK1 is inhibited by the posttranslational modification by Wee1/Myt1 kinases. Thus, data showed that there are two independent mechanisms of CDK1 activity in the game during the mitotic entry.
2. CDC6 as an Upstream Regulator of CDK1 through Its Inhibition
CDC6 is essential for the folding, unfolding, and degradation of proteins. Its 200–250 amino acid ATPase region is necessary for the assembly of pre-RCs. More specifically, CDC6 attaches to ORI (ORC-attached replication origins) on chromosomes to generate an ORI-CDC6-ORC complex, which then binds Cdt1 to recruit several MCM2-7 helicase subunits to ORI. This is powered by its ATP-hydrolase activity [16][17][18][19]. Furthermore, CDC6 has a phosphorylation site for PLK1 in its N-terminal side, and three consensus sites of phosphorylation for CDK1 and CDK2 [20]. CDC6 has a leucine zipper domain for protein–protein interaction and Cy-motif for cyclin interaction (ibid.). It also has amino-acid sequences of D- and KEN-box motifs which are necessary for ubiquitination by the anaphase-promoting complex/cyclosome (APC/C), the main mitotic ubiquitin ligase, which participates in CDC6 degradation by the proteasome in the G1/G0 phase (ibid.). It was shown that in yeast, CDC6 acts as a CDK1 inhibitor during the mitotic exit and that it inhibits CDK1-dependent histone H1 phosphorylation in vitro [21]. Moreover, together with SIC1 (Stoichiometric inhibitor of Cdk1-Clb) (an inhibitor of CDK1) and CDH1 (Cdc20 homolog 1), an activator of the APC (anaphase promoting complex), CDC6 participates in the activation of APC necessary for cyclin B degradation and mitotic exit [22]. Both SIC1 and CDC6 have been shown to associate with CDK1/cyclin B complexes with low nanomolar affinity, which is partially facilitated by the docking of the phospho-adaptor CKS1 [23][24]. The deletion of the CDC6 N-terminal CDK1-binding site increases CDK1 activity in mitosis, indicating that the CDC6 N-terminus is critical for the regulation of CDK1 activity [22]. The depletion of CDC6 causes a delay in mitotic exit. This finding was further supported by an in vitro assay showing the CDC6-dependent inhibition of CDK1 kinase activity mediated by a CDK-specific cyclin docking motif, LxF, in CDC6 and the phospho-adaptor Cks1, leading to shielding of the degron and CDC6 sequestration by CDK during mitotic exit [23].
In human cells, CDC6 also associates with and inhibits CDK1 at mitotic exit [25]. In mitosis, CDC6 is hyperphosphorylated in correlation with an increase in the level of PLK1; however, CDC6 is hypophosphorylated in PLK1-depleted cells, even when cyclins A and B are present at high levels. PLK1 phosphorylates CDC6 during mitosis, and this promotes its binding to CDK1, which in turn downregulates CDK1 activity. This results in the activation of separase, an enzyme that resolves sister chromatid cohesion during the metaphase-to-anaphase transition, leading to the mitotic exit. CDC6 depletion results in defects in chromosome segregation and cytokinesis, leading to aneuploid cells with increased CDK1 activity [25].
In addition to its role in mitotic exit in yeast and human cells [21][26][27], researchers also showed the role of CDC6 in delaying mitotic entry by inhibiting CDK1 activity in Xenopus laevis premitotic cell-free extracts [9][10][11][12][13][14]. CDC6 associated with CDK1 during the M-phase rapidly dissociates from this kinase immediately after CDK1 inactivation, suggesting that once CDK1 is fully inactivated by separation from cyclin B, the association with CDC6 becomes unnecessary. It was a surprising observation in the light of the knowledge at that time that there are suggestions of a potential role of CDC6 only in the mitotic exit, and not entry and progression [9]. The important hint to the role of CDC6 in CDK1 activation process was its abovementioned function in the M-phase exit described in yeast and human cells. Through biochemical analysis of Xenopus embryo cell-free extracts researchers showed that a recombinant CDC6 protein acts as an inhibitor of CDK1 during the first embryonic mitosis in Xenopus leavis [9][10][11][12][13][14]. Importantly, when endogenous CDC6 is depleted, the first, initial and slow phase of CDK1 activation is removed, and the kinase activation ceases to be biphasic. This, in turn, accelerates the timing of mitosis and the pic of CDK1 activity induced by the CDC25/CDK1 activation loop [9][10][11][12][13][14]. Adding a recombinant CDC6 completely reverses all these effects [14][26][28]. Additionally, exogenous cyclin A or cyclin B added to Xenopus laevis cycling extracts leads to distinct regulation of the kinetics of this mitotic kinase activity [9]. Researchers found that while adding cyclin A increases only the level of CDK1 activity, adding cyclin B increases the level of CDK1 activity and prolongs the timing of its activation. On the other hand, the absence of cyclin A causes an important delay in CDK1 activation. While adding recombinant CDC6 to cyclin A-depleted extract does not affect the timing of the delayed mitosis; however, it does inhibit the CDK1 activity. Thus, it demonstrates that the CDC6-dependent mechanism inhibits both CDK1/cyclin A and CDK1/cyclin B (ibid). This implies in turn that a CDC6-dependent mechanism of CDK1 inhibition plays a role in also delaying the initial activation of CDK1/cyclin A.
In addition to this inhibitory function of CDC6 regarding CDK1 activity, the switch from cyclin A- to cyclin B-dependent CDK1 activity at the beginning of the M-phase may also contribute to the biphasic CDK1 activation pattern during the M-phase entry. Mathematical model based on mass action kinetics shows a diauxic character of the CDK1 activity growth and allows for the simulation of changes in the dynamics of CDK1 activation in relation to changing concentrations in CDC6 and the dynamics of CDK1/cyclin B/CDC6 complexes formation [13]. The diauxic character of the process of CDK1 activation is strictly related to the presence of at least three inflection points in the curve of CDK1 activation. The two-step mode of CDK1 activation in Xenopus laevis cell-free extract resembles the dynamics of the diauxic growth of bacterial or yeast population upon the switch from one sugar to another one when the first becomes exhausted in the culture medium [13][29]. It suggested an analogic change concerning CDK1 activity, and the switch between cyclin A and cyclin B as a partner of CDK1 during the initial phases of CDK1 activation fits perfectly with this view.
In mouse zygotes, the depletion of CDC6 also causes acceleration of the entry into mitosis, similarly to the Xenopus cell-free extract. It showed the physiological relevance of CDC6 not only in the cell-free system but also in intact cells [11][14]. In addition, CDC6 regulates both meiotic entry and the metaphase-to-anaphase transition during the first meiotic division in mouse oocytes [30].
Taken together, these results show that CDC6 is involved in a CDK1 inhibitory mechanism both in vitro and in vivo during mitotic (and meiotic) entry, progression and exit.
3. Regulation of CDC6 by PP2A
The serine/threonine phosphatase PP2A is the major protein phosphatase involved in dephosphorylating CDK substrates [31]. This PP2A function is well-conserved across eukaryotes, including yeast and humans [32][33]. The majority of CDK1 substrates are dephosphorylated by PP2A [33][34][35][36][37]. Inhibiting PP2A using okadaic acid triggers premature CDK1 activation in human cells and those of Xenopus, mouse, starfish, and other organisms [38][39][40][41][42][43][44].
In yeast, the PP2A heterotrimeric complex is composed of two catalytic C subunits (Pph21 and Pph22), a regulatory B subunit (Cdc55, Rts1, or Rts3), and a scaffold A subunit (Tpd3) [45][46][47]. Among these subunits, the regulatory B subunit is responsible both for the cellular localization and PP2A substrate specificity [48]. Cdc55 (also known as B55 in mammals) and Rts1 have both been involved in mitotic progression. Cytoplasmic PP2A/Cdc55 dephosphorylates and inhibits the CDK1 inhibitor Swe1 [49][50], while nuclear PP2A/Cdc55 prevents the anaphase onset by dephosphorylating Cdc20, a co-factor of APC/C [51][52][53][54]. Furthermore, PP2A/Cdc55 also dephosphorylates Net1, an inhibitor of CDC14 phosphatase [55]. CDC14 release from the nucleolus during early anaphase is mediated by the FEAR and MEN networks and results in mitotic exit through Net1 dephosphorylation [56][57][58][59].
Recently, it has been demonstrated that CDC6 is dephosphorylated by PP2A/Cdc14phosphatases in yeast, which triggers the mitotic exit. PP2A/Cdc55 and CDC14 directly dephosphorylate CDC6 at different CDK1 sites, causing CDC6 stabilization [60][61]. These findings support a scenario in which CDC6 dephosphorylation is required to abolish the inhibition of CDK1 at the mitotic exit.
4. Role of PP2A-Greatwall/Mastl Pathway in Potential CDC6 Regulation
Greatwall/Mastl kinase is a conserved regulator of mitotic entry through PP2A phosphatase regulation [62]. There was no evidence from previous studies that PP2A-Greatwall/Mastl was part of a regulatory network during the initial stages of mitotic entry; however, it is well established now that Greatwall/Mastl kinase triggers the activation of CDK1 and other cell cycle regulators preparing the cells for mitosis [62][63][64][65][66][67][68]. Upon activation, Greatwall/Mastl takes part in the amplification cycle downstream of CDK1/Cyclin [69]. It has been shown that Greatwall/Mastl controls PP2A activity in Xenopus egg extracts [33][34][64]. Greatwall/Mastl phosphorylates the thermostable protein Arpp-19 (cAMP-regulated phosphoprotein-19) and endosulfine protein (Ensa) to inhibit the activity of PP2A-B55 and triggers mitotic entry [33][62][68].
These findings suggest an interaction between CDC6-dependent machinery and Greatwall/Mastl kinase activity. Does CDC6 inhibit Greatwall/Mastl and its substrates to delay mitosis, or is CDC6 involved in the activation of PP2A directly? Addressing these questions is of great relevance to highlight the CDC6 regulatory mechanism necessary for mitotic regulation.
5. A Model of CDC6 Involvement in CDK1 Activation
Beyond a simple mechanism of accumulation and degradation of cyclins A and B, the control of cell division is orchestrated by a complex network of regulators that include kinases and phosphatases, as well as inhibitory partners. They are required to precisely regulate CDK1 levels determining the timing of mitotic events and the timely progression of mitosis. CDC6 participates in the inhibitory mechanisms controlling CDK1/cyclin A and CDK1/cyclin B activities and regulates the time of mitosis in Xenopus laevis and mouse embryos [9][11]. This is corroborated by the data obtained in yeast and human cells [21][22][23][24][25]. This regulatory mechanism is particularly important for the coordinated activation of mitotic CDK1 and the control of both the timing and amplitude of CDK1 kinase activity during early embryo cleavage divisions, which are highly synchronous in Xenopus; therefore, this regulation is required for the coordination between cell divisions and the embryo genetic developmental program.
According to recent research [70][71][72][73], CDK1/cyclin A-B phosphorylates Bora to induce Aurora A’s phosphorylation of PLK1 to fully activate the CDK1/cyclin B amplification loop. This shows that both CDC6 and Aurora-Bora networks are crucial components of the CDK1/Cyclin B amplification mechanism and, consequently, of the control of the timing of mitosis. Researchers present a new model for the entry into mitosis, integrating CDK1, CDC6/Xic1, Wee1/Myt1 and Aurora A-Bora-PLK1 network that, together, regulate the activation of the CDK1 amplification loop (Figure 1).
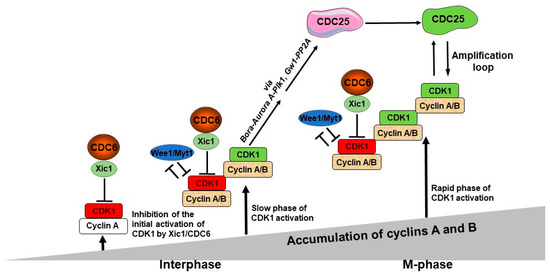
Figure 1. A model of the role of CDC6 in the control of the M-phase entry. The first, potentially active CDK1 molecules start to appear after the cyclin A level reaches a predetermined threshold, but they are actively inhibited by CDC6/Xic1. The first active CDK1/cyclin A molecules appear when they escape from CDC6-dependent inhibition, while CDK1/cyclin B complexes are inhibited by both CDC6/Xic1- and Wee1/Myt1-dependent mechanisms. This is the step of the initial activation of CDK1. In parallel, both cyclin A and B levels continue to increase with time and keep associating with CDK1. The active CDK1/cyclin A and CDK1/cyclin B complexes become more abundant, and they, or only CDK1/cyclin B, become able to phosphorylate its substrates—Bora and Gw1. Due to this activation of Plk1 via these two distinct pathways, the procedure is highly effective, which in turn promotes CDC25 activity acting as the CDK1 amplification loop.
During interphase, cyclin A accumulates and binds to CDK1. Judging by the experiments [9], researchers postulate that the initial pool of CDK1/cyclin A, which escapes from Wee1/Myt1 inhibition, is under an inhibitory control of the CDC6-dependent mechanism. Thus, CDK1/cyclin A complex is inhibited only by CDC6 and not by Wee1/Myt1 kinases, in contrast to the CDK1/cyclin B complex. Thus, the first active CDK1/cyclin A molecules appear when they escape from CDC6-dependent inhibition, while CDK1/cyclin B complexes are inhibited by both CDC6/Xic1 and Wee1/Myt1. This single inhibition of the CDK1/cyclin A pool and the double inhibition of the CDK1/cyclin B pool shows the major role of CDC6 in the control of the timing of this event. The levels of cyclin A and B continue to rise throughout time and newly synthesized cyclin molecules become progressively associated with CDK1. The active CDK1/Cyclin A and the CDK1/Cyclin B become more numerous, and they can phosphorylate their substrates, such as Bora and Greatwall/Mastl, also known as Gw1 in Xenopus. During this phase, CDK1 activity is in its slow phase of increase. Bora interacts with PLK1 and Aurora kinase A, causing PLK1 to be activated. While part of the amplification loop, PLK1 also allows for the controlled activation of CDK1 by promoting CDC6-mediated inhibition of CDK1, leading to a gradual CDK1 activity increase [11][25]. Gw1 phosphorylation protects CDC25 from dephosphorylating, enabling it to remain active and effectively activate CDK1 activity. During that period, cyclins accumulate concurrently and associate with CDK1 to produce new active CDK1 molecules. This results in a constant increase in CDK1 activity level, overcoming the CDC6/Xic1-inhibition mechanism. At that time, CDK1 activation enters the rapid phase of activation. When this occurs, CDK1 action becomes efficient in activating CDC25, and CDC25 phosphorylation continues to be further activated over time. At that point, the CDC25 and CDK1 amplification loop starts functioning efficiently, which causes the extremely rapid and massive CDK1 activation; therefore, the full activation of the CDK1 amplification loop occurs, and mitosis progresses.
References
- Murray, A.W.; Kirschner, M.W. Cyclin Synthesis Drives the Early Embryonic Cell Cycle. Nature 1989, 339, 275–280.
- Murray, A.W.; Solomon, M.J.; Kirschner, M.W. The Role of Cyclin Synthesis and Degradation in the Control of Maturation Promoting Factor Activity. Nature 1989, 339, 280–286.
- Glotzer, M.; Murray, A.W.; Kirschner, M.W. Cyclin Is Degraded by the Ubiquitin Pathway. Nature 1991, 349, 132–138.
- Hochegger, H.; Klotzbücher, A.; Kirk, J.; Howell, M.; le Guellec, K.; Fletcher, K.; Duncan, T.; Sohail, M.; Hunt, T. New B-Type Cyclin Synthesis Is Required between Meiosis I and II during Xenopus Oocyte Maturation. Development 2001, 128, 3795–3807.
- Gong, D.; Ferrell, J.E. The Roles of Cyclin A2, B1, and B2 in Early and Late Mitotic Events. Mol. Biol. Cell 2010, 21, 3149–3161.
- Pomerening, J.R.; Kim, S.Y.; Ferrell, J.E. Systems-Level Dissection of the Cell-Cycle Oscillator: Bypassing Positive Feedback Produces Damped Oscillations. Cell 2005, 122, 565–578.
- Marteil, G.; Gagné, J.P.; Borsuk, E.; Richard-Parpaillon, L.; Poirier, G.G.; Kubiak, J.Z. Proteomics Reveals a Switch in CDK1-Associated Proteins upon M-Phase Exit during the Xenopus Laevis Oocyte to Embryo Transition. Int. J. Biochem. Cell Biol. 2012, 44, 53–64.
- Sherr, C.J.; Roberts, J.M. CDK Inhibitors: Positive and Negative Regulators of G1-Phase Progression. Genes Dev. 1999, 13, 1501–1512.
- El Dika, M.; Wechselberger, L.; Djeghout, B.; Benouareth, D.E.; Jęderka, K.; Lewicki, S.; Zdanowski, R.; Prigent, C.; Kloc, M.; Kubiak, J.Z. Mitotic Timing Is Differentially Controlled by A-and B-Type Cyclins and by CDC6 Associated with a Bona Fide CDK Inhibitor Xic1 in Xenopus Laevis Cell-Free Extract. Int. J. Dev. Biol. 2021, 65, 487–496.
- El Dika, M. Régulation de La Phase M Du Cycle Cellulaire Par CDK1, PP2A et CDC6. 1. Ph.D. Thesis, University of Rennes 1, Rennes, France, 2013.
- El Dika, M.; Laskowska-Kaszub, K.; Koryto, M.; Dudka, D.; Prigent, C.; Tassan, J.P.; Kloc, M.; Polanski, Z.; Borsuk, E.; Kubiak, J.Z. CDC6 Controls Dynamics of the First Embryonic M-Phase Entry and Progression via CDK1 Inhibition. Dev. Biol. 2014, 396, 67–80.
- Debowski, M.; El Dika, M.; Malejczyk, J.; Zdanowski, R.; Prigent, C.; Tassan, J.P.; Kloc, M.; Lachowicz, M.; Kubiak, J.Z. Flexibility vs. Robustness in Cell Cycle Regulation of Timing of M-Phase Entry in Xenopus Laevis Embryo Cell-Free Extract. Int. J. Dev. Biol. 2016, 60, 305–314.
- Dębowski, M.; Szymańska, Z.; Kubiak, J.Z.; Lachowicz, M. Mathematical Model Explaining the Role of CDC6 in the Diauxic Growth of CDK1 Activity during the M-Phase of the Cell Cycle. Cells 2019, 8, 1537.
- Borsuk, E.; Jachowicz, J.; Kloc, M.; Tassan, J.P.; Kubiak, J.Z. Role of Cdc6 during Oogenesis and Early Embryo Development in Mouse and Xenopus Laevis. In Results and Problems in Cell Differentiation; Springer: Berlin/Heidelberg, Germany, 2017; Volume 59, pp. 201–211.
- Delmolino, L.M.; Saha, P.; Dutta, A. Multiple Mechanisms Regulate Subcellular Localization of Human CDC6. J. Biol. Chem. 2001, 276, 26947–26954.
- Edwards, M.C.; Tutter, A.V.; Cvetic, C.; Gilbert, C.H.; Prokhorova, T.A.; Walter, J.C. MCM2-7 Complexes Bind Chromatin in a Distributed Pattern Surrounding the Origin Recognition Complex in Xenopus Egg Extracts. J. Biol. Chem. 2002, 277, 33049–33057.
- Randell, J.C.W.; Bowers, J.L.; Rodríguez, H.K.; Bell, S.P. Sequential ATP Hydrolysis by Cdc6 and ORC Directs Loading of the Mcm2-7 Helicase. Mol. Cell 2006, 21, 29–39.
- Speck, C.; Chen, Z.; Li, H.; Stillman, B. ATPase-Dependent Cooperative Binding of ORC and Cdc6 to Origin DNA. Nat. Struct. Mol. Biol. 2005, 12, 965–971.
- Weinreich, M.; Liang, C.; Stillman, B. The Cdc6p Nucleotide-Binding Motif Is Required for Loading Mcm Proteins onto Chromatin. Proc. Natl. Acad. Sci. USA 1999, 96, 441–446.
- Petersen, B.O.; Lukas, J.; Sørensen, C.S.; Bartek, J.; Helin, K. Phosphorylation of Mammalian CDC6 by Cyclin A/CDK2 Regulates Its Subcellular Localization. EMBO J. 1999, 18, 396–410.
- Elsasser, S.; Lou, F.; Wang, B.; Campbell, J.L.; Jong, A. Interaction between Yeast Cdc6 Protein and B-Type Cyclin/Cdc28 Kinases. Mol. Biol. Cell 1996, 7, 1723–1735.
- Calzada, A.; Sacristán, M.; Sánchez, E.; Bueno, A. Cdc6 Cooperates with Sic1 and Hct1 to Inactivate Mitotic Cyclin-Dependent Kinases. Nature 2001, 412, 355–358.
- Örd, M.; Venta, R.; Möll, K.; Valk, E.; Loog, M. Cyclin-Specific Docking Mechanisms Reveal the Complexity of M-CDK Function in the Cell Cycle. Mol. Cell 2019, 75, 76–89.e3.
- Venta, R.; Valk, E.; Örd, M.; Košik, O.; Pääbo, K.; Maljavin, A.; Kivi, R.; Faustova, I.; Shtaida, N.; Lepiku, M.; et al. A Processive Phosphorylation Circuit with Multiple Kinase Inputs and Mutually Diversional Routes Controls G1/S Decision. Nat. Commun. 2020, 11, 1836.
- Yim, H.; Erikson, R.L. Cell Division Cycle 6, a Mitotic Substrate of Polo-like Kinase 1, Regulates Chromosomal Segregation Mediated by Cyclin-Dependent Kinase 1 and Separase. Proc. Natl. Acad. Sci. USA 2010, 107, 19742–19747.
- Duderstadt, K.E.; Berger, J.M. AAA+ ATPases in the Initiation of DNA Replication. Crit. Rev. Biochem. Mol. Biol. 2008, 43, 163–187.
- Archambault, V.; Li, C.X.; Tackett, A.J.; Wasch, R.; Chait, B.T.; Rout, M.P.; Cross, F.R. Genetic and Biochemical Evaluation of the Importance of Cdc6 in Regulating Mitotic Exit. Mol. Biol. Cell 2003, 14, 4592–4604.
- Parrilla, A.; Barber, M.; Majem, B.; Castellví, J.; Morote, J.; Sánchez, J.L.; Pérez-Benavente, A.; Segura, M.F.; Gil-Moreno, A.; Santamaria, A. Aurora Borealis (Bora), Which Promotes Plk1 Activation by Aurora A, Has an Oncogenic Role in Ovarian Cancer. Cancers 2020, 12, 886.
- Monod, J. The Growth of Bacterial Cultures. Annu. Rev. Microbiol. 1949, 3, 371–394.
- Yi, Z.; Meng, T.; Ma, X.; Li, J.; Zhang, C.; Ouyang, Y.; Schatten, H.; Qiao, J.; Sun, Q.; Qian, W. CDC6 Regulates Both G2/M Transition and Metaphase-to-anaphase Transition during the First Meiosis of Mouse Oocytes. J. Cell. Physiol. 2020, 235, 5541–5554.
- Virshup, D.M.; Shenolikar, S. From Promiscuity to Precision: Protein Phosphatases Get a Makeover. Mol. Cell 2009, 33, 537–545.
- Janssens, V.; Longin, S.; Goris, J. PP2A Holoenzyme Assembly: In Cauda Venenum (the Sting Is in the Tail). Trends Biochem. Sci. 2008, 33, 113–121.
- Mochida, S.; Ikeo, S.; Gannon, J.; Hunt, T. Regulated Activity of PP2A-B55 Delta Is Crucial for Controlling Entry into and Exit from Mitosis in Xenopus Egg Extracts. EMBO J. 2009, 28, 2777–2785.
- Castilho, P.V.; Williams, B.C.; Mochida, S.; Zhao, Y.; Goldberg, M.L. The M Phase Kinase Greatwall (Gwl) Promotes Inactivation of PP2A/B55delta, a Phosphatase Directed against CDK Phosphosites. Mol. Biol. Cell 2009, 20, 4777–4789.
- Ferrigno, P.; Langan, T.A.; Cohen, P. Protein Phosphatase 2A1 Is the Major Enzyme in Vertebrate Cell Extracts That Dephosphorylates Several Physiological Substrates for Cyclin-Dependent Protein Kinases. Mol. Biol. Cell 1993, 4, 669–677.
- Mayer-Jaekel, R.E.; Ohkura, H.; Gomes, R.; Sunkel, C.E.; Baumgartner, S.; Hemmings, B.A.; Glover, D.M. The 55 Kd Regulatory Subunit of Drosophila Protein Phosphatase 2A Is Required for Anaphase. Cell 1993, 72, 621–633.
- Mayer-Jaekel, R.E.; Ohkura, H.; Ferrigno, P.; Andjelkovic, N.; Shiomi, K.; Uemura, T.; Glover, D.M.; Hemmings, B.A. Drosophila Mutants in the 55 KDa Regulatory Subunit of Protein Phosphatase 2A Show Strongly Reduced Ability to Dephosphorylate Substrates of P34cdc2. J. Cell Sci. 1994, 107 Pt 9, 2609–2616.
- Goris, J.; Hermann, J.; Hendrix, P.; Ozon, R.; Merlevede, W. Okadaic Acid, a Specific Protein Phosphatase Inhibitor, Induces Maturation and MPF Formation in Xenopus Laevis Oocytes. Febs Lett. 1989, 245, 91–94.
- Karaïskou, A.; Cayla, X.; Haccard, O.; Jessus, C.; Ozon, R. MPF Amplification in Xenopus Oocyte Extracts Depends on a Two-Step Activation of Cdc25 Phosphatase. Exp. Cell Res. 1998, 244, 491–500.
- Rime, H.; Jessus, C.; Ozon, R. Estramustine Phosphate Inhibits Germinal Vesicle Breakdown and Induces Depolymerization of Microtubules in Mouse Oocyte. Reprod. Nutr. Dev. (1980) 1988, 28, 319–334.
- Picard, A.; Labbé, J.C.; Barakat, H.; Cavadore, J.C.; Dorée, M. Okadaic Acid Mimics a Nuclear Component Required for Cyclin B-Cdc2 Kinase Microinjection to Drive Starfish Oocytes into M Phase. J. Cell Biol. 1991, 115, 337–344.
- Clarke, P.R.; Hoffmann, I.; Draetta, G.; Karsenti, E. Dephosphorylation of Cdc25-C by a Type-2A Protein Phosphatase: Specific Regulation during the Cell Cycle in Xenopus Egg Extracts. Mol. Biol. Cell 1993, 4, 397–411.
- Maton, G.; Lorca, T.; Girault, J.-A.; Ozon, R.; Jessus, C. Differential Regulation of Cdc2 and Aurora-A in Xenopus Oocytes: A Crucial Role of Phosphatase 2A. J. Cell Sci. 2005, 118, 2485–2494.
- Félix, M.A.; Cohen, P.; Karsenti, E. Cdc2 H1 Kinase Is Negatively Regulated by a Type 2A Phosphatase in the Xenopus Early Embryonic Cell Cycle: Evidence from the Effects of Okadaic Acid. EMBO J. 1990, 9, 675–683.
- Healy, A.M.; Zolnierowicz, S.; Stapleton, A.E.; Goebl, M.; DePaoli-Roach, A.A.; Pringle, J.R. CDC55, a Saccharomyces Cerevisiae Gene Involved in Cellular Morphogenesis: Identification, Characterization, and Homology to the B Subunit of Mammalian Type 2A Protein Phosphatase. Mol. Cell. Biol. 1991, 11, 5767–5780.
- van Zyl, W.; Huang, W.; Sneddon, A.A.; Stark, M.; Camier, S.; Werner, M.; Marck, C.; Sentenac, A.; Broach, J.R. Inactivation of the Protein Phosphatase 2A Regulatory Subunit A Results in Morphological and Transcriptional Defects in Saccharomyces Cerevisiae. Mol. Cell. Biol. 1992, 12, 4946–4959.
- Shu, Y.; Yang, H.; Hallberg, E.; Hallberg, R. Molecular Genetic Analysis of Rts1p, a B’ Regulatory Subunit of Saccharomyces Cerevisiae Protein Phosphatase 2A. Mol. Cell. Biol. 1997, 17, 3242–3253.
- Rossio, V.; Yoshida, S. Spatial Regulation of Cdc55-PP2A by Zds1/Zds2 Controls Mitotic Entry and Mitotic Exit in Budding Yeast. J. Cell Biol 2011, 193, 445–454.
- Lin, F.; Arndt, K.T. The Role of Saccharomyces Cerevisiae Type 2A Phosphatase in the Actin Cytoskeleton and in Entry into Mitosis. EMBO J. 1995, 14, 2745–2759.
- Yang, H.; Jiang, W.; Gentry, M.; Hallberg, R.L. Loss of a Protein Phosphatase 2A Regulatory Subunit (Cdc55p) Elicits Improper Regulation of Swe1p Degradation. Mol. Cell. Biol. 2000, 20, 8143–8156.
- Rossio, V.; Michimoto, T.; Sasaki, T.; Ohbayashi, I.; Kikuchi, Y.; Yoshida, S. Nuclear PP2A-Cdc55 Prevents APC-Cdc20 Activation during the Spindle Assembly Checkpoint. J. Cell Sci. 2013, 126, 4396–4405.
- Minshull, J.; Straight, A.; Rudner, A.D.; Dernburg, A.F.; Belmont, A.; Murray, A.W. Protein Phosphatase 2A Regulates MPF Activity and Sister Chromatid Cohesion in Budding Yeast. Curr. Biol. 1996, 6, 1609–1620.
- Wang, Y.; Burke, D.J. Cdc55p, the B-Type Regulatory Subunit of Protein Phosphatase 2A, Has Multiple Functions in Mitosis and Is Required for the Kinetochore/Spindle Checkpoint in Saccharomyces Cerevisiae. Mol. Cell. Biol. 1997, 17, 620–626.
- Lianga, N.; Williams, E.C.; Kennedy, E.K.; Doré, C.; Pilon, S.; Girard, S.L.; Deneault, J.-S.; Rudner, A.D. A Wee1 Checkpoint Inhibits Anaphase Onset. J. Cell Biol. 2013, 201, 843–862.
- Queralt, E.; Lehane, C.; Novak, B.; Uhlmann, F. Downregulation of PP2A(Cdc55) Phosphatase by Separase Initiates Mitotic Exit in Budding Yeast. Cell 2006, 125, 719–732.
- Visintin, R.; Hwang, E.S.; Amon, A. Cfi1 Prevents Premature Exit from Mitosis by Anchoring Cdc14 Phosphatase in the Nucleolus. Nature 1999, 398, 818–823.
- Azzam, R.; Chen, S.L.; Shou, W.; Mah, A.S.; Alexandru, G.; Nasmyth, K.; Annan, R.S.; Carr, S.A.; Deshaies, R.J. Phosphorylation by Cyclin B-Cdk Underlies Release of Mitotic Exit Activator Cdc14 from the Nucleolus. Science 2004, 305, 516–519.
- Shou, W.; Seol, J.H.; Shevchenko, A.; Baskerville, C.; Moazed, D.; Chen, Z.W.; Jang, J.; Shevchenko, A.; Charbonneau, H.; Deshaies, R.J. Exit from Mitosis Is Triggered by Tem1-Dependent Release of the Protein Phosphatase Cdc14 from Nucleolar RENT Complex. Cell 1999, 97, 233–244.
- Tomson, B.N.; Rahal, R.; Reiser, V.; Monje-Casas, F.; Mekhail, K.; Moazed, D.; Amon, A. Regulation of Spo12 Phosphorylation and Its Essential Role in the FEAR Network. Curr. Biol. 2009, 19, 449–460.
- Zhai, Y.; Yung, P.Y.K.; Huo, L.; Liang, C. Cdc14p Resets the Competency of Replication Licensing by Dephosphorylating Multiple Initiation Proteins during Mitotic Exit in Budding Yeast. J. Cell Sci. 2010, 123, 3933–3943.
- Philip, J.; Örd, M.; Silva, A.; Singh, S.; Diffley, J.F.; Remus, D.; Loog, M.; Ikui, A.E. Cdc6 Is Sequentially Regulated by PP2A-Cdc55, Cdc14, and Sic1 for Origin Licensing in S. Cerevisiae. Elife 2022, 11, e74437.
- Gharbi-Ayachi, A.; Labbe, J.-C.; Burgess, A.; Vigneron, S.; Strub, J.-M.; Brioudes, E.; Van-Dorsselaer, A.; Castro, A.; Lorca, T. The Substrate of Greatwall Kinase, Arpp19, Controls Mitosis by Inhibiting Protein Phosphatase 2A. Science 2010, 330, 1673–1677.
- Burgess, A.; Vigneron, S.; Brioudes, E.; Labbé, J.-C.; Lorca, T.; Castro, A. Loss of Human Greatwall Results in G2 Arrest and Multiple Mitotic Defects Due to Deregulation of the Cyclin B-Cdc2/PP2A Balance. Proc. Natl. Acad. Sci. USA 2010, 107, 12564–12569.
- Vigneron, S.; Brioudes, E.; Burgess, A.; Labbé, J.-C.; Lorca, T.; Castro, A. Greatwall Maintains Mitosis through Regulation of PP2A. EMBO J. 2009, 28, 2786–2793.
- Vigneron, S.; Robert, P.; Hached, K.; Sundermann, L.; Charrasse, S.; Labbé, J.-C.; Castro, A.; Lorca, T. The Master Greatwall Kinase, a Critical Regulator of Mitosis and Meiosis. Int. J. Dev. Biol. 2016, 60, 245–254.
- Dupré, A.-I.; Haccard, O.; Jessus, C. The Greatwall Kinase Is Dominant over PKA in Controlling the Antagonistic Function of ARPP19 in Xenopus Oocytes. Cell Cycle 2017, 16, 1440–1452.
- Ma, S.; Vigneron, S.; Robert, P.; Strub, J.M.; Cianferani, S.; Castro, A.; Lorca, T. Greatwall Dephosphorylation and Inactivation upon Mitotic Exit Is Triggered by PP1. J. Cell Sci. 2016, 129, 1329–1339.
- Hached, K.; Goguet, P.; Charrasse, S.; Vigneron, S.; Sacristan, M.P.; Lorca, T.; Castro, A. ENSA and ARPP19 Differentially Control Cell Cycle Progression and Development. J. Cell Biol. 2019, 218, 541–558.
- Yu, J.; Zhao, Y.; Li, Z.; Galas, S.; Goldberg, M.L. Greatwall Kinase Participates in the Cdc2 Autoregulatory Loop in Xenopus Egg Extracts. Mol. Cell 2006, 22, 83–91.
- Tavernier, N.; Thomas, Y.; Vigneron, S.; Maisonneuve, P.; Orlicky, S.; Mader, P.; Regmi, S.; Van Hove, L.; Levinson, N.; Gasmi-Seabrook, G. Bora Phosphorylation Substitutes in Trans for T-Loop Phosphorylation in Aurora A to Promote Mitotic Entry. Nat. Commun. 2021, 12, 1899.
- Vigneron, S.; Sundermann, L.; Labbé, J.-C.; Pintard, L.; Radulescu, O.; Castro, A.; Lorca, T. Cyclin A-Cdk1-Dependent Phosphorylation of Bora Is the Triggering Factor Promoting Mitotic Entry. Dev. Cell 2018, 45, 637–650.
- Joukov, V.; De Nicolo, A. Aurora-PLK1 Cascades as Key Signaling Modules in the Regulation of Mitosis. Sci. Signal. 2018, 11, eaar4195.
- Pillan, A.; Tavernier, N.; Pintard, L. . Med. Sci. 2022, 38, 345–347.
More
Information
Subjects:
Cell Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
529
Revisions:
2 times
(View History)
Update Date:
05 Jul 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




