
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | H. K. S. de Zoysa | -- | 2901 | 2023-06-28 12:08:49 | | | |
| 2 | Jessie Wu | + 1 word(s) | 2902 | 2023-06-29 05:12:52 | | |
Video Upload Options
Fungi comprise diverse taxa that are abundant in various environments. They have crucial ecological roles as decomposers, mutualists, and disease-causing agents. The use of high-throughput sequencing (HTS) and metagenomics in analyzing DNA from environmental samples has become crucial in the identification of new fungal lineages. In addition, the development of other omics technologies interrogating different cellular components and the use of these technologies in various polyphasic methods have significantly advanced research in areas such as biodiversity, physiological ecology, environmental sciences, and natural product biosynthesis. Among these omics approaches, proteomics, transcriptomics, metatranscriptomics, and metabolomics have revolutionized the current understanding of the biological processes of fungi. In addition, more specialized omics methods such as ionomics, glycomics, glycoproteomics, glycogenomics, lipidomics, and interactomics coupled with bioinformatics can contribute to a greater understanding of fungal metabolism. The combination of omics approaches (multiomics) can be used to characterize fungal genomes and their metabolites, making multiomics approaches essential for detecting and characterizing novel metabolites with important biological properties, such as anticancer, antimicrobial, and antidiabetic for human health applications.
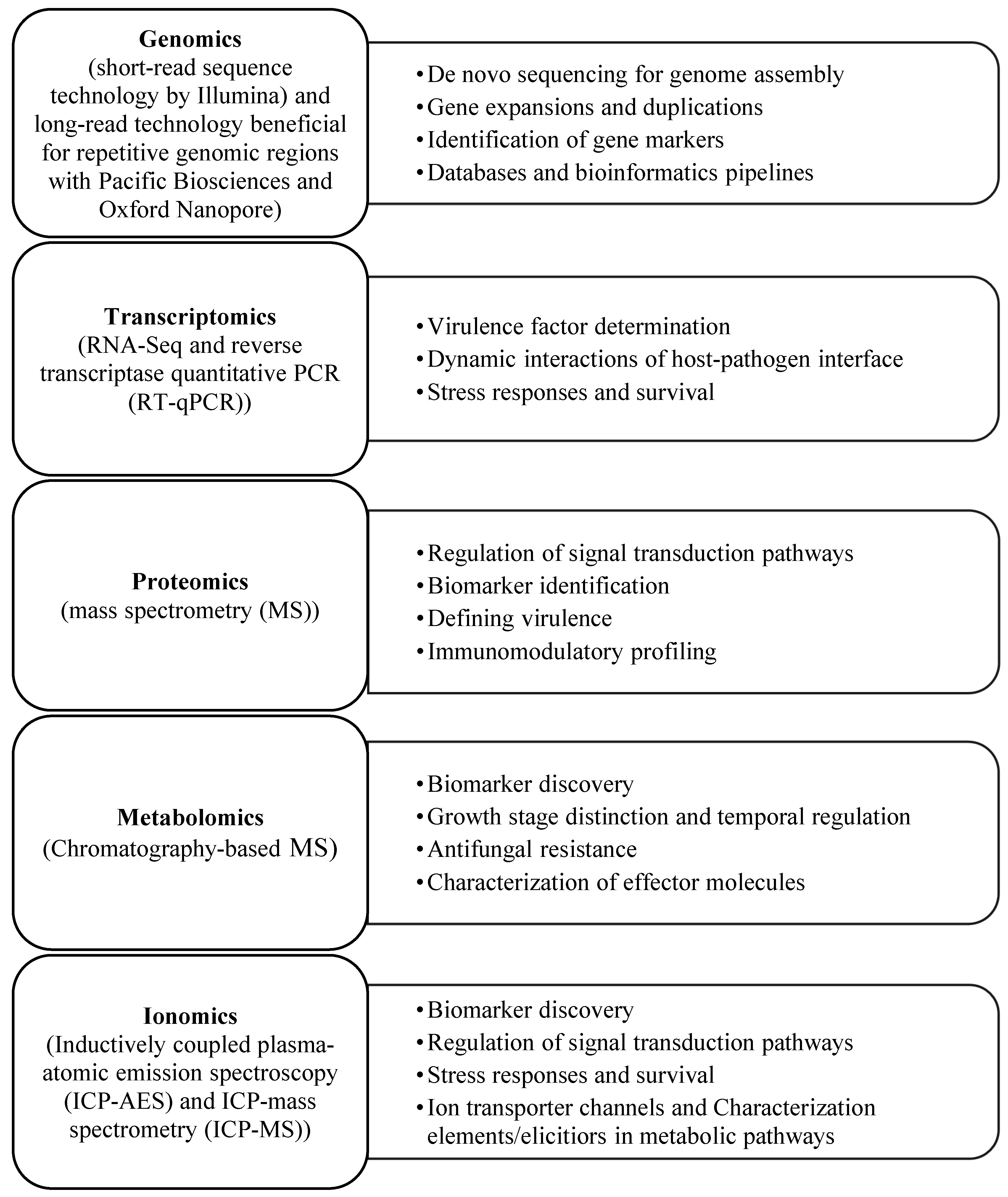
1. Food Safety and Security Based on Omics Techniques
2. The Role of Fungi in the Food Industry
3. Macrofungi and Edible Mushrooms
4. Foodomics
5. Fungal Secondary Metabolites
| Fungal Name | Pigment | Color |
|---|---|---|
| Penicillium purpurogenum | Mitorubrino Mitorubrin Purpurogenone Azaphilone |
Orange–red Yellow Yellow–orange |
| Rhodotorula glutinis | Torulene β-Carotene Torularhodin |
Red and orange |
| Thermomyces sp. | Naphthoquinone | Yellow |
| Yarrowia lipolytica | β-Carotene | Orange |
6. Mycotoxins and Fungi
7. Food Industry
8. Postharvest Losses
References
- Challa, S.; Dutta, T.; Neelapu, N.R.R. Fungal White Biotechnology Applications for Food Security: Opportunities and Challenges. In Recent Advancement in White Biotechnology through Fungi: Volume 2: Perspective for Value-Added Products and Environments; Yadav, A.N., Singh, S., Mishra, S., Gupta, A., Eds.; Fungal Biology; Springer International Publishing: Cham, Switzerland, 2019; pp. 119–148. ISBN 978-3-030-14846-1.
- Jain, S.; Rustagi, A.; Kumar, D.; Yusuf, M.A.; Shekhar, S.; Sarin, N.B. Meeting the Challenge of Developing Food Crops with Improved Nutritional Quality and Food Safety: Leveraging Proteomics and Related Omics Techniques. Biotechnol. Lett. 2019, 41, 471–481.
- Meyer, V.; Andersen, M.R.; Brakhage, A.A.; Braus, G.H.; Caddick, M.X.; Cairns, T.C.; de Vries, R.P.; Haarmann, T.; Hansen, K.; Hertz-Fowler, C.; et al. Current Challenges of Research on Filamentous Fungi in Relation to Human Welfare and a Sustainable Bio-Economy: A White Paper. Fungal Biol. Biotechnol. 2016, 3, 6.
- Xu, Y.-J. Foodomics: A Novel Approach for Food Microbiology. TrAC Trends Anal. Chem. 2017, 96, 14–21.
- Younas, A.; Rashid, M.; Riaz, N.; Munawar, M.; Fiaz, S.; Noreen, Z. Emerging Techniques to Develop Biotic Stress Resistance in Fruits and Vegetables. In Sustainable Agriculture in the Era of the OMICs Revolution; Prakash, C.S., Fiaz, S., Nadeem, M.A., Baloch, F.S., Qayyum, A., Eds.; Springer International Publishing: Cham, Switzerland, 2023; pp. 269–296. ISBN 978-3-031-15568-0.
- Josic, D.; Gašo-Sokač, D.; Šrajer Gajdošik, M.; Clifton, J. Microbial Omics for Food Safety. J. Hyg. Eng. Des. 2014, 6, 116–129.
- Ghorai, S.; Banik, S.P.; Verma, D.; Chowdhury, S.; Mukherjee, S.; Khowala, S. Fungal Biotechnology in Food and Feed Processing. Food Res. Int. 2009, 42, 577–587.
- Feeney, M.J.; Dwyer, J.; Hasler-Lewis, C.M.; Milner, J.A.; Noakes, M.; Rowe, S.; Wach, M.; Beelman, R.B.; Caldwell, J.; Cantorna, M.T.; et al. Mushrooms and Health Summit Proceedings. J. Nutr. 2014, 144, 1128S–1136S.
- Valverde, M.E.; Hernández-Pérez, T.; Paredes-López, O. Edible Mushrooms: Improving Human Health and Promoting Quality Life. Int. J. Microbiol. 2015, 2015, 376387.
- Samsudin, N.I.P.; Abdullah, N. Edible Mushrooms from Malaysia; a Literature Review on Their Nutritional and Medicinal Properties. Int. Food Res. J. 2019, 26, 11–31.
- Willis, K.J. State of the World’s Fungi 2018 Report; Royal Botanic Gardens: Kew, Australia, 2018.
- FAOSTAT. FAO’s Statistical Yearbook for 2022 Goes Live. Available online: https://www.fao.org/newsroom/detail/fao-s-statistical-yearbook-for-2022-goes-live/en (accessed on 7 March 2023).
- Pérez-Moreno, J.; Guerin-Laguette, A.; Rinaldi, A.C.; Yu, F.; Verbeken, A.; Hernández-Santiago, F.; Martínez-Reyes, M. Edible Mycorrhizal Fungi of the World: What Is Their Role in Forest Sustainability, Food Security, Biocultural Conservation and Climate Change? Plants People Planet 2021, 3, 471–490.
- Cao, L.; Zhang, Q.; Miao, R.; Lin, J.; Feng, R.; Ni, Y.; Li, W.; Yang, D.; Zhao, X. Application of Omics Technology in the Research on Edible Fungi. Curr. Res. Food Sci. 2023, 6, 100430.
- Horie, T.; Kusakabe, T.; Tsuda, M. Glutamatergic Networks in the Ciona Intestinalis Larva. J. Comp. Neurol. 2008, 508, 249–263.
- Ball, B.; Langille, M.; Geddes-McAlister, J. Fun(Gi)Omics: Advanced and Diverse Technologies to Explore Emerging Fungal Pathogens and Define Mechanisms of Antifungal Resistance. mBio 2020, 11, e01020-20.
- Pandohee, J.; Basu, R.; Dasgupta, S.; Sundarrajan, P.; Shaikh, N.; Patel, N.; Noor, A. Applications of Multi-Omics Approaches for Food and Nutritional Security. In Sustainable Agriculture in the Era of the OMICs Revolution; Prakash, C.S., Fiaz, S., Nadeem, M.A., Baloch, F.S., Qayyum, A., Eds.; Springer International Publishing: Cham, Switzerland, 2023; pp. 103–118. ISBN 978-3-031-15568-0.
- Malik, G.; Arora, R.; Chaturvedi, R.; Paul, M.S. Implementation of Genetic Engineering and Novel Omics Approaches to Enhance Bioremediation: A Focused Review. Bull. Environ. Contam. Toxicol. 2022, 108, 443–450.
- Meyer, V.; Basenko, E.Y.; Benz, J.P.; Braus, G.H.; Caddick, M.X.; Csukai, M.; de Vries, R.P.; Endy, D.; Frisvad, J.C.; Gunde-Cimerman, N.; et al. Growing a Circular Economy with Fungal Biotechnology: A White Paper. Fungal Biol. Biotechnol. 2020, 7, 5.
- Davies, H. A Role for “Omics” Technologies in Food Safety Assessment. Food Control 2010, 21, 1601–1610.
- Devi, R.; Kaur, T.; Guleria, G.; Rana, K.L.; Kour, D.; Yadav, N.; Yadav, A.N.; Saxena, A.K. Chapter 9—Fungal Secondary Metabolites and Their Biotechnological Applications for Human Health. In New and Future Developments in Microbial Biotechnology and Bioengineering; Rastegari, A.A., Yadav, A.N., Yadav, N., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 147–161. ISBN 978-0-12-820528-0.
- Manikprabhu, D.; Lingappa, K. γ Actinorhodin a Natural and Attorney Source for Synthetic Dye to Detect Acid Production of Fungi. Saudi J. Biol. Sci. 2013, 20, 163–168.
- Poorniammal, R.; Prabhu, S.; Dufossé, L.; Kannan, J. Safety Evaluation of Fungal Pigments for Food Applications. J. Fungi 2021, 7, 692.
- De Carvalho, J.C.; Oishi, B.O.; Pandey, A.; Soccol, C.R. Biopigments from Monascus: Strains Selection, Citrinin Production and Color Stability. Braz. Arch. Biol. Technol. 2005, 48, 885–894.
- Kallscheuer, N.; Classen, T.; Drepper, T.; Marienhagen, J. Production of Plant Metabolites with Applications in the Food Industry Using Engineered Microorganisms. Curr. Opin. Biotechnol. 2019, 56, 7–17.
- Gallage, N.J.; Møller, B.L. Vanilla: The Most Popular Flavour. In Biotechnology of Natural Products; Springer: Berlin/Heidelberg, Germany, 2018; pp. 3–24.
- Pozo-Bayón, M.A.; Guichard, E.; Cayot, N. Flavor Control in Baked Cereal Products. Food Rev. Int. 2006, 22, 335–379.
- Lomascolo, A.; Stentelaire, C.; Asther, M.; Lesage-Meessen, L. Basidiomycetes as New Biotechnological Tools to Generate Natural Aromatic Flavours for the Food Industry. Trends Biotechnol. 1999, 17, 282–289.
- Brochado, A.R.; Matos, C.; Møller, B.L.; Hansen, J.; Mortensen, U.H.; Patil, K.R. Improved Vanillin Production in Baker’s Yeast through in Silico Design. Microb. Cell Factories 2010, 9, 84.
- Prabhu, K.S.; Siveen, K.S.; Kuttikrishnan, S.; Iskandarani, A.N.; Khan, A.Q.; Merhi, M.; Omri, H.E.; Dermime, S.; El-Elimat, T.; Oberlies, N.H.; et al. Greensporone C, a Freshwater Fungal Secondary Metabolite Induces Mitochondrial-Mediated Apoptotic Cell Death in Leukemic Cell Lines. Front. Pharmacol. 2018, 9, 720.
- Brakhage, A.A. Regulation of Fungal Secondary Metabolism. Nat. Rev. Microbiol. 2013, 11, 21–32.
- Bigelis, R. Flavor Metabolites and Enzymes from Filamentous Fungi. In Food Technology; Springer: New York, NY, USA, 1992.
- Jòzef, S. The Use of Starch Processing Enzymes in the Food Industry. In Industrial Enzymes: Structure, Function and Applications; Polaina, J., MacCabe, A.P., Eds.; Springer: Dordrecht, The Netherlands, 2007; pp. 19–34. ISBN 978-1-4020-5377-1.
- Copetti, M.V. Fungi as Industrial Producers of Food Ingredients. Curr. Opin. Food Sci. 2019, 25, 52–56.
- Krings, U.; Berger, R.G. Dynamics of Sterols and Fatty Acids during UV-B Treatment of Oyster Mushroom. Food Chem. 2014, 149, 10–14.
- Ochsenreither, K.; Glück, C.; Stressler, T.; Fischer, L.; Syldatk, C. Production Strategies and Applications of Microbial Single Cell Oils. Front. Microbiol. 2016, 7, 1539.
- Al-Obaidi, J.R.; Jambari, N.N.; Ahmad-Kamil, E.I. Mycopharmaceuticals and Nutraceuticals: Promising Agents to Improve Human Well-Being and Life Quality. J. Fungi 2021, 7, 503.
- Gilani, G.; Lee, N. Protein, Quality. In Encyclopedia of Food Sciences and Nutrition, 2nd ed.; Academic Press Inc.: Cambridge, MA, USA, 2003.
- Jayaratne, W.M.S.C.; Abeyratne, A.H.M.A.K.; De Zoysa, H.K.S.; Dissanayake, D.M.R.B.N.; Bamunuarachchige, T.C.; Waisundara, V.Y.; Chang, S. Detection and Quantification of Aflatoxin B1 in Corn and Corn-Grown Soils in the District of Anuradhapura, Sri Lanka. Heliyon 2020, 6, e05319.
- Giacometti, J.; Josic, D. Foodomics in Microbial Safety. TrAC Trends Anal. Chem. 2013, 52, 16–22.
- Eshelli, M.; Qader, M.M.; Jambi, E.J.; Hursthouse, A.S.; Rateb, M.E. Current Status and Future Opportunities of Omics Tools in Mycotoxin Research. Toxins 2018, 10, 433.
- Chen, F.; Ma, R.; Chen, X.-L. Advances of Metabolomics in Fungal Pathogen–Plant Interactions. Metabolites 2019, 9, 169.
- Rai, P.; Singh, A.K.; Anand, K.B.; Singh, S.P.; Tomar, K. Time versus Tissue: Timely Identification of Scedosporium Rhinosinusitis in a Post-COVID-19 Case by MALDI-TOF MS Leading to Successful Management. Med. J. Armed Forces India 2022, 78, 360–364.
- Rai, N.; Keshri, P.K.; Gupta, P.; Verma, A.; Kamble, S.C.; Singh, S.K.; Gautam, V. Bioprospecting of Fungal Endophytes from Oroxylum Indicum (L.) Kurz with Antioxidant and Cytotoxic Activity. PLoS ONE 2022, 17, e0264673.
- Ali, S.; Tyagi, A.; Bae, H. Ionomic Approaches for Discovery of Novel Stress-Resilient Genes in Plants. Int. J. Mol. Sci. 2021, 22, 7182.
- Bahraminia, M.; Zarei, M.; Ronaghi, A.; Sepehri, M.; Etesami, H. Ionomic and Biochemical Responses of Maize Plant (Zea mays L.) Inoculated with Funneliformis Mosseae to Water-Deficit Stress. Rhizosphere 2020, 16, 100269.
- Soni, P.; Gangurde, S.S.; Ortega-Beltran, A.; Kumar, R.; Parmar, S.; Sudini, H.K.; Lei, Y.; Ni, X.; Huai, D.; Fountain, J.C.; et al. Functional Biology and Molecular Mechanisms of Host-Pathogen Interactions for Aflatoxin Contamination in Groundnut (Arachis hypogaea L.) and Maize (Zea mays L.). Front. Microbiol. 2020, 11, 227.
- Gupta, S.; Verma, R.; Ravi, R.K. Multiomics Approach for Crop Improvement under Climate Change. In Sustainable Agriculture in the Era of the OMICs Revolution; Prakash, C.S., Fiaz, S., Nadeem, M.A., Baloch, F.S., Qayyum, A., Eds.; Springer International Publishing: Cham, Switzerland, 2023; pp. 17–36. ISBN 978-3-031-15568-0.
- Gupta, S.M.; Arora, S.; Mirza, N.; Pande, A.; Lata, C.; Puranik, S.; Kumar, J.; Kumar, A. Finger Millet: A “Certain” Crop for an “Uncertain” Future and a Solution to Food Insecurity and Hidden Hunger under Stressful Environments. Front. Plant Sci. 2017, 8, 643.
- Abbas, A.; Shah, A.A.; Shah, A.N.; Niaz, Y.; Ahmed, W.; Ali, H.; Nawaz, M.; Hassan, M.U. CRISPR Revolution in Gene Editing: Targeting Plant Stress Tolerance and Physiology. In Sustainable Agriculture in the Era of the OMICs Revolution; Prakash, C.S., Fiaz, S., Nadeem, M.A., Baloch, F.S., Qayyum, A., Eds.; Springer International Publishing: Cham, Switzerland, 2023; pp. 315–325. ISBN 978-3-031-15568-0.
- Hussain, K.; Mahrukh; Nisa, R.T.; Zaid, A.; Mushtaq, M. The Utilization of Speed Breeding and Genome Editing to Achieve Zero Hunger. In Sustainable Agriculture in the Era of the OMICs Revolution; Prakash, C.S., Fiaz, S., Nadeem, M.A., Baloch, F.S., Qayyum, A., Eds.; Springer International Publishing: Cham, Switzerland, 2023; pp. 1–15. ISBN 978-3-031-15568-0.
- Sarfraz, S.; Ali, F.; Hameed, A.; Ahmad, Z.; Riaz, K. Sustainable Agriculture through Technological Innovations. In Sustainable Agriculture in the Era of the OMICs Revolution; Prakash, C.S., Fiaz, S., Nadeem, M.A., Baloch, F.S., Qayyum, A., Eds.; Springer International Publishing: Cham, Switzerland, 2023; pp. 223–239. ISBN 978-3-031-15568-0.
- Borges, F.; Briandet, R.; Callon, C.; Champomier-Vergès, M.-C.; Christieans, S.; Chuzeville, S.; Denis, C.; Desmasures, N.; Desmonts, M.-H.; Feurer, C.; et al. Contribution of Omics to Biopreservation: Toward Food Microbiome Engineering. Front. Microbiol. 2022, 13, 951182.
- Apaliya, M.T.; Osae, R.; Kwaw, E.; Mahunu, G.K.; Osei-Kwarteng, M.; Hardi, I.M. Omics in Traditional Fermented Foods and Beverages. In African Fermented Food Products-New Trends; Elhadi Sulieman, A.M., Adam Mariod, A., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 551–563. ISBN 978-3-030-82902-5.
- Kumari, C.; Sharma, M.; Kumar, V.; Sharma, R.; Kumar, V.; Sharma, P.; Kumar, P.; Irfan, M. Genome Editing Technology for Genetic Amelioration of Fruits and Vegetables for Alleviating Post-Harvest Loss. Bioengineering 2022, 9, 176.
- Hegazy, R. Post-Harvest Situation and Losses in India. J. Contrib. 2016, 1–21.
- Zhang, Z.-Q.; Chen, T.; Li, B.-Q.; Qin, G.-Z.; Tian, S.-P. Molecular Basis of Pathogenesis of Postharvest Pathogenic Fungi and Control Strategy in Fruits: Progress and Prospect. Mol. Hortic. 2021, 1, 2.
- Li, T.; Wu, Y.; Wang, Y.; Gao, H.; Gupta, V.K.; Duan, X.; Qu, H.; Jiang, Y. Secretome Profiling Reveals Virulence-Associated Proteins of Fusarium Proliferatum during Interaction with Banana Fruit. Biomolecules 2019, 9, 246.
- Lee, M.-H.; Chiu, C.-M.; Roubtsova, T.; Chou, C.-M.; Bostock, R.M. Overexpression of a Redox-Regulated Cutinase Gene, MfCUT1, Increases Virulence of the Brown Rot Pathogen Monilinia fructicola on Prunus Spp. Mol. Plant-Microbe Interact. 2010, 23, 176–186.
- Brito, N.; Espino, J.J.; González, C. The Endo-β-1,4-Xylanase Xyn11A Is Required for Virulence in Botrytis cinerea. Mol. Plant-Microbe Interact. 2006, 19, 25–32.
- Tian, S.; Qin, G.; Li, B. Reactive Oxygen Species Involved in Regulating Fruit Senescence and Fungal Pathogenicity. Plant Mol. Biol. 2013, 82, 593–602.
- Tian, S.; Torres, R.; Ballester, A.-R.; Li, B.; Vilanova, L.; González-Candelas, L. Molecular Aspects in Pathogen-Fruit Interactions: Virulence and Resistance. Postharvest Biol. Technol. 2016, 122, 11–21.
- Li, H.; Zhang, Z.; He, C.; Qin, G.; Tian, S. Comparative Proteomics Reveals the Potential Targets of BcNoxR, a Putative Regulatory Subunit of NADPH Oxidase of Botrytis cinerea. Mol. Plant-Microbe Interact. 2016, 29, 990–1003.
- Seo, J.-K.; Wu, J.; Lii, Y.; Li, Y.; Jin, H. Contribution of Small RNA Pathway Components in Plant Immunity. Mol. Plant-Microbe Interact. 2013, 26, 617–625.
- Weiberg, A.; Wang, M.; Lin, F.-M.; Zhao, H.; Zhang, Z.; Kaloshian, I.; Huang, H.-D.; Jin, H. Fungal Small RNAs Suppress Plant Immunity by Hijacking Host RNA Interference Pathways. Science 2013, 342, 118–123.
- Ma, H.; Sun, X.; Wang, M.; Gai, Y.; Chung, K.-R.; Li, H. The Citrus Postharvest Pathogen Penicillium Digitatum Depends on the PdMpkB Kinase for Developmental and Virulence Functions. Int. J. Food Microbiol. 2016, 236, 167–176.
- Son, H.; Seo, Y.-S.; Min, K.; Park, A.R.; Lee, J.; Jin, J.-M.; Lin, Y.; Cao, P.; Hong, S.-Y.; Kim, E.-K.; et al. A Phenome-Based Functional Analysis of Transcription Factors in the Cereal Head Blight Fungus, Fusarium graminearum. PLoS Pathog. 2011, 7, e1002310.




