
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Shin Kwak | -- | 2729 | 2023-06-19 10:45:52 | | | |
| 2 | Lindsay Dong | -1 word(s) | 2728 | 2023-06-20 02:54:35 | | |
Video Upload Options
Amyotrophic lateral sclerosis (ALS) is an incurable motor neuron disease caused by upper and lower motor neuron death. As aging is a major risk factor for ALS, age-related molecular changes may provide clues for the development of new therapeutic strategies. Dysregulation of age-dependent RNA metabolism plays a pivotal role in the pathogenesis of ALS. In addition, failure of RNA editing at the glutamine/arginine (Q/R) site of GluA2 mRNA causes excitotoxicity due to excessive Ca2+ influx through Ca2+-permeable α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptors, which is recognized as an underlying mechanism of motor neuron death in ALS. Circular RNAs (circRNAs), a circular form of cognate RNA generated by back-splicing, are abundant in the brain and accumulate with age. Hence, they are assumed to play a role in neurodegeneration. Emerging evidence has demonstrated that age-related dysregulation of RNA editing and changes in circRNA expression are involved in ALS pathogenesis.
1. Introduction
2. Age-Related Changes of circRNAs and RNA Editing
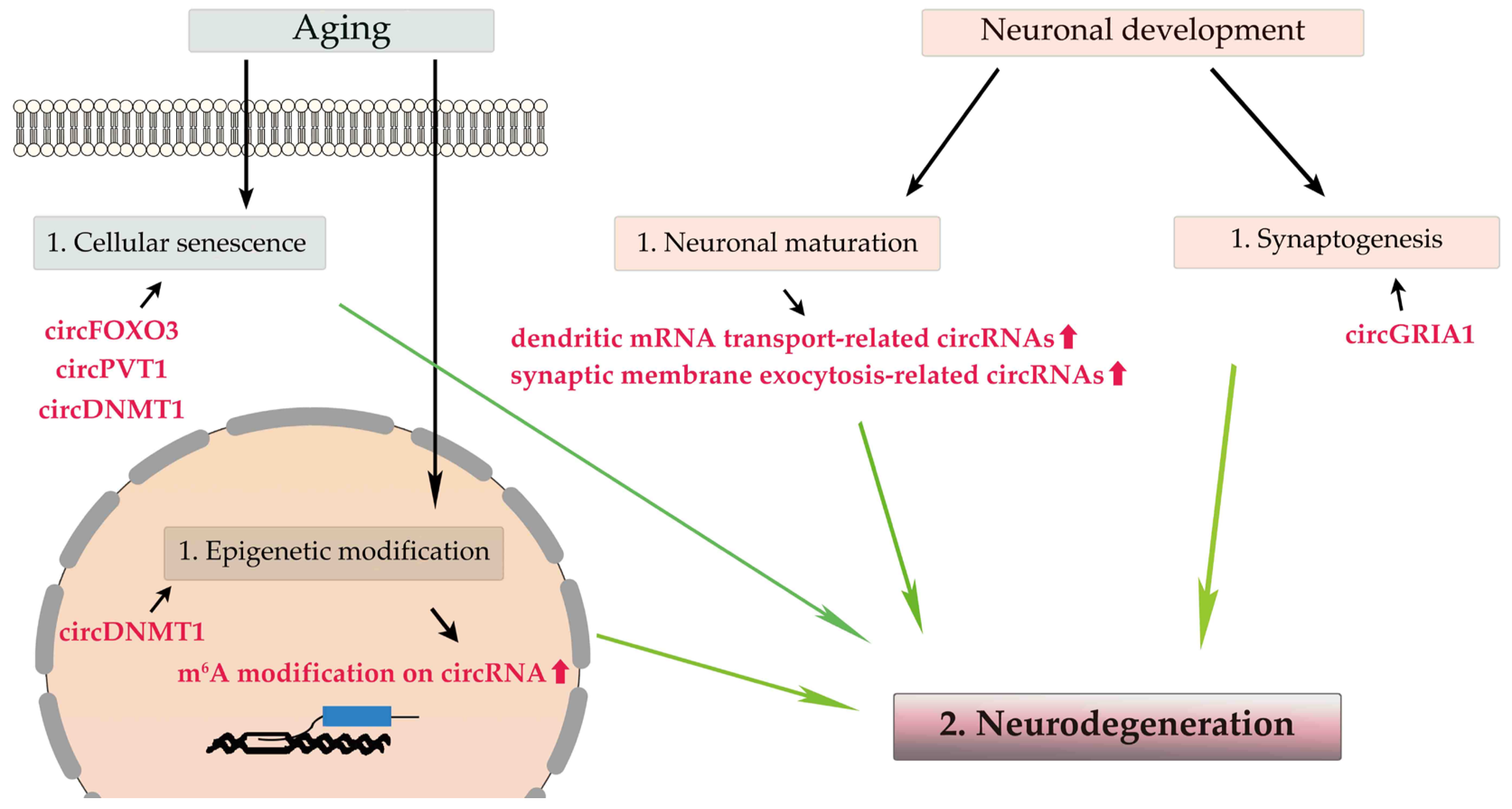
2.1. Age-Related circRNAs in the Brain (Figure 1)
2.2. Age-Related RNA Editing
3. ALS-Related Changes of circRNAs and Dysregulation of RNA Editing
3.1. ALS-Related circRNA
3.2. Dysregulation of RNA Editing in ALS Motor Neurons
3.3. Aging, circRNAs, and RNA Editing in ALS
4. CircRNAs and the Dysregulation of RNA Editing as Potential Biomarkers and Therapeutic Targets in ALS
4.1. Potential Biomarker Candidates for ALS
4.2. Therapeutic Targets for ALS
References
- Rowland, L.P.; Shneider, N.A. Amyotrophic Lateral Sclerosis. N. Engl. J. Med. 2001, 344, 1688–1700.
- Feldman, E.L.; Goutman, S.A.; Petri, S.; Mazzini, L.; Savelieff, M.G.; Shaw, P.J.; Sobue, G. Amyotrophic Lateral Sclerosis. Lancet 2022, 400, 1363–1380.
- Arthur, K.C.; Calvo, A.; Price, T.R.; Geiger, J.T.; Chiò, A.; Traynor, B.J. Projected Increase in Amyotrophic Lateral Sclerosis from 2015 to 2040. Nat. Commun. 2016, 7, 12408.
- Brown, R.H.; Al-Chalabi, A. Amyotrophic Lateral Sclerosis. N. Engl. J. Med. 2017, 377, 162–172.
- Goutman, S.A.; Hardiman, O.; Al-Chalabi, A.; Chió, A.; Savelieff, M.G.; Kiernan, M.C.; Feldman, E.L. Emerging Insights into the Complex Genetics and Pathophysiology of Amyotrophic Lateral Sclerosis. Lancet Neurol. 2022, 21, 465–479.
- Butti, Z.; Patten, S.A. RNA Dysregulation in Amyotrophic Lateral Sclerosis. Front. Genet. 2018, 9, 712.
- Braun, S.; Domdey, H.; Wiebauer, K. Inverse Splicing of a Discontinuous Pre-mRNA Intron Generates a Circular Exon in a HeLa Cell Nuclear Extract. Nucleic Acids Res. 1996, 24, 4152–4157.
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA Biogenesis Competes with Pre-mRNA Splicing. Mol. Cell 2014, 56, 55–66.
- Meng, S.; Zhou, H.; Feng, Z.; Xu, Z.; Tang, Y.; Li, P.; Wu, M. CircRNA: Functions and Properties of a Novel Potential Biomarker for Cancer. Mol. Cancer 2017, 16, 94.
- Ren, S.; Lin, P.; Wang, J.; Yu, H.; Lv, T.; Sun, L.; Du, G. Circular RNAs: Promising Molecular Biomarkers of Human Aging-Related Diseases via Functioning as An miRNA Sponge. Mol. Ther. Methods Clin. Dev. 2020, 18, 215–229.
- Chen, Y.; Yao, L.; Tang, Y.; Jhong, J.H.; Wan, J.; Chang, J.; Cui, S.; Luo, Y.; Cai, X.; Li, W.; et al. CircNet 2.0: An Updated Database for Exploring Circular RNA Regulatory Networks in Cancers. Nucleic Acids Res. 2022, 50, D93–D101.
- Mahmoudi, E.; Cairns, M.J. Circular RNAs Are Temporospatially Regulated Throughout Development and Ageing in the Rat. Sci. Rep. 2019, 9, 2564.
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs Are Abundant, Conserved, and Associated with ALU Repeats. RNA 2013, 19, 141–157.
- Holdt, L.M.; Stahringer, A.; Sass, K.; Pichler, G.; Kulak, N.A.; Wilfert, W.; Kohlmaier, A.; Herbst, A.; Northoff, B.H.; Nicolaou, A.; et al. Circular Non-coding RNA ANRIL Modulates Ribosomal RNA Maturation and Atherosclerosis in Humans. Nat. Commun. 2016, 7, 12429.
- Rybak-Wolf, A.; Stottmeister, C.; Glažar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R.; et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol. Cell 2015, 58, 870–885.
- Vo, J.N.; Cieslik, M.; Zhang, Y.; Shukla, S.; Xiao, L.; Zhang, Y.; Wu, Y.M.; Dhanasekaran, S.M.; Engelke, C.G.; Cao, X.; et al. The Landscape of Circular RNA in Cancer. Cell 2019, 176, 869–881.e13.
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-Intron Circular RNAs Regulate Transcription in the Nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264.
- Liu, Y.; Su, H.; Zhang, J.; Liu, Y.; Feng, C.; Han, F. Back-Spliced RNA from Retrotransposon Binds to Centromere and Regulates Centromeric Chromatin Loops in Maize. PLoS Biol. 2020, 18, e3000582.
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA Circles Function as Efficient microRNA Sponges. Nature 2013, 495, 384–388.
- Du, W.W.; Yang, W.; Liu, E.; Yang, Z.; Dhaliwal, P.; Yang, B.B. Foxo3 Circular RNA Retards Cell Cycle Progression via Forming Ternary Complexes with p21 and CDK2. Nucleic Acids Res. 2016, 44, 2846–2858.
- Nishikura, K. A-to-I Editing of Coding and Non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 2016, 17, 83–96.
- Rueter, S.M.; Dawson, T.R.; Emeson, R.B. Regulation of Alternative Splicing by RNA Editing. Nature 1999, 399, 75–80.
- Heraud-Farlow, J.E.; Walkley, C.R. What Do Editors Do? Understanding the Physiological Functions of A-to-I RNA Editing by Adenosine Deaminase Acting on RNAs. Open Biol. 2020, 10, 200085.
- Yang, W.; Chendrimada, T.P.; Wang, Q.; Higuchi, M.; Seeburg, P.H.; Shiekhattar, R.; Nishikura, K. Modulation of microRNA Processing and Expression Through RNA Editing by ADAR Deaminases. Nat. Struct. Mol. Biol. 2006, 13, 13–21.
- Ivanov, A.; Memczak, S.; Wyler, E.; Torti, F.; Porath, H.T.; Orejuela, M.R.; Piechotta, M.; Levanon, E.Y.; Landthaler, M.; Dieterich, C.; et al. Analysis of Intron Sequences Reveals Hallmarks of Circular RNA Biogenesis in Animals. Cell Rep. 2015, 10, 170–177.
- Guo, C.; Ma, Y.Y. Calcium Permeable-AMPA Receptors and Excitotoxicity in Neurological Disorders. Front. Neural Circuits 2021, 15, 711564.
- Greger, I.H.; Khatri, L.; Ziff, E.B. RNA Editing at arg607 Controls AMPA Receptor Exit from the Endoplasmic Reticulum. Neuron 2002, 34, 759–772.
- Baker, D.J.; Petersen, R.C. Cellular Senescence in Brain Aging and Neurodegenerative Diseases: Evidence and Perspectives. J. Clin. Investig. 2018, 128, 1208–1216.
- Azam, S.; Haque, M.E.; Balakrishnan, R.; Kim, I.S.; Choi, D.K. The Ageing Brain: Molecular and Cellular Basis of Neurodegeneration. Front. Cell Dev. Biol. 2021, 9, 683459.
- Hou, Y.; Dan, X.; Babbar, M.; Wei, Y.; Hasselbalch, S.G.; Croteau, D.L.; Bohr, V.A. Ageing as a Risk Factor for Neurodegenerative Disease. Nat. Rev. Neurol. 2019, 15, 565–581.
- Du, S.; Zheng, H. Role of FoxO Transcription Factors in Aging and Age-Related Metabolic and Neurodegenerative Diseases. Cell Biosci. 2021, 11, 188.
- Du, W.W.; Yang, W.; Chen, Y.; Wu, Z.K.; Foster, F.S.; Yang, Z.; Li, X.; Yang, B.B. Foxo3 Circular RNA Promotes Cardiac Senescence by Modulating Multiple Factors Associated with Stress and Senescence Responses. Eur. Heart J. 2017, 38, 1402–1412.
- Haque, S.; Ames, R.M.; Moore, K.; Pilling, L.C.; Peters, L.L.; Bandinelli, S.; Ferrucci, L.; Harries, L.W. circRNAs Expressed in Human Peripheral Blood Are Associated with Human Aging Phenotypes, Cellular Senescence and Mouse Lifespan. GeroScience 2020, 42, 183–199.
- Roush, S.; Slack, F.J. The Let-7 Family of microRNAs. Trends Cell Biol. 2008, 18, 505–516.
- Lehmann, S.M.; Krüger, C.; Park, B.; Derkow, K.; Rosenberger, K.; Baumgart, J.; Trimbuch, T.; Eom, G.; Hinz, M.; Kaul, D.; et al. An Unconventional Role for miRNA: Let-7 Activates Toll-Like Receptor 7 and Causes Neurodegeneration. Nat. Neurosci. 2012, 15, 827–835.
- Pang, Y.; Lin, W.; Zhan, L.; Zhang, J.; Zhang, S.; Jin, H.; Zhang, H.; Wang, X.; Li, X. Inhibiting Autophagy Pathway of PI3K/AKT/mTOR Promotes Apoptosis in SK-N-SH Cell Model of Alzheimer’s Disease. J. Healthc. Eng. 2022, 2022, 6069682.
- Panda, A.C.; Grammatikakis, I.; Kim, K.M.; De, S.; Martindale, J.L.; Munk, R.; Yang, X.; Abdelmohsen, K.; Gorospe, M. Identification of Senescence-Associated Circular RNAs (SAC-RNAs) Reveals Senescence Suppressor CircPVT1. Nucleic Acids Res. 2017, 45, 4021–4035.
- Han, W.; Tao, X.; Weng, T.; Chen, L. Circular RNA PVT1 Inhibits Tendon Stem/Progenitor Cell Senescence by Sponging microRNA-199a-5p. Toxicol. Vitr. 2022, 79, 105297.
- Jęśko, H.; Wencel, P.; Strosznajder, R.P.; Strosznajder, J.B. Sirtuins and Their Roles in Brain Aging and Neurodegenerative Disorders. Neurochem. Res. 2017, 42, 876–890.
- Sousa, C.; Mendes, A.F. Monoterpenes as Sirtuin-1 Activators: Therapeutic Potential in Aging and Related Diseases. Biomolecules 2022, 12, 921.
- Boulias, K.; Greer, E.L. Biological Roles of Adenine Methylation in RNA. Nat. Rev. Genet. 2023, 24, 143–160.
- Xu, T.; He, B.; Sun, H.; Xiong, M.; Nie, J.; Wang, S.; Pan, Y. Novel Insights into the Interaction Between N6-Methyladenosine Modification and Circular RNA. Mol. Ther. Nucleic Acids 2022, 27, 824–837.
- Zhang, X.; Yang, S.; Han, S.; Sun, Y.; Han, M.; Zheng, X.; Li, F.; Wei, Y.; Wang, Y.; Bi, J. Differential Methylation of circRNA m6A in an APP/PS1 Alzheimer’s Disease Mouse Model. Mol. Med. Rep. 2023, 27, 1–8.
- Bayer, C.; Pitschelatow, G.; Hannemann, N.; Linde, J.; Reichard, J.; Pensold, D.; Zimmer-Bensch, G. DNA Methyltransferase 1 (DNMT1) Acts on Neurodegeneration by Modulating Proteostasis-Relevant Intracellular Processes. Int. J. Mol. Sci. 2020, 21, 5420.
- D’Anca, M.; Buccellato, F.R.; Fenoglio, C.; Galimberti, D. Circular RNAs: Emblematic Players of Neurogenesis and Neurodegeneration. Int. J. Mol. Sci. 2022, 23, 4134.
- You, X.; Vlatkovic, I.; Babic, A.; Will, T.; Epstein, I.; Tushev, G.; Akbalik, G.; Wang, M.; Glock, C.; Quedenau, C.; et al. Neural Circular RNAs Are Derived from Synaptic Genes and Regulated by Development and Plasticity. Nat. Neurosci. 2015, 18, 603–610.
- Yang, Y.; Okada, S.; Sakurai, M. Adenosine-to-Inosine RNA Editing in Neurological Development and Disease. RNA Biol. 2021, 18, 999–1013.
- Eisenberg, E.; Levanon, E.Y. A-to-I RNA Editing—Immune Protector and Transcriptome Diversifier. Nat. Rev. Genet. 2018, 19, 473–490.
- Lorenzini, I.; Moore, S.; Sattler, R. RNA Editing Deficiency in Neurodegeneration. Adv. Neurobiol. 2018, 20, 63–83.
- Solomon, O.; Di Segni, A.; Cesarkas, K.; Porath, H.T.; Marcu-Malina, V.; Mizrahi, O.; Stern-Ginossar, N.; Kol, N.; Farage-Barhom, S.; Glick-Saar, E.; et al. RNA Editing by ADAR1 Leads to Context-Dependent Transcriptome-Wide Changes in RNA Secondary Structure. Nat. Commun. 2017, 8, 1440.
- Montano, M.; Long, K. RNA Surveillance-an Emerging Role for RNA Regulatory Networks in Aging. Ageing Res. Rev. 2011, 10, 216–224.
- Wahlstedt, H.; Daniel, C.; Ensterö, M.; Ohman, M. Large-Scale mRNA Sequencing Determines Global Regulation of RNA Editing During Brain Development. Genome Res. 2009, 19, 978–986.
- Shtrichman, R.; Germanguz, I.; Mandel, R.; Ziskind, A.; Nahor, I.; Safran, M.; Osenberg, S.; Sherf, O.; Rechavi, G.; Itskovitz-Eldor, J. Altered A-to-I RNA Editing in Human Embryogenesis. PLoS ONE 2012, 7, e41576.
- Hosaka, T.; Tsuji, H.; Kwak, S. RNA Editing: A New Therapeutic Target in Amyotrophic Lateral Sclerosis and Other Neurological Diseases. Int. J. Mol. Sci. 2021, 22, 10958.
- Slotkin, W.; Nishikura, K. Adenosine-to-Inosine RNA Editing and Human Disease. Genome Med. 2013, 5, 105.
- Dube, U.; Del-Aguila, J.L.; Li, Z.; Budde, J.P.; Jiang, S.; Hsu, S.; Ibanez, L.; Fernandez, M.V.; Farias, F.; Norton, J.; et al. An Atlas of Cortical Circular RNA Expression in Alzheimer Disease Brains Demonstrates Clinical and Pathological Associations. Nat. Neurosci. 2019, 22, 1903–1912.
- Aquilina-Reid, C.; Brennan, S.; Curry-Hyde, A.; Teunisse, G.M.; The Nygc Als Consortium; Janitz, M. Circular RNA Expression and Interaction Patterns Are Perturbed in Amyotrophic Lateral Sclerosis. Int. J. Mol. Sci. 2022, 23, 14665.
- Tsitsipatis, D.; Mazan-Mamczarz, K.; Si, Y.; Herman, A.B.; Yang, J.H.; Guha, A.; Piao, Y.; Fan, J.; Martindale, J.L.; Munk, R.; et al. Transcriptomic Analysis of Human ALS Skeletal Muscle Reveals a Disease-Specific Pattern of Dysregulated circRNAs. Aging 2022, 14, 9832–9859.
- Assoni, A.F.; Foijer, F.; Zatz, M. Amyotrophic Lateral Sclerosis, FUS and Protein Synthesis Defects. Stem Cell Rev. Rep. 2023, 19, 625–638.
- Errichelli, L.; Dini Modigliani, S.; Laneve, P.; Colantoni, A.; Legnini, I.; Capauto, D.; Rosa, A.; De Santis, R.; Scarfò, R.; Peruzzi, G.; et al. FUS Affects Circular RNA Expression in Murine Embryonic Stem Cell-Derived Motor Neurons. Nat. Commun. 2017, 8, 14741.
- Colantoni, A.; Capauto, D.; Alfano, V.; D’Ambra, E.; D’Uva, S.; Tartaglia, G.G.; Morlando, M. FUS Alters circRNA Metabolism in Human Motor Neurons Carrying the ALS-Linked P525L Mutation. Int. J. Mol. Sci. 2023, 24, 3181.
- DeJesus-Hernandez, M.; Mackenzie, I.R.; Boeve, B.F.; Boxer, A.L.; Baker, M.; Rutherford, N.J.; Nicholson, A.M.; Finch, N.A.; Flynn, H.; Adamson, J.; et al. Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron 2011, 72, 245–256.
- Renton, A.E.; Majounie, E.; Waite, A.; Simón-Sánchez, J.; Rollinson, S.; Gibbs, J.R.; Schymick, J.C.; Laaksovirta, H.; van Swieten, J.C.; Myllykangas, L.; et al. A Hexanucleotide Repeat Expansion in C9ORF72 Is the Cause of Chromosome 9p21-Linked ALS-FTD. Neuron 2011, 72, 257–268.
- Rohrer, J.D.; Isaacs, A.M.; Mizielinska, S.; Mead, S.; Lashley, T.; Wray, S.; Sidle, K.; Fratta, P.; Orrell, R.W.; Hardy, J.; et al. C9orf72 Expansions in Frontotemporal Dementia and Amyotrophic Lateral Sclerosis. Lancet Neurol. 2015, 14, 291–301.
- Belzil, V.V.; Katzman, R.B.; Petrucelli, L. ALS and FTD: An Epigenetic Perspective. Acta Neuropathol. 2016, 132, 487–502.
- Leighton, G.O.; Irvin, E.M.; Kaur, P.; Liu, M.; You, C.; Bhattaram, D.; Piehler, J.; Riehn, R.; Wang, H.; Pan, H.; et al. Densely Methylated DNA Traps Methyl-CpG-Binding Domain Protein 2 but Permits Free Diffusion by Methyl-CpG-Binding Domain Protein 3. J. Biol. Chem. 2022, 298, 102428.
- Liu, D.; Liu, W.; Chen, X.; Yin, J.; Ma, L.; Liu, M.; Zhou, X.; Xian, L.; Li, P.; Tan, X.; et al. circKCNN2 Suppresses the Recurrence of Hepatocellular Carcinoma at Least Partially via Regulating miR-520c-3p/methyl-DNA-binding Domain Protein 2 Axis. Clin. Transl. Med. 2022, 12, e662.
- Cervera-Carles, L.; Dols-Icardo, O.; Molina-Porcel, L.; Alcolea, D.; Cervantes-Gonzalez, A.; Muñoz-Llahuna, L.; Clarimon, J. Assessing Circular RNAs in Alzheimer’s Disease and Frontotemporal Lobar Degeneration. Neurobiol. Aging 2020, 92, 7–11.
- Van Damme, P.; Dewil, M.; Robberecht, W.; Van Den Bosch, L. Excitotoxicity and Amyotrophic Lateral Sclerosis. Neurodegener. Dis. 2005, 2, 147–159.
- King, A.E.; Woodhouse, A.; Kirkcaldie, M.T.; Vickers, J.C. Excitotoxicity in ALS: Overstimulation, or Overreaction? Exp. Neurol. 2016, 275, 162–171.
- Diering, G.H.; Huganir, R.L. The AMPA Receptor Code of Synaptic Plasticity. Neuron 2018, 100, 314–329.
- Gregory, J.M.; Livesey, M.R.; McDade, K.; Selvaraj, B.T.; Barton, S.K.; Chandran, S.; Smith, C. Dysregulation of AMPA Receptor Subunit Expression in Sporadic ALS Post-Mortem Brain. J. Pathol. 2020, 250, 67–78.
- Udagawa, T.; Fujioka, Y.; Tanaka, M.; Honda, D.; Yokoi, S.; Riku, Y.; Ibi, D.; Nagai, T.; Yamada, K.; Watanabe, H.; et al. FUS Regulates AMPA Receptor Function and FTLD/ALS-Associated Behaviour via GluA1 mRNA Stabilization. Nat. Commun. 2015, 6, 7098.
- Capauto, D.; Colantoni, A.; Lu, L.; Santini, T.; Peruzzi, G.; Biscarini, S.; Morlando, M.; Shneider, N.A.; Caffarelli, E.; Laneve, P.; et al. A Regulatory Circuitry Between Gria2, miR-409, and miR-495 Is Affected by ALS FUS Mutation in ESC-Derived Motor Neurons. Mol. Neurobiol. 2018, 55, 7635–7651.
- Selvaraj, B.T.; Livesey, M.R.; Zhao, C.; Gregory, J.M.; James, O.T.; Cleary, E.M.; Chouhan, A.K.; Gane, A.B.; Perkins, E.M.; Dando, O.; et al. C9ORF72 Repeat Expansion Causes Vulnerability of Motor Neurons to Ca2+-Permeable AMPA Receptor-Mediated Excitotoxicity. Nat. Commun. 2018, 9, 347.
- Dafinca, R.; Barbagallo, P.; Farrimond, L.; Candalija, A.; Scaber, J.; Ababneh, N.A.; Sathyaprakash, C.; Vowles, J.; Cowley, S.A.; Talbot, K. Impairment of Mitochondrial Calcium Buffering Links Mutations in C9ORF72 and TARDBP in iPS-Derived Motor Neurons from Patients with ALS/FTD. Stem Cell Rep. 2020, 14, 892–908.
- Xu, K.; Zhang, Y.; Xiong, W.; Zhang, Z.; Wang, Z.; Lv, L.; Liu, C.; Hu, Z.; Zheng, Y.T.; Lu, L.; et al. CircGRIA1 Shows an Age-Related Increase in Male Macaque Brain and Regulates Synaptic Plasticity and Synaptogenesis. Nat. Commun. 2020, 11, 3594.
- Markovinovic, A.; Greig, J.; Martín-Guerrero, S.M.; Salam, S.; Paillusson, S. Endoplasmic Reticulum-Mitochondria Signaling in Neurons and Neurodegenerative Diseases. J. Cell Sci. 2022, 135, jcs248534.
- Watanabe, S.; Ilieva, H.; Tamada, H.; Nomura, H.; Komine, O.; Endo, F.; Jin, S.; Mancias, P.; Kiyama, H.; Yamanaka, K. Mitochondria-Associated Membrane Collapse Is a Common Pathomechanism in SIGMAR1- and SOD1-Linked ALS. EMBO Mol. Med. 2016, 8, 1421–1437.
- Lau, D.H.W.; Hartopp, N.; Welsh, N.J.; Mueller, S.; Glennon, E.B.; Mórotz, G.M.; Annibali, A.; Gomez-Suaga, P.; Stoica, R.; Paillusson, S.; et al. Disruption of ER-Mitochondria Signalling in Fronto-temporal Dementia and Related Amyotrophic Lateral Sclerosis. Cell Death Dis. 2018, 9, 327.
- Sakai, S.; Watanabe, S.; Komine, O.; Sobue, A.; Yamanaka, K. Novel Reporters of Mitochondria-Associated Membranes (MAM), MAMtrackers, Demonstrate MAM Disruption as a Common Pathological Feature in Amyotrophic Lateral Sclerosis. FASEB J. 2021, 35, e21688.
- Al-Saif, A.; Al-Mohanna, F.; Bohlega, S. A Mutation in Sigma-1 Receptor Causes Juvenile Amyotrophic Lateral Sclerosis. Ann. Neurol. 2011, 70, 913–919.
- Tagashira, H.; Shinoda, Y.; Shioda, N.; Fukunaga, K. Methyl Pyruvate Rescues Mitochondrial Damage Caused by SIGMAR1 Mutation Related to Amyotrophic Lateral Sclerosis. Biochim. Biophys. Acta 2014, 1840, 3320–3334.
- Luty, A.A.; Kwok, J.B.; Dobson-Stone, C.; Loy, C.T.; Coupland, K.G.; Karlström, H.; Sobow, T.; Tchorzewska, J.; Maruszak, A.; Barcikowska, M.; et al. Sigma Nonopioid Intracellular Receptor 1 Mutations Cause Frontotemporal Lobar Degeneration-Motor Neuron Disease. Ann. Neurol. 2010, 68, 639–649.
- Dreser, A.; Vollrath, J.T.; Sechi, A.; Johann, S.; Roos, A.; Yamoah, A.; Katona, I.; Bohlega, S.; Wiemuth, D.; Tian, Y.; et al. The ALS-Linked E102Q Mutation in Sigma Receptor-1 Leads to ER Stress-Mediated Defects in Protein Homeostasis and Dysregulation of RNA-Binding Proteins. Cell Death Differ. 2017, 24, 1655–1671.
- Plaitakis, A.; Constantakakis, E.; Smith, J. The Neuroexcitotoxic Amino Acids Glutamate and Aspartate Are Altered in the Spinal Cord and Brain in Amyotrophic Lateral Sclerosis. Ann. Neurol. 1988, 24, 446–449.
- Rothstein, J.D.; Tsai, G.; Kuncl, R.W.; Clawson, L.; Cornblath, D.R.; Drachman, D.B.; Pestronk, A.; Stauch, B.L.; Coyle, J.T. Abnormal Excitatory Amino Acid Metabolism in Amyotrophic Lateral Sclerosis. Ann. Neurol. 1990, 28, 18–25.
- Shaw, P.J.; Forrest, V.; Ince, P.G.; Richardson, J.P.; Wastell, H.J. CSF and Plasma Amino Acid Levels in Motor Neuron Disease: Elevation of CSF Glutamate in a Subset of Patients. Neurodegeneration 1995, 4, 209–216.
- Spreux-Varoquaux, O.; Bensimon, G.; Lacomblez, L.; Salachas, F.; Pradat, P.F.; Le Forestier, N.; Marouan, A.; Dib, M.; Meininger, V. Glutamate Levels in Cerebrospinal Fluid in Amyotrophic Lateral Sclerosis: A Reappraisal Using a New HPLC Method with Coulometric Detection in a Large Cohort of Patients. J. Neurol. Sci. 2002, 193, 73–78.
- Rothstein, J.D.; Martin, L.J.; Kuncl, R.W. Decreased Glutamate Transport by the Brain and Spinal Cord in Amyotrophic Lateral Sclerosis. N. Engl. J. Med. 1992, 326, 1464–1468.
- Lacomblez, L.; Bensimon, G.; Leigh, P.N.; Guillet, P.; Meininger, V. Dose-Ranging Study of Riluzole in Amyotrophic Lateral Sclerosis. Amyotrophic Lateral Sclerosis/Riluzole Study Group II. Lancet 1996, 347, 1425–1431.
- Martin, D.; Thompson, M.A.; Nadler, J.V. The Neuroprotective Agent Riluzole Inhibits Release of Glutamate and Aspartate from Slices of Hippocampal Area CA1. Eur. J. Pharmacol. 1993, 250, 473–476.
- Bensimon, G.; Lacomblez, L.; Meininger, V. A Controlled Trial of Riluzole in Amyotrophic Lateral Sclerosis. ALS/Riluzole Study Group. N. Engl. J. Med. 1994, 330, 585–591.
- Fang, T.; Al Khleifat, A.; Meurgey, J.H.; Jones, A.; Leigh, P.N.; Bensimon, G.; Al-Chalabi, A. Stage at Which Riluzole Treatment Prolongs Survival in Patients with Amyotrophic Lateral Sclerosis: A Retrospective Analysis of Data from a Dose-Ranging Study. Lancet Neurol. 2018, 17, 416–422.
- Kawahara, Y.; Ito, K.; Sun, H.; Aizawa, H.; Kanazawa, I.; Kwak, S. Glutamate Receptors: RNA Editing and Death of Motor Neurons. Nature 2004, 427, 801.
- Hideyama, T.; Yamashita, T.; Aizawa, H.; Tsuji, S.; Kakita, A.; Takahashi, H.; Kwak, S. Profound Downregulation of the RNA Editing Enzyme ADAR2 in ALS Spinal Motor Neurons. Neurobiol. Dis. 2012, 45, 1121–1128.
- Hideyama, T.; Yamashita, T.; Suzuki, T.; Tsuji, S.; Higuchi, M.; Seeburg, P.H.; Takahashi, R.; Misawa, H.; Kwak, S. Induced Loss of ADAR2 Engenders Slow Death of Motor Neurons from Q/R Site-Unedited GluR2. J. Neurosci. 2010, 30, 11917–11925.
- Yamashita, T.; Hideyama, T.; Hachiga, K.; Teramoto, S.; Takano, J.; Iwata, N.; Saido, T.C.; Kwak, S. A Role for Calpain-Dependent Cleavage of TDP-43 in Amyotrophic Lateral Sclerosis Pathology. Nat. Commun. 2012, 3, 1307.
- Yamashita, T.; Kwak, S. The Molecular Link Between Inefficient GluA2 Q/R Site-RNA Editing and TDP-43 Pathology in Motor Neurons of Sporadic Amyotrophic Lateral Sclerosis Patients. Brain Res. 2014, 1584, 28–38.
- Kawahara, Y.; Sun, H.; Ito, K.; Hideyama, T.; Aoki, M.; Sobue, G.; Tsuji, S.; Kwak, S. Underediting of GluR2 mRNA, a Neuronal Death Inducing Molecular Change in Sporadic ALS, Does Not Occur in Motor Neurons in ALS1 or SBMA. Neurosci. Res. 2006, 54, 11–14.
- Zhang, Y.; Xue, W.; Li, X.; Zhang, J.; Chen, S.; Zhang, J.L.; Yang, L.; Chen, L.L. The Biogenesis of Nascent Circular RNAs. Cell Rep. 2016, 15, 611–624.
- Zhang, X.O.; Wang, H.B.; Zhang, Y.; Lu, X.; Chen, L.L.; Yang, L. Complementary Sequence-Mediated Exon Circularization. Cell 2014, 159, 134–147.
- Chung, H.; Calis, J.J.A.; Wu, X.; Sun, T.; Yu, Y.; Sarbanes, S.L.; Dao Thi, V.L.; Shilvock, A.R.; Hoffmann, H.H.; Rosenberg, B.R.; et al. Human ADAR1 Prevents Endogenous RNA from Triggering Translational Shutdown. Cell 2018, 172, 811–824.e14.
- Aktaş, T.; Avşar Ilık, İ.; Maticzka, D.; Bhardwaj, V.; Pessoa Rodrigues, C.; Mittler, G.; Manke, T.; Backofen, R.; Akhtar, A. DHX9 Suppresses RNA Processing Defects Originating from the Alu Invasion of the Human Genome. Nature 2017, 544, 115–119.
- Ma, C.; Wang, X.; Yang, F.; Zang, Y.; Liu, J.; Wang, X.; Xu, X.; Li, W.; Jia, J.; Liu, Z. Circular RNA hsa_circ_0004872 Inhibits Gastric Cancer Progression via the miR-224/Smad4/ADAR1 Successive Regulatory Circuit. Mol. Cancer 2020, 19, 157.
- Omata, Y.; Okawa, M.; Haraguchi, M.; Tsuruta, A.; Matsunaga, N.; Koyanagi, S.; Ohdo, S. RNA Editing Enzyme ADAR1 Controls miR-381-3p-mediated Expression of Multidrug Resistance Protein MRP4 via Regulation of circRNA in Human Renal Cells. J. Biol. Chem. 2022, 298, 102184.
- Hosaka, T.; Yamashita, T.; Teramoto, S.; Hirose, N.; Tamaoka, A.; Kwak, S. ADAR2-Dependent A-to-I RNA Editing in the Extracellular Linear and Circular RNAs. Neurosci. Res. 2019, 147, 48–57.
- Kim, E.; Kim, Y.K.; Lee, S.V. Emerging Functions of Circular RNA in Aging. Trends Genet. 2021, 37, 819–829.
- Kokot, K.E.; Kneuer, J.M.; John, D.; Rebs, S.; Möbius-Winkler, M.N.; Erbe, S.; Müller, M.; Andritschke, M.; Gaul, S.; Sheikh, B.N.; et al. Reduction of A-to-I RNA Editing in the Failing Human Heart Regulates Formation of Circular RNAs. Basic Res. Cardiol. 2022, 117, 32.
- Dolinar, A.; Koritnik, B.; Glavač, D.; Ravnik-Glavač, M. Circular RNAs as Potential Blood Biomarkers in Amyotrophic Lateral Sclerosis. Mol. Neurobiol. 2019, 56, 8052–8062.
- Campos-Melo, D.; Droppelmann, C.A.; He, Z.; Volkening, K.; Strong, M.J. Altered microRNA Expression Profile in Amyotrophic Lateral Sclerosis: A Role in the Regulation of NFL mRNA Levels. Mol. Brain 2013, 6, 26.
- Vrabec, K.; Boštjančič, E.; Koritnik, B.; Leonardis, L.; Dolenc Grošelj, L.; Zidar, J.; Rogelj, B.; Glavač, D.; Ravnik-Glavač, M. Differential Expression of Several miRNAs and the Host Genes AATK and DNM2 in Leukocytes of Sporadic ALS Patients. Front. Mol. Neurosci. 2018, 11, 106.
- Ravnik-Glavač, M.; Glavač, D. Circulating RNAs as Potential Biomarkers in Amyotrophic Lateral Sclerosis. Int. J. Mol. Sci. 2020, 21, 1714.
- Wu, D.P.; Zhao, Y.D.; Yan, Q.Q.; Liu, L.L.; Wei, Y.S.; Huang, J.L. Circular RNAs: Emerging Players in Brain Aging and Neurodegenerative Diseases. J. Pathol. 2023, 259, 1–9.
- Armakola, M.; Higgins, M.J.; Figley, M.D.; Barmada, S.J.; Scarborough, E.A.; Diaz, Z.; Fang, X.; Shorter, J.; Krogan, N.J.; Finkbeiner, S.; et al. Inhibition of RNA Lariat Debranching Enzyme Suppresses TDP-43 Toxicity in ALS Disease Models. Nat. Genet. 2012, 44, 1302–1309.
- Martin, L.J.; Adams, D.A.; Niedzwiecki, M.V.; Wong, M. Aberrant DNA and RNA Methylation Occur in Spinal Cord and Skeletal Muscle of Human SOD1 Mouse Models of ALS and in Human ALS: Targeting DNA Methylation Is Therapeutic. Cells 2022, 11, 3448.
- Yang, L.; Han, B.; Zhang, Z.; Wang, S.; Bai, Y.; Zhang, Y.; Tang, Y.; Du, L.; Xu, L.; Wu, F.; et al. Extracellular Vesicle-Mediated Delivery of Circular RNA SCMH1 Promotes Functional Recovery in Rodent and Nonhuman Primate Ischemic Stroke Models. Circulation 2020, 142, 556–574.
- Yu, X.; Bai, Y.; Han, B.; Ju, M.; Tang, T.; Shen, L.; Li, M.; Yang, L.; Zhang, Z.; Hu, G.; et al. Extracellular Vesicle-Mediated Delivery of circDYM Alleviates CUS-Induced Depressive-Like Behaviours. J. Extracell. Vesicles 2022, 11, e12185.
- Yamashita, T.; Chai, H.L.; Teramoto, S.; Tsuji, S.; Shimazaki, K.; Muramatsu, S.; Kwak, S. Rescue of Amyotrophic Lateral Sclerosis Phenotype in a Mouse Model by Intravenous AAV9-ADAR2 Delivery to Motor Neurons. EMBO Mol. Med. 2013, 5, 1710–1719.




