
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Alberto Falcó | -- | 3747 | 2023-06-16 14:16:56 | | | |
| 2 | Lindsay Dong | -29 word(s) | 3718 | 2023-06-19 03:05:10 | | |
Video Upload Options
The slow discovery of new antibiotics combined with the alarming emergence of antibiotic-resistant bacteria underscores the need for alternative treatments. In this regard, fish skin mucus has been demonstrated to contain a diverse array of bioactive molecules with antimicrobial properties, including peptides, proteins, and other metabolites. This entry aims to provide an overview of the antimicrobial molecules found in fish skin mucus and its reported in vitro antimicrobial capacity against bacteria, fungi, and viruses. Additionally, the different methods of mucus extraction, which can be grouped as aqueous, organic, and acidic extractions, are presented. Finally, omic techniques (genomics, transcriptomics, proteomics, metabolomics, and multiomics) are described as key tools for the identification and isolation of new antimicrobial compounds.
1. Introduction
2. Fish Skin Mucus as a Promising Source of Antimicrobials
3. Composition of Fish Skin Mucus in Innate Immunity Antimicrobial Molecules
3.1. Antimicrobial Peptides (AMPs)
3.2. Proteins
Animal mucosa, in a broad sense, is characterized by the presence of mucins, which are glycosylated proteins responsible for providing viscoelastic and rheological properties, as well as trapping pathogens and contributing to cell surface signaling. Other types of glycoproteins have been found in fish skin mucus. For example, Ebran et al., (2000) [54] isolated and characterized glycoproteins from rainbow trout (Oncorhynchus mykiss), European eel (Anguilla anguilla), and tench (Tinca tinca) skin mucus. These proteins possess both α-helix and random coil structures and show antibacterial activity correlated with pore-forming properties. Transferrin glycoprotein has also been isolated from Atlantic cod [55] and Atlantic salmon (Salmo salar) [56] skin mucus. Transferrin is responsible for iron transporting in absorption, storage, and disposal sites in vertebrates.
Lectins are a diverse class of highly specific carbohydrate-binding proteins [57]. They have been found in the skin mucus of fish, where they provide an external defense mechanism via the agglutination process to stop pathogen penetration and colonization [58]. There are several types of different lectins depending on their structure; for example, C-type lectins, whose binding is dependent on Ca2+, F-type lectins or fucolectins, which are distinguished by their α-l-fucose recognition domain, galectin family or S-type, which require thiol, and pentraxins or pentameric lectins, or P-type lectins, which target glycoproteins containing mannose 6-phosphate [57]. The isolation of C-type lectins has been described in cichlid (Symphysodon aequifasciata) skin mucus [59].
3.3. Other Components
4. Antimicrobial Activity of Fish Skin Mucus
The humoral component of fish skin mucus has been extensively studied for its high content in molecules endowed with antimicrobial properties and, thus, its potential for implementation in biomedical and veterinary applications. Such a variety of compounds has also necessitated the use of different molecular extraction approaches in mucus samples (Figure 1).
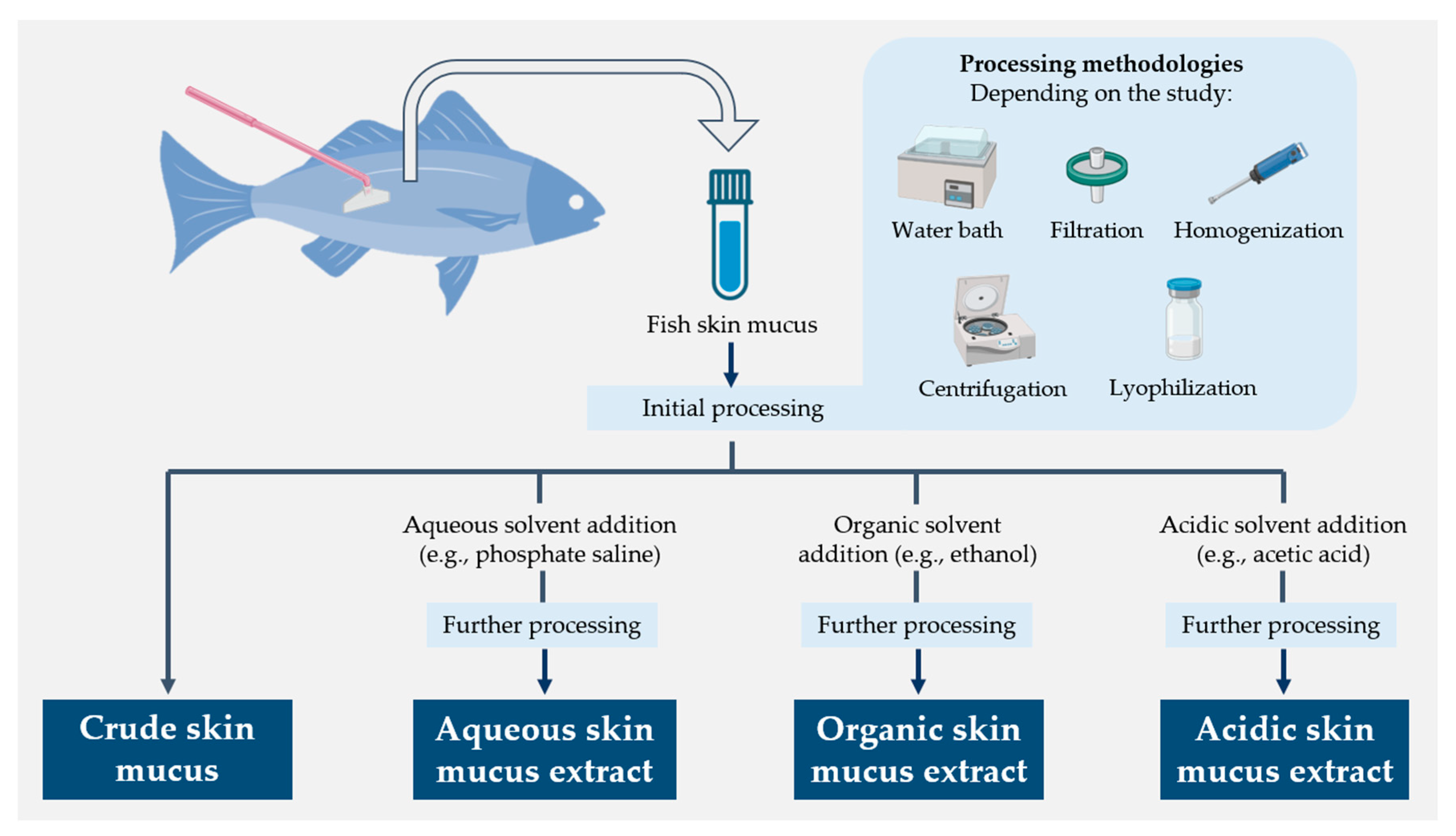
Figure 1. Schematic diagram summarizing the different molecular extraction approaches used for fish skin mucus samples
4.1. Antibacterial Activity of Fish Skin Mucus Extracts
4.1.1. Aqueous Extractions
The most frequently used extraction method was the aqueous one. The most commonly employed solvents in these studies, listed in order of frequency of use, were physiological saline, water, ammonium bicarbonate, and Tris-buffered saline.
Although aqueous extraction was the most popular extraction method, it also showed the least antibacterial activity. This was particularly evident in those experiments where different extraction methods were compared. In some experiments, aqueous extracts did not show any antibacterial activity [72][73][74][75][76][77][78]. For example, Subramanian et al., (2008) [76] found antimicrobial agents, such as lysozyme, cathepsin B, and trypsin-like proteases, in the aqueous skin mucus extracts of several fish species, but they did not exert any antimicrobial activity. Al-Rashed et al., (2018) [72] used aqueous and acidic extracts of the skin mucus of the climbing perch (Anabas testudineus), but only found antibacterial activity in the latter. Similarly, Hellio et al., (2002) [74] performed aqueous and organic extractions of the skin mucus of the ballan wrasse (L. bergylta), but only observed antibacterial activity with the organic extracts. Subhashini et al., (2013) [78] did not find antimicrobial activity in the aqueous extract of the skin mucus of tinfoil barb fish (Barbonymus Schwanenfeldii), even if the amount of protein was higher than in the organic extracts obtained in parallel.
4.1.2. Organic Extractions
For the organic extractions, the most used solvents have been ethanol and dichloromethane. Some authors performed alcoholic extractions and then partitioned distilled water with dichloromethane to obtain aqueous and organic phases [74][78]. High antibacterial activity has been reported for organic fish skin mucus extracts. Indeed, in some studies, all bacteria tested were inhibited, both gram-positive and gram-negative [74][78][79][80]. The main reasons explaining such activity are that (i) the presence of hydrophobic groups is often a common feature of antimicrobial molecules because of their affinity for membranes and their ability to disrupt them [81]; and (ii) these extracts are enriched in hydrophobic molecules because organic solvents favor their isolation by reducing the interactions between hydrophobic groups, which hinders their aggregation [82]. In fact, Mahadevan et al., (2019) [80] obtained greater inhibitory activity against gram-negative and gram-positive bacteria using organic mucus extracts compared to aqueous ones. Hellio et al., (2002) [74] correlated high antimicrobial activity with low polarity of the solvents used; they also showed that extracts from the dichloromethane phase were more active than those from the aqueous phase. In a study by Bergsson et al., (2005) [79], an organic (acetonitrile (ACN) + 1% trifluoroacetic acid (TFA)) extract of cod skin mucus exhibited high antimicrobial activity against gram-positive and gram-negative bacteria. In these extracts, they also identified four peptides with known antimicrobial activity, i.e., those derived from the histone H2B and the 60S ribosomal proteins L40, L36A, and L35.
4.1.3. Acidic Extractions
4.1.4. Crude Mucus
4.2. Antifungal Activity of Fish Skin Mucus Extracts
Several studies have produced mixed results using crude fish skin mucus. On the one hand, the antifungal activity of crude skin mucus of catla, mrigal carp, and European eel inhibited the growth of Aspergillus awamori, Colletotrichum falcatum, and Fusarium oxysporum in the study by Pethkar et al., (2017) [91]. Fuochi et al., (2017) [90] also found that the crude skin mucus of the common stingray was active against Candida albicans, Candida glabrata, and Candida tropicalis. On the other hand, Hisar et al., (2014) [92] tested the crude skin mucus of rainbow trout against C. albicans and Candida parapsilosis, but no antifungal activity was observed. Ikram et al., (2013) [93] screened the antifungal activity of crude and aqueous (i.e., PBS and water) skin mucus of Asian swamp eel (Monopterus albus) against C. albicans, Candida krusei, Cryptococcus neoformans, and Fusarium spp., but only the water extract revealed an inhibitory effect, with activity against all the fungi tested and mostly against Fusarium spp.
4.3. Antiviral Activity of Fish Skin Mucus
5. Omics Techniques as a Promising Tool in Fish Skin Mucus Research
6. Conclusion
Besides being a key component in several physiological functions, fish skin mucus provides an effective chemical and physical barrier against pathogens. The performance of this activity is highly dependent on mucus composition. Therefore, the choice of a suitable molecular extraction method is crucial for its antimicrobial use in other applications. Indeed, notable differences in antimicrobial activity have been shown for the different types of extracts, which is particularly relevant in those studies comparing different extraction methods on the same samples. In general, acidic extracts, followed by organic ones, showed the highest antimicrobial activity. This may be because these procedures favor the isolation of cationic and/or amphipathic antimicrobial compounds, such as AMPs, their enrichment in the final extracts and, apparently, the minimization of molecular inactivation events.
7. Recommendations and Future Perspectives
References
- Spellberg, B.; Guidos, R.; Gilbert, D.; Bradley, J.; Boucher, H.W.; Scheld, W.M.; Bartlett, J.G.; Edwards, J., Jr.; Infectious Diseases Society of America. The epidemic of antibiotic-resistant infections: A call to action for the medical community from the Infectious Diseases Society of America. Clin. Infect. Dis. 2008, 46, 155–164.
- Perlin, D.S.; Rautemaa-Richardson, R.; Alastruey-Izquierdo, A. The global problem of antifungal resistance: Prevalence, mechanisms, and management. Lancet Infect. Dis. 2017, 17, e383–e392.
- Mason, S.; Devincenzo, J.P.; Toovey, S.; Wu, J.Z.; Whitley, R.J. Comparison of antiviral resistance across acute and chronic viral infections. Antivir. Res. 2018, 158, 103–112.
- Butler, M.S.; Paterson, D.L. Antibiotics in the clinical pipeline in October 2019. J. Antibiot. 2020, 73, 329–364.
- Bassetti, M.; Merelli, M.; Temperoni, C.; Astilean, A. New antibiotics for bad bugs: Where are we? Ann. Clin. Microbiol. Antimicrob. 2013, 12, 22.
- Ivanov, M.; Ćirić, A.; Stojković, D. Emerging antifungal targets and strategies. Int. J. Mol. Sci. 2022, 23, 2756.
- Tompa, D.R.; Immanuel, A.; Srikanth, S.; Kadhirvel, S. Trends and strategies to combat viral infections: A review on FDA approved antiviral drugs. Int. J. Biol. Macromol. 2021, 172, 524–541.
- Baker, R.E.; Mahmud, A.S.; Miller, I.F.; Rajeev, M.; Rasambainarivo, F.; Rice, B.L.; Takahashi, S.; Tatem, A.J.; Wagner, C.E.; Wang, L.-F. Infectious disease in an era of global change. Nat. Rev. Microbiol. 2022, 20, 193–205.
- Pawlowski, A.C.; Johnson, J.W.; Wright, G.D. Evolving medicinal chemistry strategies in antibiotic discovery. Curr. Opin. Biotechnol. 2016, 42, 108–117.
- Miethke, M.; Pieroni, M.; Weber, T.; Brönstrup, M.; Hammann, P.; Halby, L.; Arimondo, P.B.; Glaser, P.; Aigle, B.; Bode, H.B. Towards the sustainable discovery and development of new antibiotics. Nat. Rev. Chem. 2021, 5, 726–749.
- Falco, A.; Adamek, M.; Pereiro, P.; Hoole, D.; Encinar, J.A.; Novoa, B.; Mallavia, R. The Immune System of Marine Organisms as Source for Drugs against Infectious Diseases. Mar. Drugs 2022, 20, 363.
- Hagström, A.k.; Pommier, T.; Rohwer, F.; Simu, K.; Stolte, W.; Svensson, D.; Zweifel, U.L. Use of 16S ribosomal DNA for delineation of marine bacterioplankton species. Appl. Environ. Microbiol. 2002, 68, 3628–3633.
- Margulis, L.; Chapman, M.J. Kingdoms and Domains: An Illustrated Guide to the Phyla of Life on Earth; Academic Press: Cambridge, MA, USA, 2009.
- Kong, D.-X.; Jiang, Y.-Y.; Zhang, H.-Y. Marine natural products as sources of novel scaffolds: Achievement and concern. Drug Discov. Today 2010, 15, 884–886.
- Harizani, M.; Ioannou, E.; Roussis, V. The Laurencia paradox: An endless source of chemodiversity. Prog. Chem. Org. Nat. Prod. 2016, 102, 91–252.
- Suttle, C.A. Marine viruses—Major players in the global ecosystem. Nat. Rev. Microbiol. 2007, 5, 801–812.
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2019, 36, 122–173.
- Ntie-Kang, F.; Svozil, D. An enumeration of natural products from microbial, marine and terrestrial sources. Phys. Sci. Rev. 2020, 5.
- Principe, P.P.; Fisher, W.S. Spatial distribution of collections yielding marine natural products. J. Nat. Prod. 2018, 81, 2307–2320.
- Agrawal, S.; Adholeya, A.; Deshmukh, S.K. The pharmacological potential of non-ribosomal peptides from marine sponge and tunicates. Front. Pharmacol. 2016, 7, 333.
- Brinchmann, M.F. Immune relevant molecules identified in the skin mucus of fish using -omics technologies. Mol. BioSyst. 2016, 12, 2056–2063.
- Reverter, M.; Tapissier-Bontemps, N.; Lecchini, D.; Banaigs, B.; Sasal, P. Biological and Ecological Roles of External Fish Mucus: A Review. Fishes 2018, 3, 41.
- Tiralongo, F.; Messina, G.; Lombardo, B.M.; Longhitano, L.; Li Volti, G.; Tibullo, D. Skin mucus of marine fish as a source for the development of antimicrobial agents. Front. Mar. Sci. 2020, 7, 760.
- Azam, F.; Malfatti, F. Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 2007, 5, 782–791.
- Bergh, Ø.; Børsheim, K.Y.; Bratbak, G.; Heldal, M. High abundance of viruses found in aquatic environments. Nature 1989, 340, 467–468.
- Ducklow, H. Bacterial production and biomass in the oceans. Microb. Ecol. Oceans 2000, 1, 85–120.
- Grossart, H.-P.; Van den Wyngaert, S.; Kagami, M.; Wurzbacher, C.; Cunliffe, M.; Rojas-Jimenez, K. Fungi in aquatic ecosystems. Nat. Rev. Microbiol. 2019, 17, 339–354.
- Castro, R.; Tafalla, C. Overview of fish immunity. In Mucosal Health in Aquaculture; Elsevier: Amsterdam, The Netherlands, 2015; pp. 3–54.
- Ponnappan, N.; Budagavi, D.P.; Yadav, B.K.; Chugh, A. Membrane-active peptides from marine organisms-antimicrobials, cell-penetrating peptides and Peptide toxins: Applications and prospects. Probiotics Antimicrob. Proteins 2015, 7, 75–89.
- Litman, G.W.; Rast, J.P.; Fugmann, S.D. The origins of vertebrate adaptive immunity. Nat. Rev. Immunol. 2010, 10, 543–553.
- Esteban, M.Á.; Cerezuela, R. Fish mucosal immunity: Skin. In Mucosal Health in Aquaculture; Elsevier: Amsterdam, The Netherlands, 2015; pp. 67–92.
- Salinas, I. The mucosal immune system of teleost fish. Biology 2015, 4, 525–539.
- Ellis, A. Innate host defense mechanisms of fish against viruses and bacteria. Dev. Comp. Immunol. 2001, 25, 827–839.
- Gomez, D.; Sunyer, J.O.; Salinas, I. The mucosal immune system of fish: The evolution of tolerating commensals while fighting pathogens. Fish Shellfish Immunol. 2013, 35, 1729–1739.
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial peptides: An emerging category of therapeutic agents. Front. Cell. Infect. Microbiol. 2016, 6, 194.
- Hancock, R.E.; Haney, E.F.; Gill, E.E. The immunology of host defence peptides: Beyond antimicrobial activity. Nat. Rev. Immunol. 2016, 16, 321.
- Schroeder, B.O.; Wu, Z.; Nuding, S.; Groscurth, S.; Marcinowski, M.; Beisner, J.; Buchner, J.; Schaller, M.; Stange, E.F.; Wehkamp, J. Reduction of disulphide bonds unmasks potent antimicrobial activity of human β-defensin 1. Nature 2011, 469, 419–423.
- Haag, A.F.; Kerscher, B.; Dall’Angelo, S.; Sani, M.; Longhi, R.; Baloban, M.; Wilson, H.M.; Mergaert, P.; Zanda, M.; Ferguson, G.P. Role of cysteine residues and disulfide bonds in the activity of a legume root nodule-specific, cysteine-rich peptide. J. Biol. Chem. 2012, 287, 10791–10798.
- Falco, A.; Ortega-Villaizan, M.; Chico, V.; Brocal, I.; Perez, L.; Coll, J.M.; Estepa, A. Antimicrobial peptides as model molecules for the development of novel antiviral agents in aquaculture. Mini Rev. Med. Chem. 2009, 9, 1159–1164.
- Masso-Silva, J.A.; Diamond, G. Antimicrobial peptides from fish. Pharmaceuticals 2014, 7, 265–310.
- Robinette, D.; Wada, S.; Arroll, T.; Levy, M.G.; Miller, W.L.; Noga, E.J. Antimicrobial activity in the skin of the channel catfish Ictalurus punctatus: Characterization of broad-spectrum histone-like antimicrobial proteins. Cell. Mol. Life Sci. CMLS 1998, 54, 467–475.
- Kim, H.S.; Park, C.B.; Kim, M.S.; Kim, S.C. cDNA cloning and characterization of buforin I, an antimicrobial peptide: A cleavage product of histone H2A. Biochem. Biophys. Res. Commun. 1996, 229, 381–387.
- Fernandes, J.M.O.; Kemp, G.D.; Molle, M.G.; Smith, V.J. Anti-microbial properties of histone H2A from skin secretions of rainbow trout, Oncorhynchus mykiss. Biochem. J. 2002, 368, 611–620.
- Birkemo, G.A.; Lüders, T.; Andersen, Ø.; Nes, I.F.; Nissen-Meyer, J. Hipposin, a histone-derived antimicrobial peptide in Atlantic halibut (Hippoglossus hippoglossus L.). Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2003, 1646, 207–215.
- Park, I.Y.; Park, C.B.; Kim, M.S.; Kim, S.C. Parasin I, an antimicrobial peptide derived from histone H2A in the catfish, Parasilurus asotus. FEBS Lett. 1998, 437, 258–262.
- Cho, J.H.; Park, I.Y.; Kim, H.S.; Lee, W.T.; Kim, M.S.; Kim, S.C. Cathepsin D produces antimicrobial peptide parasin I from histone H2A in the skin mucosa of fish. FASEB J. 2002, 16, 429–431.
- Valero, Y.; Chaves-Pozo, E.; Meseguer, J.; Esteban, M.; Cuesta, A. Biologial Role of Fish Antimicrobial Peptides. In Antimicrobial Peptides; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2013; pp. 31–60.
- Fernandes, J.M.O.; Saint, N.; Kemp, G.D.; Smith, V.J. Oncorhyncin III: A potent antimicrobial peptide derived from the non-histone chromosomal protein H6 of rainbow trout, Oncorhynchus mykiss. Biochem. J. 2003, 373, 621–628.
- Subramanian, S.; Ross, N.W.; MacKinnon, S.L. Myxinidin, A Novel Antimicrobial Peptide from the Epidermal Mucus of Hagfish, Myxine glutinosa L. Mar. Biotechnol. 2009, 11, 748.
- Lazarovici, P.; Primor, N.; Loew, L.M. Purification and pore-forming activity of two hydrophobic polypeptides from the secretion of the Red Sea Moses sole (Pardachirus marmoratus). J. Biol. Chem. 1986, 261, 16704–16713.
- Oren, Z.; Shai, Y. A Class of Highly Potent Antibacterial Peptides Derived from Pardaxin, A Pore-Forming Peptide Isolated from Moses Sole Fish Pardachirus marmoratus. Eur. J. Biochem. 1996, 237, 303–310.
- Su, Y. Isolation and identification of pelteobagrin, a novel antimicrobial peptide from the skin mucus of yellow catfish (Pelteobagrus fulvidraco). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2011, 158, 149–154.
- Bae, J.-S.; Jung, J.-M.; An, C.M.; Kim, J.-W.; Hwang, S.D.; Kwon, M.-G.; Park, M.-A.; Kim, M.-C.; Park, C.-I. Piscidin: Antimicrobial peptide of rock bream, Oplegnathus fasciatus. Fish Shellfish Immunol. 2016, 51, 136–142.
- Ebran, N.; Julien, S.; Orange, N.; Auperin, B.; Molle, G. Isolation and characterization of novel glycoproteins from fish epidermal mucus: Correlation between their pore-forming properties and their antibacterial activities. Biochim. Biophys. Acta (BBA)-Biomembr. 2000, 1467, 271–280.
- Easy, R.H.; Trippel, E.A.; Burt, M.D.B.; Cone, D.K. Identification of transferrin in Atlantic cod Gadus morhua epidermal mucus. J. Fish Biol. 2012, 81, 2059–2063.
- Ræder, I.L.U.; Paulsen, S.M.; Smalås, A.O.; Willassen, N.P. Effect of fish skin mucus on the soluble proteome of Vibrio salmonicida analysed by 2-D gel electrophoresis and tandem mass spectrometry. Microb. Pathog. 2007, 42, 36–45.
- Raposo, C.D.; Canelas, A.B.; Barros, M.T. Human Lectins, Their Carbohydrate Affinities and Where to Find Them. Biomolecules 2021, 11, 188.
- Suzuki, Y.; Tasumi, S.; Tsutsui, S.; Okamoto, M.; Suetake, H. Molecular diversity of skin mucus lectins in fish. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2003, 136, 723–730.
- Chong, K.; Joshi, S.; Jin, L.T.; Shu-Chien, A.C. Proteomics profiling of epidermal mucus secretion of a cichlid (Symphysodon aequifasciata) demonstrating parental care behavior. Proteomics 2006, 6, 2251–2258.
- Lazado, C.C.; Skov, P.V. Secretory Proteins in the Skin Mucus of Nile Tilapia (Oreochromis niloticus) are Modulated Temporally by Photoperiod and Bacterial Endotoxin Cues. Fishes 2019, 4, 57.
- Nigam, A.K.; Kumari, U.; Mittal, S.; Mittal, A.K. Comparative analysis of innate immune parameters of the skin mucous secretions from certain freshwater teleosts, inhabiting different ecological niches. Fish Physiol. Biochem. 2012, 38, 1245–1256.
- Ren, T.; Koshio, S.; Ishikawa, M.; Yokoyama, S.; Micheal, F.R.; Uyan, O.; Tung, H.T. Influence of dietary vitamin C and bovine lactoferrin on blood chemistry and non-specific immune responses of Japanese eel, Anguilla japonica. Aquaculture 2007, 267, 31–37.
- Subramanian, S.; MacKinnon, S.L.; Ross, N.W. A comparative study on innate immune parameters in the epidermal mucus of various fish species. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2007, 148, 256–263.
- Esteban, M.Á. An Overview of the Immunological Defenses in Fish Skin. ISRN Immunol. 2012, 2012, 853470.
- Hjelmeland, K.; Christie, M.; Raa, J. Skin mucus protease from rainbow trout, Salmo gairdneri Richardson, and its biological significance. J. Fish Biol. 1983, 23, 13–22.
- Aranishi, F.; Nakane, M. Epidermal proteases of the Japanese eel. Fish Physiol. Biochem. 1997, 16, 471–478.
- Aranishi, F.; Nakane, M. Epidermal Proteinases in the European Eel. Physiol. Zool. 1997, 70, 563–570.
- Firth, K.; Johnson, S.; Ross, N. Characterisation of proteases in the skin mucus of Atlantic Salmon (Salmo salar) infected with the Salmon louse (Lepeophtheirus salmonis) and in whole-body louse homogenate. J. Parasitol. 2001, 86, 1199–1205.
- Balasubramanian, S.; Gunasekaran, G. Fatty acids and amino acids composition in skin epidermal mucus of selected fresh water fish mugil cephalus. World J. Pharm. Pharm. Sci. 2015, 4, 1275–1287.
- Torrecillas, S.; Montero, D.; Domínguez, D.; Robaina, L.; Izquierdo, M. Skin Mucus Fatty Acid Composition of Gilthead Sea Bream (Sparus Aurata): A Descriptive Study in Fish Fed Low and High Fish Meal Diets. Fishes 2019, 4, 15.
- Lewis, R.W. Fish cutaneous mucus: A new source of skin surface lipid. Lipids 1970, 5, 947–949.
- Al-Rasheed, A.; Handool, K.O.; Garba, B.; Noordin, M.M.; Bejo, S.K.; Kamal, F.M.; Daud, H.H.M. Crude extracts of epidermal mucus and epidermis of climbing perch Anabas testudineus and its antibacterial and hemolytic activities. Egypt. J. Aquat. Res. 2018, 44, 125–129.
- Manikantan, G.; Lyla, S.; Khan, S.A.; Vijayanand, P.; Edward, G.; Jothi, G. Bioactive potency of epidermal mucus extracts from greasy grouper, Epinephelus tauvina (Forsskal, 1775). J. Coast. Life Med. 2016, 4, 510–520.
- Hellio, C.; Pons, A.M.; Beaupoil, C.; Bourgougnon, N.; Gal, Y.L. Antibacterial, antifungal and cytotoxic activities of extracts from fish epidermis and epidermal mucus. Int. J. Antimicrob. Agents 2002, 20, 214–219.
- Katra, N.; Hisar, O.; Karatas, S.; Turgay, E.; Sarvan, C.; KATRA, N. In vitro antimicrobial activities of extracts from ballan wrasse (Labrus bergylta) skin mucus. Mar. Sci. Technol. Bull. 2016, 5, 13–15.
- Subramanian, S.; Ross, N.W.; MacKinnon, S.L. Comparison of antimicrobial activity in the epidermal mucus extracts of fish. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2008, 150, 85–92.
- García-Marciano, M.; Apún-Molina, J.P.; Sainz-Hernández, J.C.; Santamaría-Miranda, A.; Medina-Godoy, S.; Aguiñaga-Cruz, J.A. Antibacterial activity evaluation of the nile tilapia Oreochromis niloticus (Linnaeus, 1758) skin mucus, against vibrio bacteria affecting the white shrimp Penaeus vannamei. Lat. Am. J. Aquat. Res. 2019, 47, 580–585.
- Subhashini, S.; Lavanya, J.; Jain, S.; Agihotri, T. Screening of Antibacterial and Cytotoxic Activity of Extracts from Epidermis and Epidermal Mucus of Barbonymus Schwanenfeldii (Tinfoil Barb Fish). Int. J. Res. Eng. Technol. 2013, 2, 492–497.
- Bergsson, G.; Agerberth, B.; Jörnvall, H.; Gudmundsson, G.H. Isolation and identification of antimicrobial components from the epidermal mucus of Atlantic cod (Gadus morhua). FEBS J. 2005, 272, 4960–4969.
- Mahadevan, G.; Mohan, K.; Vinoth, J.; Ravi, V. Biotic potential of mucus extracts of giant mudskipper Periophthalmodon schlosseri (Pallas, 1770) from Pichavaram, southeast coast of India. J. Basic Appl. Zool. 2019, 80, 13.
- Yount, N.Y.; Bayer, A.S.; Xiong, Y.Q.; Yeaman, M.R. Advances in antimicrobial peptide immunobiology. Pept. Sci. Orig. Res. Biomol. 2006, 84, 435–458.
- Afkarian, M.; Bhasin, M.; Dillon, S.T.; Guerrero, M.C.; Nelson, R.G.; Knowler, W.C.; Thadhani, R.; Libermann, T.A. Optimizing a proteomics platform for urine biomarker discovery. Mol. Cell. Proteom. 2010, 9, 2195–2204.
- Patel, M.; Ashraf, M.S.; Siddiqui, A.J.; Ashraf, S.A.; Sachidanandan, M.; Snoussi, M.; Adnan, M.; Hadi, S. Profiling and Role of Bioactive Molecules from Puntius sophore (Freshwater/Brackish Fish) Skin Mucus with Its Potent Antibacterial, Antiadhesion, and Antibiofilm Activities. Biomolecules 2020, 10, 920.
- Wei, O.Y.; Xavier, R.; Marimuthu, K. Screening of antibacterial activity of mucus extract of snakehead fish, Channa striatus. Eur. Rev. Med. Pharmacol. Sci. 2010, 14, 675–681.
- Lirio, G.A.C.; De Leon, J.A.A.; Villafuerte, A.G. Antimicrobial Activity of Epidermal Mucus from Top Aquaculture Fish Species against Medically-Important Pathogens. Walailak J. Sci. Technol. 2018, 16, 329–340.
- Kumari, U.; Nigam, A.K.; Mittal, S.; Mittal, A.K. Antibacterial properties of the skin mucus of the freshwater fishes, Rita rita and Channa punctatus. Eur. Rev. Med. Pharmacol. Sci. 2011, 15, 781–786.
- Nigam, A.K.; Kumari, U.; Mittal, S.; Mittal, A.K. Evaluation of antibacterial activity and innate immune components in skin mucus of Indian major carp, Cirrhinus mrigala. Aquac. Res. 2017, 48, 407–418.
- Ming, L.; Xiaoling, P.; Yan, L.; Lili, W.; Qi, W.; Xiyong, Y.; Boyao, W.; Ning, H. Purification of antimicrobial factors from human cervical mucus. Hum. Reprod. 2007, 22, 1810–1815.
- Sanahuja, I.; Fernández-Alacid, L.; Ordóñez-Grande, B.; Sánchez-Nuño, S.; Ramos, A.; Araujo, R.M.; Ibarz, A. Comparison of several non-specific skin mucus immune defences in three piscine species of aquaculture interest. Fish Shellfish Immunol. 2019, 89, 428–436.
- Fuochi, V.; Li Volti, G.; Camiolo, G.; Tiralongo, F.; Giallongo, C.; Distefano, A.; Petronio Petronio, G.; Barbagallo, I.; Viola, M.; Furneri, P.M.; et al. Antimicrobial and Anti-Proliferative Effects of Skin Mucus Derived from Dasyatis pastinaca (Linnaeus, 1758). Mar. Drugs 2017, 15, 342.
- Pethkar, M.R.; Lokhande, M.V. Antifungal activity of skin mucus of three cultivable fish species. Int. J. Zool. Stud. 2017, 2, 1–3.
- Hisar, O.; Hisar, S.A.; Uyanik, M.H.; Sahin, T.; Cakir, F.; Yilmaz, S. In vitro antimicrobial and antifungal activities of aqueous skin mucus from rainbow trout (Oncorhynchus mykiss) on human pathogens. Mar. Sci. Technol. Bull. 2014, 3, 19–22.
- Ikram, M.; Ridzwan, B.H. A preliminary screening of antifungal activities from skin mucus extract of Malaysian local swamp eel (Monopterus albus). Int. Res. J. Pharm. 2013, 3, 1–8.
- Raj, V.S.; Fournier, G.; Rakus, K.; Ronsmans, M.; Ouyang, P.; Michel, B.; Delforges, C.; Costes, B.; Farnir, F.; Leroy, B. Skin mucus of Cyprinus carpio inhibits cyprinid herpesvirus 3 binding to epidermal cells. Vet. Res. 2011, 42, 1–10.
- Valero, Y.; Arizcun, M.; Cortes, J.; Ramirez-Cepeda, F.; Guzman, F.; Mercado, L.; Esteban, M.Á.; Chaves-Pozo, E.; Cuesta, A. NK-lysin, dicentracin and hepcidin antimicrobial peptides in European sea bass. Ontogenetic development and modulation in juveniles by nodavirus. Dev. Comp. Immunol. 2020, 103, 103516.
- Falco, A.; Medina-Gali, R.M.; Poveda, J.A.; Bello-Perez, M.; Novoa, B.; Encinar, J.A. Antiviral activity of a Turbot (Scophthalmus maximus) NK-lysin peptide by inhibition of low-pH virus-induced membrane fusion. Mar. Drugs 2019, 17, 87.
- Casadei, E.; Wang, T.; Zou, J.; Vecino, J.L.G.; Wadsworth, S.; Secombes, C.J. Characterization of three novel β-defensin antimicrobial peptides in rainbow trout (Oncorhynchus mykiss). Mol. Immunol. 2009, 46, 3358–3366.
- Falco, A.; Chico, V.; Marroqui, L.; Perez, L.; Coll, J.; Estepa, A. Expression and antiviral activity of a β-defensin-like peptide identified in the rainbow trout (Oncorhynchus mykiss) EST sequences. Mol. Immunol. 2008, 45, 757–765.
- Rodrigues, P.M.; Silva, T.S.; Dias, J.; Jessen, F. PROTEOMICS in aquaculture: Applications and trends. J. Proteom. 2012, 75, 4325–4345.
- Ao, J.; Mu, Y.; Xiang, L.-X.; Fan, D.; Feng, M.; Zhang, S.; Shi, Q.; Zhu, L.-Y.; Li, T.; Ding, Y.; et al. Genome sequencing of the perciform fish Larimichthys crocea provides insights into molecular and genetic mechanisms of stress adaptation. PLoS Genet. 2015, 11, e1005118.
- Greer, J.B.; Andrzejczyk, N.E.; Mager, E.M.; Stieglitz, J.D.; Benetti, D.; Grosell, M.; Schlenk, D. Whole-Transcriptome Sequencing of Epidermal Mucus as a Novel Method for Oil Exposure Assessment in Juvenile Mahi-Mahi (Coryphaena hippurus). Environ. Sci. Technol. Lett. 2019, 6, 538–544.
- Nissa, M.U.; Pinto, N.; Parkar, H.; Goswami, M.; Srivastava, S. Proteomics in fisheries and aquaculture: An approach for food security. Food Control 2021, 127, 108125.
- Jurado, J.; Fuentes-Almagro, C.A.; Guardiola, F.A.; Cuesta, A.; Esteban, M.Á.; Prieto-Álamo, M.-J. Proteomic profile of the skin mucus of farmed gilthead seabream (Sparus aurata). J. Proteom. 2015, 120, 21–34.
- Rajan, B.; Fernandes, J.M.O.; Caipang, C.M.A.; Kiron, V.; Rombout, J.H.W.M.; Brinchmann, M.F. Proteome reference map of the skin mucus of Atlantic cod (Gadus morhua) revealing immune competent molecules. Fish Shellfish Immunol. 2011, 31, 224–231.
- Minniti, G.; Rød Sandve, S.; Padra, J.T.; Heldal Hagen, L.; Lindén, S.; Pope, P.B.; Arntzen, M.Ø.; Vaaje-Kolstad, G. The Farmed Atlantic Salmon (Salmo salar) Skin-Mucus Proteome and Its Nutrient Potential for the Resident Bacterial Community. Genes 2019, 10, 515.
- Liu, H.-h.; Sun, Q.; Jiang, Y.-t.; Fan, M.-h.; Wang, J.-x.; Liao, Z. In-depth proteomic analysis of Boleophthalmus pectinirostris skin mucus. J. Proteom. 2019, 200, 74–89.
- Chong, K.; Sock Ying, T.; Foo, J.; Toong Jin, L.; Chong, A. Characterisation of proteins in epidermal mucus of discus fish (Symphysodon spp.) during parental phase. Aquaculture 2005, 249, 469–476.
- Cordero, H.; Brinchmann, M.F.; Cuesta, A.; Meseguer, J.; Esteban, M.A. Skin mucus proteome map of European sea bass (Dicentrarchus labrax). Proteomics 2015, 15, 4007–4020.
- Patel, D.M.; Brinchmann, M.F. Skin mucus proteins of lumpsucker (Cyclopterus lumpus). Biochem. Biophys. Rep. 2017, 9, 217–225.
- Cordero, H.; Brinchmann, M.F.; Cuesta, A.; Esteban, M.A. Chronic wounds alter the proteome profile in skin mucus of farmed gilthead seabream. BMC Genom. 2017, 18, 939.
- Rajan, B.; Lokesh, J.; Kiron, V.; Brinchmann, M.F. Differentially expressed proteins in the skin mucus of Atlantic cod (Gadus morhua) upon natural infection with Vibrio anguillarum. BMC Vet. Res. 2013, 9, 103.
- Xiong, Y.; Dan, C.; Ren, F.; Su, Z.; Zhang, Y.; Mei, J. Proteomic profiling of yellow catfish (Pelteobagrus fulvidraco) skin mucus identifies differentially-expressed proteins in response to Edwardsiella ictaluri infection. Fish Shellfish Immunol. 2020, 100, 98–108.
- Fernández-Montero, Á.; Torrecillas, S.; Montero, D.; Acosta, F.; Prieto-Álamo, M.-J.; Abril, N.; Jurado, J. Proteomic profile and protease activity in the skin mucus of greater amberjack (Seriola dumerili) infected with the ectoparasite Neobenedenia girellae—An immunological approach. Fish Shellfish Immunol. 2021, 110, 100–115.
- Pérez-Sánchez, J.; Terova, G.; Simó-Mirabet, P.; Rimoldi, S.; Folkedal, O.; Calduch-Giner, J.A.; Olsen, R.E.; Sitjà-Bobadilla, A. Skin Mucus of Gilthead Sea Bream (Sparus aurata L.). Protein Mapping and Regulation in Chronically Stressed Fish. Front. Physiol. 2017, 8, 34.
- Fæste, C.K.; Tartor, H.; Moen, A.; Kristoffersen, A.B.; Dhanasiri, A.K.S.; Anonsen, J.H.; Furmanek, T.; Grove, S. Proteomic profiling of salmon skin mucus for the comparison of sampling methods. J. Chromatogr. B 2020, 1138, 121965.
- Goodacre, R.; Vaidyanathan, S.; Dunn, W.B.; Harrigan, G.G.; Kell, D.B. Metabolomics by numbers: Acquiring and understanding global metabolite data. Trends Biotechnol. 2004, 22, 245–252.
- Samuelsson, L.M.; Larsson, D.G.J. Contributions from metabolomics to fish research. Mol. BioSyst. 2008, 4, 974–979.





