Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Peter N. Lipke | -- | 3100 | 2023-06-15 21:19:15 | | | |
| 2 | Lindsay Dong | Meta information modification | 3100 | 2023-06-16 05:07:33 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Lipke, P.N.; Ragonis-Bachar, P. Multifunctionality in Microbial Adhesins. Encyclopedia. Available online: https://encyclopedia.pub/entry/45682 (accessed on 07 February 2026).
Lipke PN, Ragonis-Bachar P. Multifunctionality in Microbial Adhesins. Encyclopedia. Available at: https://encyclopedia.pub/entry/45682. Accessed February 07, 2026.
Lipke, Peter N., Peleg Ragonis-Bachar. "Multifunctionality in Microbial Adhesins" Encyclopedia, https://encyclopedia.pub/entry/45682 (accessed February 07, 2026).
Lipke, P.N., & Ragonis-Bachar, P. (2023, June 15). Multifunctionality in Microbial Adhesins. In Encyclopedia. https://encyclopedia.pub/entry/45682
Lipke, Peter N. and Peleg Ragonis-Bachar. "Multifunctionality in Microbial Adhesins." Encyclopedia. Web. 15 June, 2023.
Copy Citation
Microbial adhesins have multiple functions, and these activities are all evolved and selected. Adhesins can act as enzymes, as assembly scaffolds and components of complex nano-machines. Sometimes, these activities are called secondary because they were discovered secondarily. For instance, microbial type IV pili were first called adhesive. In contrast, phosphoglycerate kinase has its well-known enzymatic activity, but in fungi it also moonlights as an extracellular adhesin.
bacterial cell wall
fungal cell wall
koff
multidomain protein
moonlighting protein
1. Multiple Activities of Microbial Adhesins
1.1. What Is an Adhesin?
This question has a simple answer: a cell surface protein that binds one cell to another or binds a cell to a substrate. This definition encompasses two types of proteins: professional, dedicated adhesins and ‘moonlighting’ proteins that are displayed on a cell surface and happen to bind to a partner on another cell. Both types of adhesin are multifunctional and are found on the surface of both prokaryotic and eukaryotic microbes. For microbes, many of these adhesins are involved in microbe-host interactions, in biofilm formation and maintenance, or both. This essay concentrates on those adhesins that have cell–cell interaction activity. Nevertheless, many of these adhesins also bind microbes to biotic and abiotic surfaces.
1.2. Biofilms
Biofilms are often considered to be the natural state of microbial growth and maintenance. They are organized communities in which cells are differentiated and adapted to specific metabolic and adhesive roles. Cells in both bacterial and fungal biofilms are also characterized by high resistance to antibiotics and antifungals [1][2]. Biofilms are also resistant to nematode predation, a property conferred by the adhesins [3][4]. Biofilms include at least four different morphologies: amorphous layers on surfaces; surface layers with perpendicular towers or mushroom-shaped constructs; well-organized mats of cells with regular wrinkles and crenellations; and pellicles, biofilms formed at air-water interfaces [1][5]. Within each biofilm morphology, individual cells have different specialized roles dependent on specific patterns of gene expression [6][7][8].
1.3. Multiple Activities of Microbial Adhesins
Minimally, each adhesin has three activities (Figure 1):
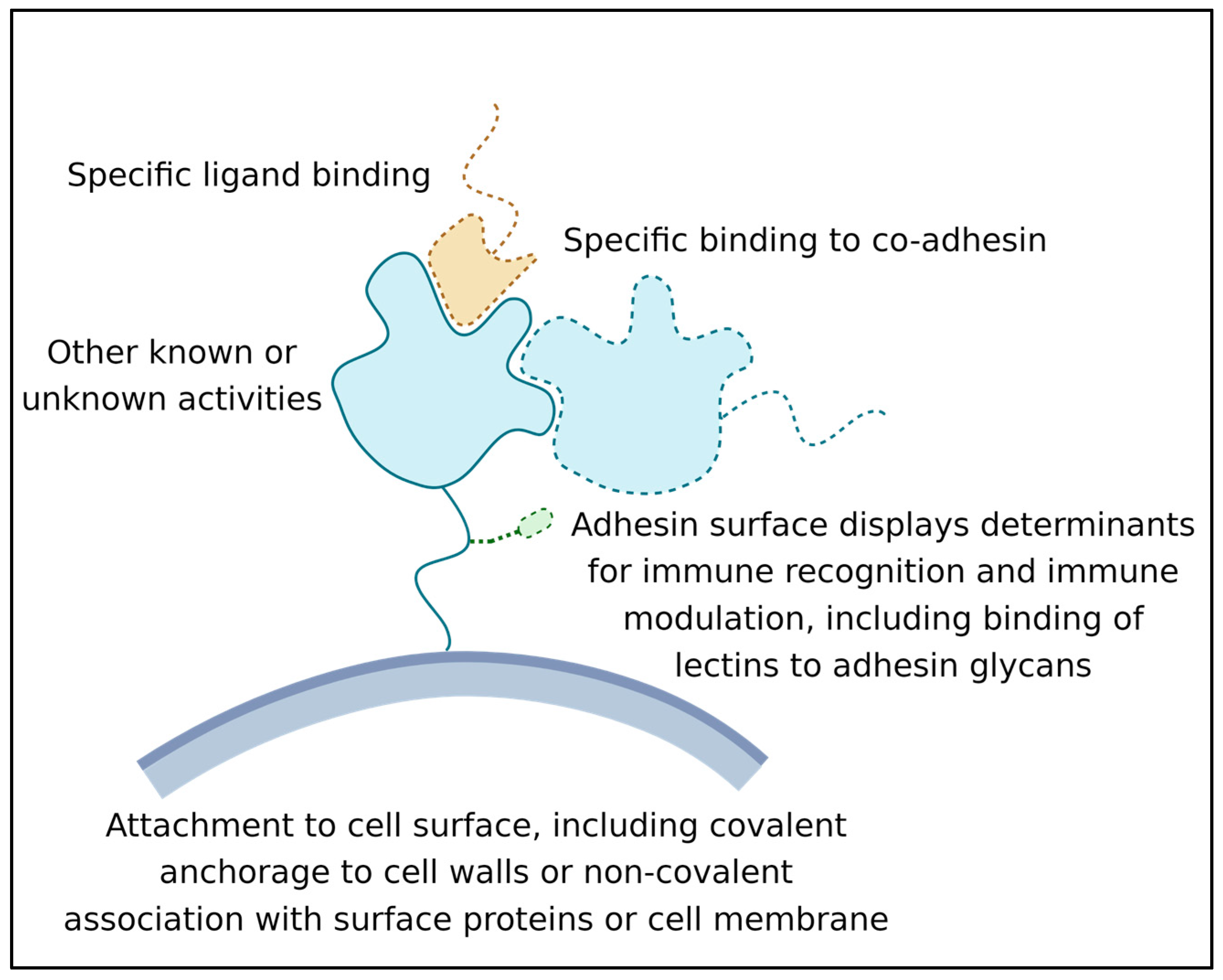
Figure 1. Some general features and activities of microbial adhesins. Adhesins (blue with a green glycosylation) are bound to cell wall (grey). The ligand is shown in tan.
-
Its ability to recognize and bind a ligand or other binding partner. This binding must last long enough for the adhesion to be biologically relevant; often this means that the adhesive bond lasts for hours or even years [9];
-
Its attachment to the surface of the expressing cell. Attachment can be through covalent binding to the cell wall, membrane embedment, or association with another wall-attached protein [10];
-
Its activity as a surface marker: This activity is crucial in microbe-host interactions, and many adhesins are also immune modulators. Adhesin structures are bound by immune effectors and by other cell signal receptors. This binding leads to the modulation of host responses. In addition, biofilm adhesins mediate microbe–microbe associations, which induce changes in microbial cellular physiology [6][7][11]. This activity can be due to direct signaling between adhesins, or indirect due to adhesin-induced long-term increases in population density [12][13].
1.4. Time and Adhesin Activity
A simple definition of an adhesin is a protein that mediates direct binding of one cell to another. By convention, the definition excludes many intercellular signaling complexes that induce cellular differentiation or alter cellular activity within a few seconds. The timing is somewhat flexible: for instance, lectin binding is brief and allows lymphocytes to roll along the endothelium as successive molecules bind and dissociate [14]; on the other hand, the adhesins that mediate bacterial mating are active for 20 min to an hour [15]. There are also instances where continued adhesion leads to continuing signaling that alters cell fate: in yeast mating, responses to sex pheromone include changes in gene expression after a few seconds of exposure, but the ability to elicit a mating process requires at least an hour of response to high pheromone concentration, and this response requires continued cell–cell adhesion [16][17].
1.5. Adhesion Assays
The gold standard assays show that a specific protein is both necessary and sufficient for the adhesion of cells to each other or to a substrate. Genetically, these properties are often established by showing that adhesion is lost when a specific gene is mutated (necessity demonstrated by loss of function, LoF), and, conversely, that expression of the protein in a heterologous system confers adhesion activity (sufficiency shown by gain of function) [18]. In practice, LoF mutations are often used to identify putative adhesins, but they can be confounded by other activities of the proteins (for instance, several putative adhesins are transcription factors, or inducers of transcription regulation, and deletions cause failure to express other cell-surface adhesins [19]).
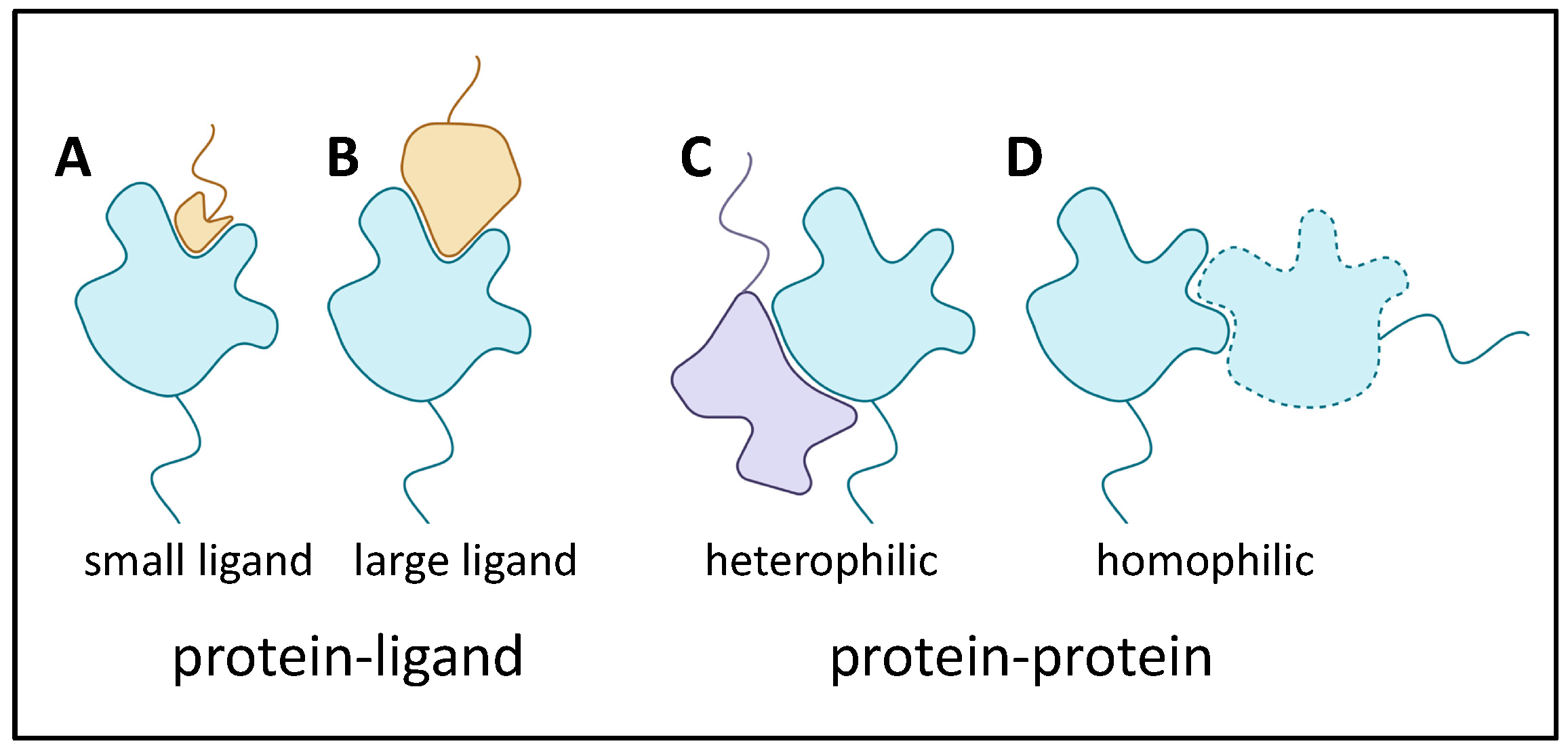
1.6. Characteristics of Adhesin Binding
1.6.1. Adhesin Ligands and Partners
There are many ways that adhesins bind to target proteins. In fact, the standard ligand receptor terminology is often inappropriate when both binders are adhesins that contribute large binding surfaces. Thus, Figure 2 shows several examples. In (A), a protein with a well-folded domain incorporating a well-defined pocket binds to a small segment of a heterologous adhesin. This cartoon is like a classic receptor-ligand model, and an example is the Saccharomyces cerevisiae mating adhesin alpha-agglutinin, which binds the C-terminal residues of its partner, a-agglutinin subunit Aga2. The existence of such binding pockets is, of course, analogous to active sites on most enzymes. In (B) and (C), heterologous adhesins bind through interactions of larger portions of the domain surfaces. Finally, in (D), there is extensive interaction between two identical adhesins to show homophilic binding.

Figure 2. Adhesin binding to ligands and binding partners. Colors are the same Figure 1, with a heterophilic binding partner (lavender in C).
1.6.2. Adhesin Bonding
The vast majority of adhesin interactions are non-covalent. These events include all of the modes of protein-protein binding including Van der Waals, polar, and ionic interactions, as well as the hydrophobic effect between complementary surfaces. An exception is the Candida albicans adhesin Hwp1, one of a class of adhesins covalently bound to fungal cell surface through glycosyl phosphatidyl inositol (GPI) links to cell wall glucans. Hwp1 is a substrate for transglutaminase on mammalian cell surface, and the enzyme crosslinks Gln residues in the N-terminal region of Hwp1 to Lys residues on the surface of mammalian keratinocytes [20][21].
2. Intracellular Proteins That Moonlight as Adhesins
In both bacteria and fungi, highly expressed cytoplasmic and mitochondrial proteins have often been identified as adhesins. Among those most often identified are the glycolytic enzymes enolase and glyceraldehyde phosphate dehydrogenase (GAPDH), along with cytoplasmic chaperones. These proteins are consistently identified as cell wall components, despite their lack of secretion signals. In bacteria the mechanism of externalization is still not clear [22]. In fungi they are unconventionally secreted through the endosomal/vacuolar system [23]. Such moonlighters have often been identified as receptors for plasma and extracellular matrix (ECM) proteins.
3. Multifunctional Bacterial Adhesins
3.1. Bacillus subtilis TasA
This ~30 kD protein has a wide variety of effects. It was discovered as a component of spore coats. There are at least two forms of the protein: a soluble monomeric α/β globular form that forms amyloid-like fibers in vivo, which, however, lack canonical cross-β structure [13]. Fibrous TasA is a major component of biofilm matrix, where it contributes to biofilm wrinkling (thought to be a strategy for making nutrient channels within the biofilm [24]. Unsurprisingly, TasA determines biofilm hydrophobicity and cohesion under stress. It also contributes to antibiotic resistance of biofilms. However, TasA also acts intracellularly upstream of SinR to regulate cell fate. Mutant tasA cells form biofilms with fewer motile cells and more cells that excrete extracellular matrix components [25].
3.2. Pili Adhesins
Bacterial pili and fimbriae often have adhesion as a primary function [26][27]. The adhesins are the tip subunits in some cases, or in other cases adhesion activity is a property of the major pilin structural subunits themselves. The curli pili of gram-negative bacteria assemble in amyloid-like structures, and greatly contribute to biofilm structure and adhesiveness and antibiotic resistance. There is extensive literature on the biosynthesis, assembly, and consequences of these structures [28][29].
3.3. MSCRAMMS
Gram-positive cocci like Staphylococcus and Streptococcus express MSCRAMMS (microbial surface components recognizing adhesive matrix molecules) [30]. These adhesins are cross-linked through their C-terminal residues to the wall peptidoglycan by transpeptidylation during wall synthesis [31]. Various MSCRAMMS bind to a large variety of mammalian ECM components including fibrinogen, collagens, fibronectin, cytokeratins, complement factors, and others. Most MSCRAMMS are 500–1000 amino acids. They have tandem Ig-like β-sandwich domains near the N-terminus. A primary binding site is in the linear cleft or trench between the domains. Ligand peptides dock in the cleft through sidechain interactions. Then, an unstructured segment of the protein immediately C-terminal to the second Ig-like domain locks the ligand in place by crossing on top of it. This action forms a ‘latch’ onto the first Ig-like domain by constituting an additional β-strand at the edge of one β-sheet. This dock, lock, and latch mechanism (DLL) results in an extremely strong adhesive bond. In the collagen-binding MSCRAMM CNA from Staph. aureus, the Ig-like domains are connected by a flexible linker. The two domains then close around the fiber, and the latch peptide adds stability by β-strand addition to domain I, as in DLL binding [30].
4. General Characteristics of Fungal Adhesins
Each fungal adhesin has different specificity for binding, as well as a characteristic profile for expression time in the growth cycle in the various fungal morphs and growth phases. Nevertheless, they share certain molecular features, including heavy glycosylation, hydrophobicity, secretion signals, C-terminal modified glycosyl phosphatidyl inositol (GPI) anchors covalently bound to cell wall glucans, amyloid core sequences, and often Cys-rich motifs and dibasic motifs [10]. In a few cases, some of the resulting adhesion partners have been identified. Simple examples include the S. cerevisiae Flo adhesins. These adhesins facilitate formation and maintenance mats, a type of biofilm that looks like a large, structured colony on an agar plate [3][32][33][34]. The FLO1 family encodes 3 adhesins, and each is a lectin with a glycan binding site specific for α-mannosyl or α-glucosyl residues [35]. The binding has moderate affinity, and is non-specific, considering the prevalence of these residues on fungal and animal cell surfaces. In addition, each Flo lectin has multiple amyloid core sequences that can form homotypic cross-β structures on cell surfaces to cluster the adhesins and increase avidity of the interactions. The formation of cross-β aggregates increases the strength of adhesive bonds. The adhesin Flo11 is not homologous to the Flo lectins and does not have glycan binding activity. Instead, its interactions include homotypic association between Trp residues arrayed in a ring around the surface of the well-folded N-terminal domain. The Trp indole and Tyr phenol rings stack to generate electron delocalization similar to that of base stacking in a DNA double helix. Trp and Tyr residues also contribute hydrophobic effect interactions [36].
5. Candida albicans Int1
This protein is large and highly multifunctional. Although its structure is largely unknown, it has low homology to mammalian integrins, and includes a region similar to the Saccharomyces cerevisiae Bud4 GTPase. Accordingly, it has intracellular functions including interaction with septin ring proteins. Its roles include ploidy stabilization, bud site selection, and in hyphal morphogenesis under some conditions [37][38]. A poly Gln sequence at the C-terminal is characteristic of proteins that regulate cellular responses through the formation of reversible membrane-less RNA-protein aggregates [39]. Despite its lack of a signal sequence, it can be detected on the cell surface, so it is presumably unconventionally secreted [23]. Int1 mediates binding to epithelial cells as well as binding of complement protein iC3b [37][40]. Thus, Int1 has evolved to function in many key features of cellular morphogenesis, and its adhesin activity is perhaps moonlighting.
6. Multiple Functions of Yeast Mating Adhesins
The S. cerevisiae mating process illustrates the multiple roles for adhesins. The classic adhesin Sag1 expressed by mating type α (MATα) cells binds to the C-terminal residues of Aga2, which is expressed only in mating type a (MATa) cells [17][41][42]. By homology arguments, this binding is similar to the Candida albicans Als adhesin family [43], as discussed below. S. cerevisiae Aga2 is the only known Sag1 ligand in the biosphere; thus, Sag1 and Aga2 have only a single known activity. However, Aga2 is a 69-amino acid mannoprotein, and is anchored to the cell surface through disulfide bonding to Aga1, a GPI-crosslinked cell wall protein. In contrast, Aga1, which appears to be an unstructured mannoprotein, is expressed on the surface of both mating types [44]. In matings on solid substrates, Sag1 and Aga2 are not essential, and at least in these mating conditions cell adhesion is mediated by Aga1 binding to its distant homolog Fig2 [44]. Thus, Aga1 is at least a bifunctional adhesin. Fig2 can also bind to itself to facilitate mating on surfaces. Binding Aga1-Fig2 and Fig2-Fig2 binding require conserved Cys residues in WCPY and CX4C motifs in both partners. Thus, it is possible but untested that these adhesins form intermolecular disulfide bonds, and such bonds would constitute intercellular disulfide bonds, during mating. All of these adhesins are required for normal morphology of the zygotes [45]. These adhesins Aga1 and Fig2 thus illustrate adhesins with multiple binding partners, and Aga1 has the additional role in disulfide binding to Aga2.
7. Multiple Binding Modes and Multiple Functions in Candida adhesins
Dozens of adhesins have been identified in C. albicans and C. glabrata, human commensals and opportunistic pathogens. C. albicans is the most frequent agent for human fungal infections and the cause of most fungal-induced mortality [46]. C. glabrata is more closely related to baker’s yeast S. cerevisiae than to C. albicans, and it is highly resistant to many antifungals. Therefore, C. glabrata is now the second most frequent fungal disease agent. Each of these fungi expresses specific adhesins differentially in different growth conditions and in different fungal morphs and phases. These adhesins share certain characteristics, including heavy glycosylation, hydrophobicity, GPI-mediated bonds to wall glucans, and amyloid core sequences [10]. In C. glabrata, two large adhesin families, EPA and AWP, have been well-studied [35][47]. The EPA family encodes many glycoprotein lectins that bind galactose and related saccharides in mammalian glycocalyx. In each adhesin, an N-terminal C-type lectin domain is attached to a long, Ser/Thr-rich glycosylated stalk and a modified GPI anchor covalently bonded to cell wall glucan. Epa family adhesins do not appear to use amyloid-mediated interactions, a characteristic of S. cerevisiae Flo family lectins. Therefore, EPA family lectins are likely to mediate short-lived interactions, like the lectins that mediate rolling of lymphocytes along the endothelium [14].
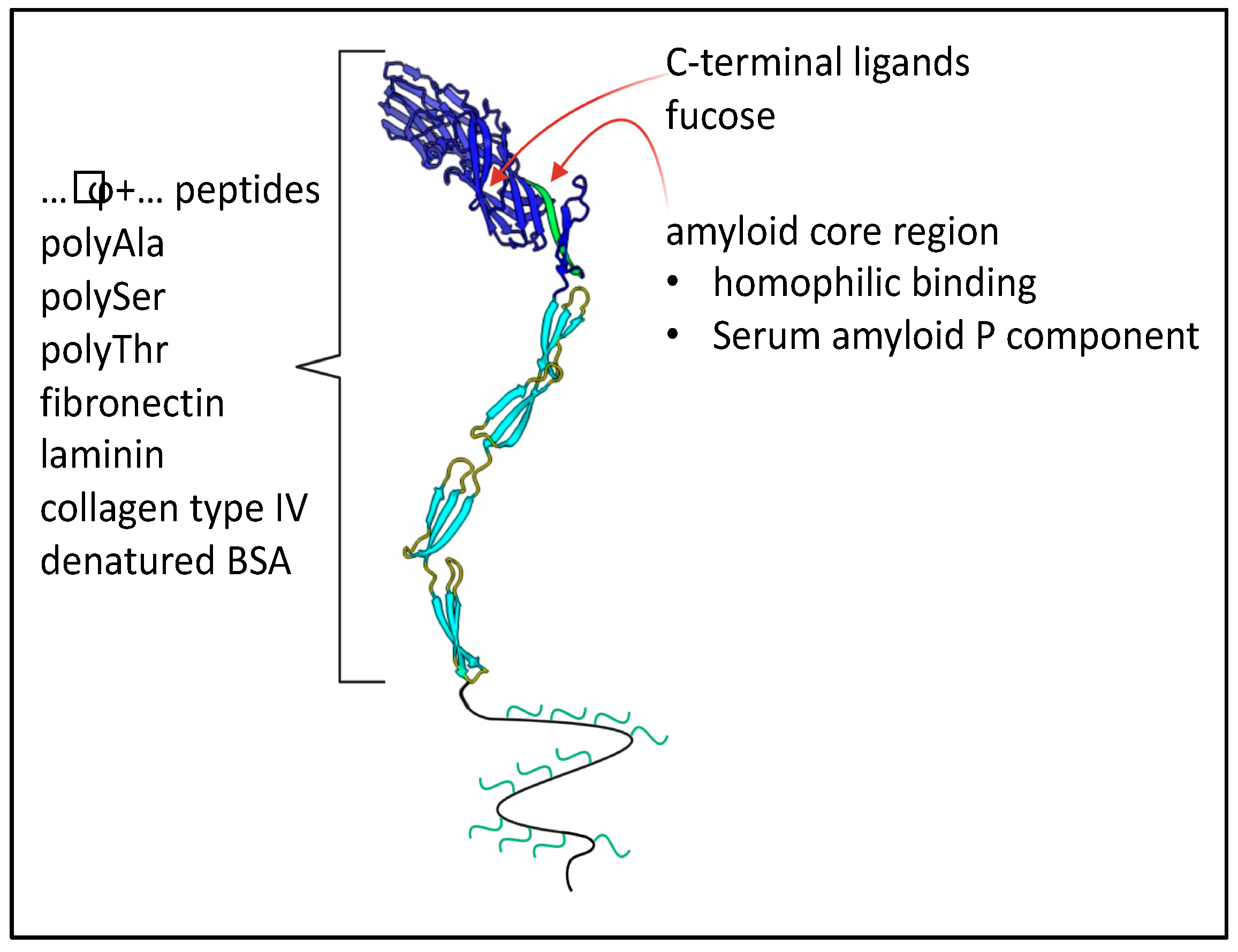
8. C. albicans Als Family Adhesins
In C. albicans eight Als adhesins are encoded at separate loci, ALS1 through ALS7 and ALS9. These loci are usually heteroallelic (i.e., the diploid alleles have different sequences, so 16 different gene products are encoded) [48][49]. More diversity is generated by the ambiguity of the CUG codon, which is translated as Ser 97% of the time, and as Leu 3% of the time. CUG encodes 2–18 surface-exposed residues in different Als proteins [50]. Even at this low amount of ambiguity, many adhesin molecules transcribed from the same gene display non-identical peptide sequences; individual molecules also vary in glycosylation patterns. This heterogeneity generates adhesins with different binding characteristics. Nevertheless, all members of the gene family share the same basic architecture (Figure 6). Each adhesin has a tandem pair of N-terminal Ig-like invasin domains, and there is a peptide binding cleft at the interface between the domains. In each adhesin, there is a sequence-conserved amyloid core sequence immediately following this region. This sequence is initially folded at the interface between the highly conserved T region and the Ig-like domains [51]. C-terminal to the T region are 5–35 copies of a 36-amino acid tandem repeat with a three-stranded antiparallel β-sheet structure. The tandem repeats are followed by a flexible but unstructured stalk region of 600–1000 amino acids with 30–50% glycosylated residues, then the modified GPI anchor that mediates attachment to the cell wall glucan. Similar adhesins are encoded in many diverse yeasts, including S. cerevisiae α-agglutinin (Sag1).

Figure 3. Binding partners of C. albicans Als adhesins. The model shows the two N-terminal Ig-like domains in dark blue, with the binding cleft between them. Three tandem repeats in the T region and two of many repeats in the TR regions are shown in light blue. The unstructured C-terminal stalk region is shown as a wavy line with attached glycans in green. This region typically consists of 600–1000 residues.
9. The Remarkable Similarity of Bacterial MSCRAMMs and Fungal Als Adhesins
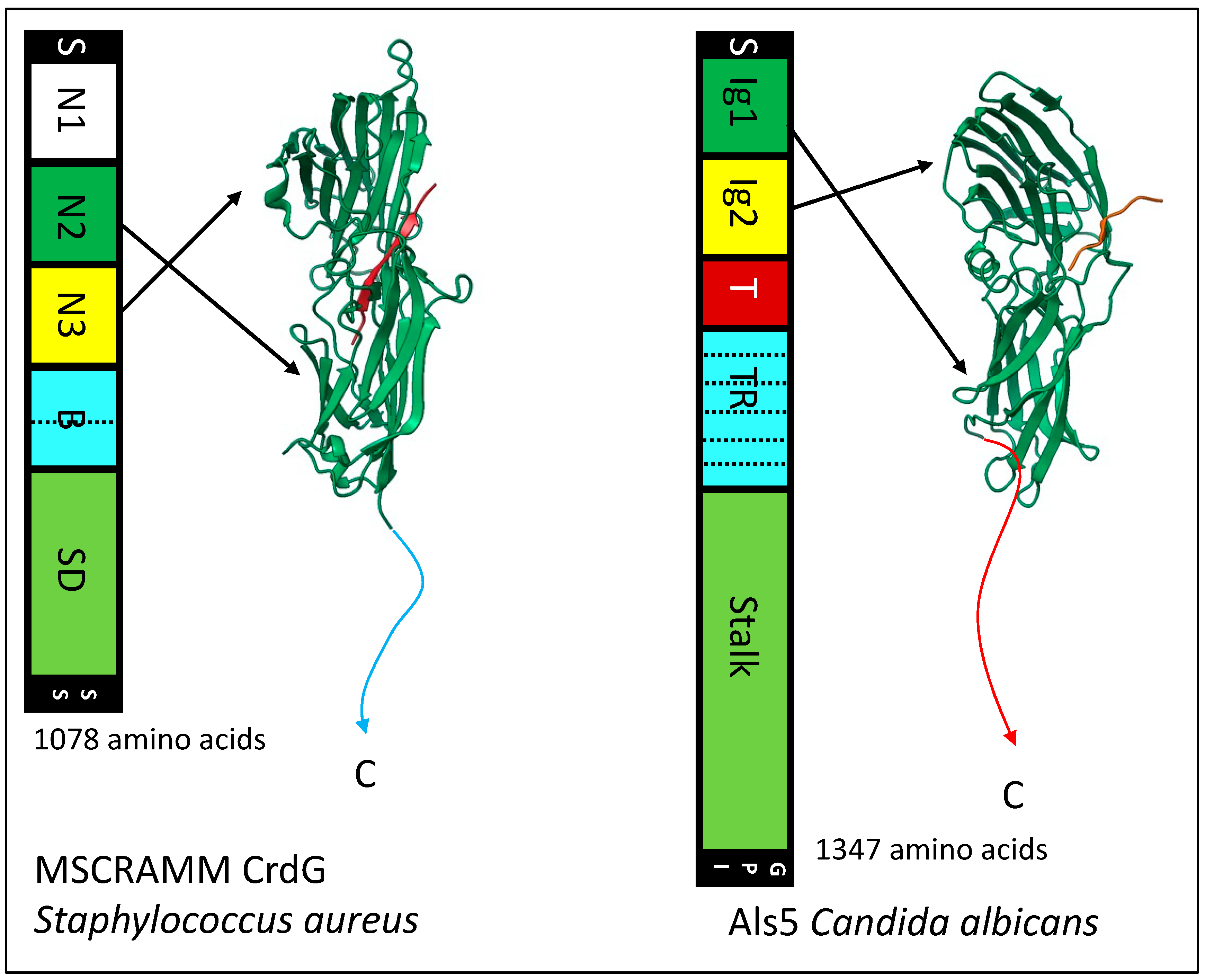
The N-terminal region Ig-like domains region of Als adhesins are structurally similar to the Ig-like domains of MSCRAMMs and Als3 (Figure 4) [30][49][52][53]. Als proteins bind C-terminal peptides in the cleft between the domain, like the binding trench in MSCRAMMs. In each of these adhesin families, the Ig-like domains are followed in sequences by tandem β-sheet domains, a long unstructured stalk region that holds the adhesin domains away from the cell surface, and a covalent bond to the microbe wall. There is the sortase-dependent transpeptidylation of MSCRAMMs to the wall peptidoglycan, and in the fungi there is the GPI-dependent transglycosylation of adhesins to the wall β-glucans. In each family, the peptide immediately C-terminal to Ig-like domain 2 has a non-structured to structured transition: in MSCRAMMs it is the DLL lock and latch mechanism. In Als proteins this peptide forms amyloid-like cross-β bonds that cluster the adhesins to increase avidity. In each family, at least some members can form these cross-β amyloids between cells.

Figure 4. Similarities between a bacterial MSCRAMM and a fungal Als adhesin. In the maps, regions with analogous structures are similarly colored: N-terminal secretion signals (S) and C-terminal wall anchorage signals (SS, bacterial sortase; GPI glycosyl phosphatidyl inositol) are black, Ig-like domains are green and yellow, tandem repeats are light blue, and low-complexity stalk regions are light green (SD, (SerAsp)n repeats). The structural models show the Ig-like domains with the ligand peptides in orange. The CrdG structure is from pdb file 1R17 and the Als3 structure from file 4LEB.
The similarities extend to multifunctional properties of these adhesins. Both families mediate microbial binding to abiotic and biological substrata, including indwelling medical devices. Each family shows broad binding to mammalian ECM components, and each family includes members that act as epithelial invasins. In each family, there are secondary binding sites for other ligands, and these secondary sites are used to form homophilic bonds. These bonds cluster the adhesins on the cell surface to greatly increase the avidity of binding to complex substrata. These bonds are amyloid-like in Als proteins and MSCRAMM properties are consistent with that model. Als and MSCRAMMs also form amyloid-like cross-β bonds between kindred cells to strengthen biofilms under shear stress [54][55]. Both adhesin families show flow-dependent strengthening of the adhesive bonds (catch bonding), and in Als proteins this is due to the shear-dependent unfolding of the amyloid-containing peptide. The structures of MSCRAMMs and Als adhesins therefore appear to be similar, and they mediate similar multifunctional activities. Thus, there are adhesins with similar multifunctionality in bacteria and eukaryotic fungi.
References
- Blankenship, J.R.; Mitchell, A.P. How to build a biofilm: A fungal perspective. Curr. Opin. Microbiol. 2006, 9, 588–594.
- O′Toole, G.; Kaplan, H.B.; Kolter, R. Biofilm formation as microbial development. Annu. Rev. Microbiol. 2000, 54, 49–79.
- Chow, J.; Dionne, H.M.; Prabhakar, A.; Mehrotra, A.; Somboonthum, J.; Gonzalez, B.; Edgerton, M.; Cullen, P.J. Aggregate Filamentous Growth Responses in Yeast. mSphere 2019, 4, e00702-18.
- DePas, W.H.; Syed, A.K.; Sifuentes, M.; Lee, J.S.; Warshaw, D.; Saggar, V.; Csankovszki, G.; Boles, B.R.; Chapman, M.R. Biofilm formation protects Escherichia coli against killing by Caenorhabditis elegans and Myxococcus xanthus. Appl. Environ. Microbiol. 2014, 80, 7079–7087.
- Andersson, S.; Kuttuva Rajarao, G.; Land, C.J.; Dalhammar, G. Biofilm formation and interactions of bacterial strains found in wastewater treatment systems. FEMS Microbiol. Lett. 2008, 283, 83–90.
- Serra, D.O.; Hengge, R. Bacterial Multicellularity: The Biology of Escherichia coli Building Large-Scale Biofilm Communities. Annu. Rev. Microbiol. 2021, 75, 22.
- Palkova, Z.; Vachova, L. Spatially structured yeast communities: Understanding structure formation and regulation with omics tools. Comput. Struct. Biotechnol. J. 2021, 19, 5613–5621.
- Vachova, L.; Palkova, Z. Diverse roles of Tup1p and Cyc8p transcription regulators in the development of distinct types of yeast populations. Curr. Genet. 2019, 65, 147–151.
- Friedmann, E.I. Endolithic microorganisms in the Antarctic cold desert. Science 1982, 215, 1045–1053.
- Lipke, P.N. What We Do Not Know about Fungal Cell Adhesion Molecules. J. Fungi 2018, 4, 59.
- Vachova, L.; Stovicek, V.; Hlavacek, O.; Chernyavskiy, O.; Stepanek, L.; Kubinova, L.; Palkova, Z. Flo11p, drug efflux pumps, and the extracellular matrix cooperate to form biofilm yeast colonies. J. Cell Biol. 2011, 194, 679–687.
- Wang, Y.; Bian, Z.; Wang, Y. Biofilm formation and inhibition mediated by bacterial quorum sensing. Appl. Microbiol. Biotechnol. 2022, 106, 6365–6381.
- Salinas, N.; Povolotsky, T.L.; Landau, M.; Kolodkin-Gal, I. Emerging roles of functional bacterial amyloids in gene regulation, toxicity, and immunomodulation. Microbiol. Mol. Biol. Rev. 2021, 85, e00062-20.
- Salas, A.; Shimaoka, M.; Phan, U.; Kim, M.; Springer, T.A. Transition from rolling to firm adhesion can be mimicked by extension of integrin alphaLbeta2 in an intermediate affinity state. J. Biol. Chem. 2006, 281, 10876–10882.
- Couturier, A.; Virolle, C.; Goldlust, K.; Berne-Dedieu, A.; Reuter, A.; Nolivos, S.; Yamaichi, Y.; Bigot, S.; Lesterlin, C. Real-time visualisation of the intracellular dynamics of conjugative plasmid transfer. Nat. Commun. 2023, 14, 294.
- Baffi, R.A.; Shenbagamurthi, P.; Terrance, K.; Becker, J.M.; Naider, F.; Lipke, P.N. Different structure-function relationships for α-factor-induced morphogenesis and agglutination in Saccharomyces cerevisiae. J. Bacteriol. 1984, 158, 1152–1156.
- Lipke, P.N.; Kurjan, J. Sexual agglutination in budding yeasts: Structure, function, and regulation of adhesion glycoproteins. Microbiol. Rev. 1992, 56, 180–194.
- Hajra, K.M.; Fearon, E.R. Cadherin and catenin alterations in human cancer. Genes Chromosomes Cancer 2002, 34, 255–268.
- Arvizu-Rubio, V.; García-Carnero, L.C.; Mora-Montes, H. Moonlighting proteins in medically relevant fungi. PeerJ 2022, 10, e14001.
- Staab, J.F.; Bradway, S.D.; Fidel, P.L.; Sundstrom, P. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 1999, 283, 1535–1538.
- Staab, J.F.; Bahn, Y.S.; Tai, C.H.; Cook, P.F.; Sundstrom, P. Expression of transglutaminase substrate activity on Candida albicans germ tubes through a coiled, disulfide-bonded N-terminal domain of Hwp1 requires C-terminal glycosylphosphatidylinositol modification. J. Biol. Chem. 2004, 279, 40737–40747.
- Kopeckova, M.; Pavkova, I.; Stulik, J. Diverse Localization and Protein Binding Abilities of Glyceraldehyde-3-Phosphate Dehydrogenase in Pathogenic Bacteria: The Key to its Multifunctionality? Front. Cell. Infect. Microbiol. 2020, 10, 89.
- Cohen, M.J.; Chirico, W.J.; Lipke, P.N. Through the back door: Unconventional protein secretion. Cell Surf. 2020, 6, 100045.
- Ghrayeb, M.; Hayet, S.; Lester-Zer, N.; Levi-Kalisman, Y.; Chai, L. Fibrilar polymorphism of the bacterial extracellular matrix protein tasa. Microorganisms 2021, 9, 529.
- Romero, D.; Aguilar, C.; Losick, R.; Kolter, R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. USA 2010, 107, 2230–2234.
- Lukaszczyk, M.; Pradhan, B.; Remaut, H. The Biosynthesis and Structures of Bacterial Pili. Bact. Cell Walls Membr. 2019, 92, 45.
- Ligthart, K.; Belzer, C.; de Vos, W.M.; Tytgat, H.L.P. Bridging Bacteria and the Gut: Functional Aspects of Type IV Pili. Trends Microbiol. 2020, 28, 340–348.
- Mignolet, J.; Panis, G.; Viollier, P.H. More than a Tad: Spatiotemporal control of Caulobacter pili. Curr. Opin. Microbiol. 2018, 42, 79–86.
- Sønderby, T.V.; Najarzadeh, Z.; Otzen, D.E. Functional Bacterial Amyloids: Understanding Fibrillation, Regulating Biofilm Fibril Formation and Organizing Surface Assemblies. Molecules 2022, 27, 4080.
- Foster, T.J. The MSCRAMM Family of Cell-Wall-Anchored Surface Proteins of Gram-Positive Cocci. Trends Microbiol. 2019, 27, 927–941.
- Cossart, P.; Jonquieres, R. Sortase, a universal target for therapeutic agents against Gram-positive bacteria? Proc. Natl. Acad. Sci. USA 2000, 97, 5013–5015.
- Reynolds, T.B.; Fink, G.R. Bakers′ yeast, a model for fungal biofilm formation. Science 2001, 291, 878–881.
- Beauvais, A.; Loussert, C.; Prevost, M.C.; Verstrepen, K.; Latge, J.P. Characterization of a biofilm-like extracellular matrix in FLO1-expressing Saccharomyces cerevisiae cells. FEMS Yeast Res. 2009, 9, 411–419.
- Chow, J.; Starr, I.; Jamalzadeh, S.; Muniz, O.; Kumar, A.; Gokcumen, O.; Ferkey, D.M.; Cullen, P.J. Filamentation Regulatory Pathways Control Adhesion-Dependent Surface Responses in Yeast. Genetics 2019, 212, 667–690.
- Willaert, R.G.; Kayacan, Y.; Devreese, B. The Flo Adhesin Family. Pathogens 2021, 10, 1397.
- Kraushaar, T.; Bruckner, S.; Veelders, M.; Rhinow, D.; Schreiner, F.; Birke, R.; Pagenstecher, A.; Mosch, H.U.; Essen, L.O. Interactions by the Fungal Flo11 Adhesin Depend on a Fibronectin Type III-like Adhesin Domain Girdled by Aromatic Bands. Structure 2015, 23, 1005–1017.
- Gale, C.A.; Bendel, C.M.; McClellan, M.; Hauser, M.; Becker, J.M.; Berman, J.; Hostetter, M.K. Linkage of adhesion, filamentous growth, and virulence in Candida albicans to a single gene, INT1. Science 1998, 279, 1355–1358.
- Orellana-Munoz, S.; Duenas-Santero, E.; Arnaiz-Pita, Y.; Del Rey, F.; Correa-Bordes, J.; Vazquez de Aldana, C.R. The anillin-related Int1 protein and the Sep7 septin collaborate to maintain cellular ploidy in Candida albicans. Sci. Rep. 2018, 8, 2257–2259.
- Chen, C.; Ding, X.; Akram, N.; Xue, S.; Luo, S. Fused in Sarcoma: Properties, Self-Assembly and Correlation with Neurodegenerative Diseases. Molecules 2019, 24, 1622.
- Hostetter, M.K. The iC3b receptor of Candida albicans and its roles in pathogenesis. Vaccine 2008, 26 (Suppl. S8), 108.
- Shen, Z.M.; Wang, L.; Pike, J.; Jue, C.K.; Zhao, H.; de Nobel, H.; Kurjan, J.; Lipke, P.N. Delineation of functional regions within the subunits of the Saccharomyces cerevisiae cell adhesion molecule a-agglutinin. J. Biol. Chem. 2001, 276, 15768–15775.
- Cappellaro, C.; Baldermann, C.; Rachel, R.; Tanner, W. Mating type-specific cell-cell recognition of Saccharomyces cerevisiae: Cell wall attachment and active sites of a- and alpha-agglutinin. EMBO J. 1994, 13, 4737–4744.
- Oh, S.-H.; Hoyer, L.L. Assessing Als3 Peptide-Binding Cavity and Amyloid-Forming Region Contributions to Candida albicans Invasion of Human Oropharyngeal Epithelial Cells. Front. Cell. Infect. Microbiol. 2022, 12, 571.
- Huang, G.; Dougherty, S.D.; Erdman, S.E. Conserved WCPL and CX4C domains mediate several mating adhesin interactions in Saccharomyces cerevisiae. Genetics 2009, 182, 173–189.
- Zhang, M.; Bennett, D.; Erdman, S.E. Maintenance of mating cell integrity requires the adhesin Fig2p. Eukaryot. Cell 2002, 1, 811–822.
- Pappas, P.G.; Lionakis, M.S.; Arendrup, M.C.; Ostrosky-Zeichner, L.; Kullberg, B.J. Invasive candidiasis. Nat. Rev. Dis. Prim. 2018, 4, 18026.
- Timmermans, B.; Peñas, A.D.L.; Castaño, I.; Van Dijck, P. Adhesins in Candida glabrata. J. Fungi 2018, 4, 60.
- Hoyer, L.L. The ALS gene family of Candida albicans. Trends Microbiol. 2001, 9, 176–180.
- Hoyer, L.L.; Cota, E. Candida albicans Agglutinin-Like Sequence (Als) Family Vignettes: A Review of Als Protein Structure and Function. Front. Microbiol. 2016, 7, 280.
- Miranda, I.; Silva-Dias, A.; Rocha, R.; Teixeira-Santos, R.; Coelho, C.; Gonçalves, T.; Santos, M.A.S.; Pina-Vaz, C.; Solis, N.V.; Filler, S.G.; et al. Candida albicans CUG mistranslation is a mechanism to create cell surface variation. mBio 2013, 4, e00285-13.
- Lipke, P.N.; Klotz, S.A.; Dufrene, Y.F.; Jackson, D.N.; Garcia-Sherman, M.C. Amyloid-Like beta-Aggregates as Force-Sensitive Switches in Fungal Biofilms and Infections. Microbiol. Mol. Biol. Rev. 2017, 82, e00035-17.
- Salgado, P.S.; Yan, R.; Taylor, J.D.; Burchell, L.; Jones, R.; Hoyer, L.L.; Matthews, S.J.; Simpson, P.J.; Cota, E. Structural basis for the broad specificity to host-cell ligands by the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci. USA 2011, 108, 15775–15779.
- Sheppard, D.C.; Yeaman, M.R.; Welch, W.H.; Phan, Q.T.; Fu, Y.; Ibrahim, A.S.; Filler, S.G.; Zhang, M.; Waring, A.J.; Edwards, E.J., Jr. Functional and structural diversity in the Als protein family of Candida albicans. J. Biol. Chem. 2004, 279, 30480–30489.
- Chan, C.X.; El-Kirat-Chatel, S.; Joseph, I.G.; Jackson, D.N.; Ramsook, C.B.; Dufrene, Y.F.; Lipke, P.N. Force Sensitivity in Saccharomyces cerevisiae Flocculins. mSphere 2016, 1, e00128-16.
- Foster, T.J. Surface Proteins of Staphylococcus epidermidis. Front. Microbiol. 2020, 11, 1829.
More
Information
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
808
Revisions:
2 times
(View History)
Update Date:
16 Jun 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




