Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Haruka Mitsuhashi | -- | 2592 | 2023-06-15 20:40:21 | | | |
| 2 | Sirius Huang | Meta information modification | 2592 | 2023-06-16 03:45:47 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Mitsuhashi, H.; Nagy, C. N6-Methyladenosine in the Brain. Encyclopedia. Available online: https://encyclopedia.pub/entry/45681 (accessed on 07 February 2026).
Mitsuhashi H, Nagy C. N6-Methyladenosine in the Brain. Encyclopedia. Available at: https://encyclopedia.pub/entry/45681. Accessed February 07, 2026.
Mitsuhashi, Haruka, Corina Nagy. "N6-Methyladenosine in the Brain" Encyclopedia, https://encyclopedia.pub/entry/45681 (accessed February 07, 2026).
Mitsuhashi, H., & Nagy, C. (2023, June 15). N6-Methyladenosine in the Brain. In Encyclopedia. https://encyclopedia.pub/entry/45681
Mitsuhashi, Haruka and Corina Nagy. "N6-Methyladenosine in the Brain." Encyclopedia. Web. 15 June, 2023.
Copy Citation
RNA modifications known as epitranscriptomics have emerged as a novel layer of transcriptomic regulation. Like the well-studied epigenetic modifications characterized in DNA and on histone-tails, they have been shown to regulate activity-dependent gene expression and play a vital role in shaping synaptic connections in response to external stimuli. Among the hundreds of known RNA modifications, N6-methyladenosine (m6A) is the most abundant mRNA modification in eukaryotes. Through recognition of its binding proteins, m6A can regulate various aspects of mRNA metabolism and is essential for maintaining higher brain functions.
N6-methyladenosine
epitranscriptomics
Brain
1. Introduction
Epitranscriptomics, also known as RNA modifications, refers to the study of post-transcriptional modifications of RNA molecules. Although chemical modifications of RNA have been described for a half-century, only recently with the advancement of technology have we started to elucidate their functions [1]. To date, over 170 post-transcriptional base modifications have been identified. In addition, RNA modifications have been characterized not only in abundant non-coding RNA, such as transfer RNA (tRNA), ribosomal RNA (rRNA), and small nuclear RNA (snRNAs), but also in messenger RNA (mRNA) [2]. The post-transcriptional modifications of mRNA, which include N6-methyladenosine (m6A), N1-methyladenosine (m1A), 5-methylcytidine (m5C), N7-methylguanosine (m7G), and N6,2-O-dimethyladenosine (m6Am) add a new layer to regulating mRNA metabolism and gene expression [2]. Among them, m6A is one of the most abundant modifications of the mRNA in eukaryotes and the best-studied modification so far [3]. Given its diverse roles in mRNA metabolism and gene regulation, altered m6A profiles have been linked to various illnesses, including cancers and psychiatric disorders [4].
2. m6A in the Brain
Methylation at the N6 position of adenosine is referred to as m6A. It is the most abundant RNA modification, with approximately 25% of mammalian messenger RNAs (mRNA) bearing the mark [3]. m6A is enriched in conserved regions, namely within the 3′ untranslated regions (UTRs) and near the stop codons of transcripts [5]. It has a known consensus motif RRACH (R represents A or G, and H represents A, C, or U) and, most importantly, it is dynamic and highly reversible [3][5][6]. m6A is known to be regulated at three complementary levels through proteins that act as writers, erasers, and readers [4][7] (Figure 1). Methylation is deposited by a multicomponent methyltransferase complex (“writers”) consisting of a core writer complex and an interacting complex. In the core complex, methyltransferase-like 3 (METTL3) is the catalytic component, methyltransferase-like 14 (METTL14) liaises with METTL3 to recognize the substrate, and WTAP guides METTL3/14 heterodimer [7]. The interacting complex contains the RNA binding motif protein 15/15B (RBM15/15B), vir-like m6A methyltransferase associated (VIRMA), zinc finger CCCH-type containing 13 (ZC3H13), and HAKAI which support the functioning and positioning of a writer complex. VIRMA interacts with WTAP and mediates selective methylation in the 3′UTR and near the stop codon [8]. RBM15/15B mediate the binding of a writer complex to the U enriched region on mRNA and recruit writer complexes to specific sites [9]. ZC3H13 mediates the nuclear localization of writer complexes [10][11]. Conversely, m6A is removed by demethylases (“erasers”), which include the fat mass and obesity-associated protein (FTO) and ALKBB homolog 5 (ALKBH5). For the mark to elicit its various effects, “reader” proteins must recognize and bind m6A, thereby regulating gene expression through diverse mechanisms such as mRNA stability, splicing, nuclear export, and translation efficiency [12]. Among the known m6A readers, the YTH family of proteins, with its conserved YTH-domain, is the best studied. Each member of the YTH family is reported to have a unique function; for example, YTHDC1 is predominantly found in the nucleus and promotes exon inclusion by selectively recruiting pre-mRNA splicing factor SRSF3 [13]. Further, YTHDC1 facilitates the nuclear export of methylated transcripts by interacting with nuclear transport receptors [14]. On the other hand, YTHDC2 and YTHDF1/2/3 are found in the cytoplasm. YTHDC2 has been shown to facilitate translation by resolving secondary structures or to promote mRNA degradation by interacting with 5′-3′ exoribonuclease [15][16]. In contrast, YTHDF2 destabilizes the target transcripts by recruiting a deadenylase complex [17]. YTHDF1 and YTHDF3 modulate translation efficiency [18][19]. However, recent studies propose that YTHDF1/2/3 bind to the same target transcripts and act redundantly to influence mRNA degradation [20]. This may not be surprising given that the YTHDF family has high sequence homology, sharing close to 85% of their sequence across the family [21]. Other readers include the fragile X mental retardation protein (FMRP), heterogeneous nuclear ribonucleoproteins (HNRNPs), and insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs). FMRP facilitates the nuclear export of the target transcripts through interactions with mRNA nuclear export factors or a selective mRNA export pathway mediated by chromosomal maintenance 1 (CRM1) [22]. HNRNPC and HNRNPG modulate pre-mRNA processing and pre-mRNA alternative splicing, respectively [23]. In addition, HNRNPA2B1 is known to regulate alternative splicing and primary microRNA processing [24]. IGF2BP1-3 stabilizes the target transcripts and promotes the storage of mRNA by recruiting stabilizers [25]. More studies are needed to better understand the selective binding of m6A readers to different m6A sites; nonetheless, these findings support diverse molecular outcomes driven by how the m6A is read, functionally altering the levels of transcripts present in the cell.
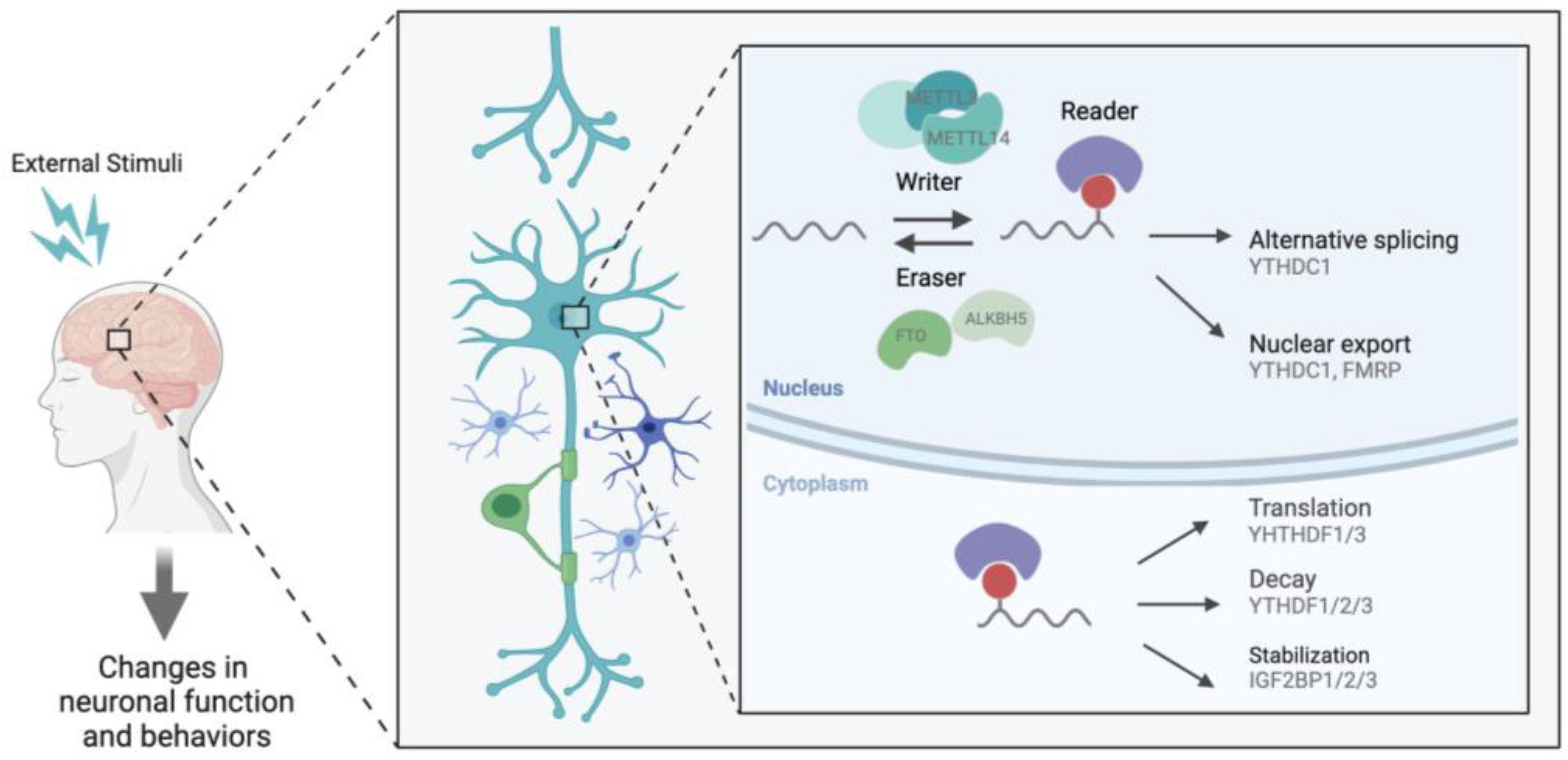
Figure 1. The suggested regulation of transcript by m6A in the nervous system. m6A is catalyzed by a writer complex consisting of METTL3, METTL14, WTAP, RBM15/15B, ZC3H13, VRMA, and HAKAI. m6A is demethylated by FTO or ALKBH5. m6A regulates the variety of mRNA metabolism in the nucleus and cytoplasm through recognition by readers, YTHDF1/2/3, YTHDC1,2, FMRP, HNRNPs, and IGFBP1/2/3. m6A has been shown to affect translation, degradation, splicing, and nuclear export and, therefore, could regulate the nervous system.
Interestingly, m6A is highly enriched in the brain, with more than 30% of the transcripts in the brain harboring this modification [26]. Indeed, m6A profiles in the human brain show functional enrichment in synaptic and neuronal pathways for genes harboring brain-specific m6A [27]. Additionally, m6A writers, erasers, and readers are widely expressed throughout the brain, supporting the argument that m6A plays a vital role in various aspects of brain functions.
Although cell type-specific m6A profiles in the brain have not yet been generated at large, there is evidence to support that the proteins related to m6A are expressed across all cell types, albeit more highly expressed in the neurons compared to the glial cells [26]. However, given that non-neuronal cell types amount for at least half of all the cells in the brain, the roles of m6A in non-neuronal cell types cannot be overlooked. Indeed, studies have demonstrated the dynamic regulatory role of m6A in the oligodendrocyte lineage—a subclass of glial cells, including both oligodendrocyte precursor cells—and mature oligodendrocytes [28]. Thousands of transcripts are differentially methylated between the two cell types, implying that m6A may underlie cell-specific functions in the brain [28][29]. Taken together, this suggests a fundamental role for m6A methylation in the process of cellular differentiation.
3. Activity-Dependent Role of m6A
3.1. m6A Localizes Transcripts to the Synapse
The delivery of select transcripts to distal compartments, such as axons and dendrites, is critical in neurons where thousands of transcripts are locally regulated, influencing the production of proteins that affect synaptic organization and transmission. Recent research has shown that m6A modifies mRNAs destined for subcellular transport, specifically those targeted to the synapse. m6A profiles from the synaptosomes isolated from mouse forebrains found that m6A transcripts are enriched for pathways associated with synaptic function [30]. Specifically, mRNAs from the synaptosomes with high m6A levels were significantly enriched for synaptic functions compared to mRNAs with low methylation, which were more associated with cellular metabolic processes [30]. Moreover, m6A writers, erasers, and readers were found at dendrites and adjacent to synapses, suggesting that subcellular modification or the recruitment of modified mRNAs may play a role in synaptic activity [31]. Accordingly, the m6A profiling of neuronal subcellular compartments revealed that thousands of m6A transcripts are enriched in dendrites and axons compared to cell bodies [32]. Likewise, it has been found that the m6A sites within the 3′UTR promote the localization of a subset of transcripts to dendrites and axons in cultured mouse hippocampal neurons [32]. Furthermore, deleting the m6A writer METTL3 resulted in the altered localization of hundreds of transcripts [32]. Notably, the neurite-depleted transcripts corresponded to protein-coding genes associated with synaptic function and structure and neuronal projections. In line with this, knocking down the m6A readers YTHDF1 or YTHDF3 in cultured hippocampal pyramidal neurons decreased the translation of the dendritically localized mRNA and resulted in impaired synaptic transmission and abnormal dendrite spite morphology [30]. Similarly, YTHDF1 depletion was shown to impair hippocampal synaptic transmission and long-term transmission with a reduction in dendritic spine density [33]. It appears that readers are required to maintain translational efficiency at the presynaptic terminal.
Another mechanism by which local translation is regulated is through m6A eraser function within the axon, the effects of which have been associated with axon guidance, growth, and regeneration [34][35]. The eraser FTO is highly expressed in axons compared to other m6A machinery and has been shown to demethylate the local axonal mRNA [34]. The axon-specific loss of function of FTO inhibited the demethylation of the growth-associated protein-43 (GAP-43), which is required for axonal elongation [34]. The inhibition of demethylation increased m6A levels and, in turn, inhibited the local translation of GAP-43. Together these data suggest that m6A contributes to axon elongation by regulating the local translation of GAP-43 [34]. Additionally, m6A regulates axonal guidance by facilitating mRNA translation in the spinal cord. For example, for spinal commissural axons to cross the midline the m6A reader YTHDF1 must be inhibited, resulting in the decrease of axon guidance receptor Robo3.1 [35].
Taken together, the current literature suggests that m6A participates in the localization of target transcripts within neurons. Moreover, those targeted transcripts include the precursors for synaptic structural proteins which are important for synaptic communication. This implies that m6A may be one of the mediators in synaptic communication.
3.2. m6A and Synaptic Plasticity from Learning and Memory Studies
Synaptic plasticity is an essential part of the mechanism of neuronal adaptation. There is growing evidence to suggest that mice with mutations of m6A machinery genes affect the learning and memory processes. For example, the deletion of the m6A writer METTL3 in the hippocampus shows normal synaptic transmission and short-term memory but deficits in long-term memory consolidation [36]. At the molecular levels, METTL3 promotes the translation efficacy of immediate early genes (IEGS), which are induced rapidly by experience-triggered neuronal activity and are necessary for long-term memory [36]. Consistent with this finding, m6A writer METTL14 is required for long-term memory formation and neuronal excitability [37].
Likewise, the downregulation of the m6A eraser FTO and, thus, the increase of global m6A methylation level has been linked to learning and memory. At the basal levels, memory formation causes a short-term reduction in the abundance of FTO, preferentially at the synapse [38]. Moreover, FTO knockdown in the hippocampus before learning and memory training in mice enhanced memory formation in the contextual fear conditioning task [38]. Similarly, FTO knockdown in the mouse prefrontal cortex led to enhanced memory consolidation [37]. In a like manner, the depletion of FTO led to the upregulation of synaptic plasticity-related transcripts after an auditory fear conditioning behavioural task [39]. Similar to fear memory, the loss of FTO impaired working memory but did not influence long-term memory [40].
Readers, such as YTHDF1, have been shown to mediate the effect of m6A in learning and memory, most likely through the regulation of translation efficiency. For example, deleting YTHDF1 resulted in learning and memory defects, impaired hippocampal synaptic transmission, and long-term potentiation by promoting the translation of neuronal transcripts [33]. Conversely, restoring YTHDF1 in the hippocampus of Ythdf1-knockout mice rescued the behavioural and synaptic defects [33]. This change was accompanied by a decreased abundance of transcripts related to synaptic plasticity, including glutamate receptors and calcium calmodulin-dependent kinase (CaMK2a) [33]. Altogether, these data suggest that altered FTO expression in response to external stimuli plays a role in learning and memory, influencing the transcript level of gene-associated synaptic plasticity.
3.3. m6A Regulates Pathways Implicated in Psychiatric Disorders
Synaptic communication, within and across brain regions, is critical for mood regulation and cognitive function and alterations to synaptic function are a key feature of psychiatric disorders, including MDD. Emerging evidence implicates m6A in the molecular mechanisms closely associated with synaptic connectivity and psychiatric disorders. Indeed, recent studies have shown altered m6A profiles in neurodegenerative and psychiatric disorders, including Alzheimer’s disease [41][42][43], Parkinson’s disease [44], Huntington’s disease [45], alcohol use disorder [46], and post-traumatic stress disorder (PTSD) [47] in mice and human post-mortem brains [48][49].
As described above, m6A regulation has an important influence on synaptic plasticity, which has a direct impact on learning and memory, a system highly impacted by depression [50][51]. Other important biological systems implicated in depression, such as neurogenesis, HPA axis, inflammatory response, and neurotropic factors, are also shown to be regulated by m6A and will be discussed below.
While adult hippocampal neurogenesis (AHN) remains a controversial topic in humans, some studies have nonetheless shown an effect of AHN on psychiatric disorders with evidence that suggests m6A plays a role. The m6A eraser FTO is highly expressed in adult neural stem cells (aNSCs) and its loss reduces proliferation and neuronal differentiation in mice [40][52][53]. These changes are accompanied by an alteration in the methylation status of genes related to the brain-derived neurotrophic factor (BDNF) signaling pathway, which is known to promote the proliferation and differentiation of NSCs [52]. Another study showed the loss of FTO led to the disrupted precursor BDNF and mature BDNF [54]. BDNF is highly crucial for synaptic neuropil outgrowth, suggesting that m6A could be indirectly mediating synaptic deficits. Another study demonstrated that the METTL3-mediated m6A regulates neurogenesis and neuronal development by modulating the expression of histone methyltransferase Ezh2 [55]. METTL3 depletion inhibits the proliferation and cell cycle progression of aNSCs, with lineage commitment more toward glia during differentiation in vitro. Moreover, m6A is uniquely tagged under either proliferation or differentiation stages, suggesting that m6A may correlate with its functions in aNSCs and regulates neurogenesis at a normal state. Given the roles of m6A in adult neurogenesis, m6A may contribute to the development of MDD by regulating adult neurogenesis.
Dysregulation of the immune and inflammatory response, both peripherally and centrally, is commonly found across psychiatric disorders. Several studies have identified links between m6A and the inflammatory responses of microglia and macrophage, particularly in the polarization toward different phenotypes and inflammation: for example, genes related to microglia phenotypes. Pro-inflammatory-like M1-like, anti-inflammatory M2-like, and unstimulated M0-like are represented by state-specific methylation patterns, suggesting a role for m6A in the pro- and anti-inflammatory responses of the brain [56]. In macrophages, FTO knockdown led to changes in the gene expression of transcription factors essential for macrophage polarization and inhibited the nuclear factor-kappa B (NFkB) signaling pathway, thereby regulating macrophage activation [57]. In another study, METTL3 knockdown was shown to inhibit M1 polarization but enhance M2 polarization [58]. Similarly, YTHDF2 was shown to participate in the inflammatory response of macrophages by stabilizing the expression of inflammatory-related transcription factors and activating the MAPK and NFkB signaling pathways [59]. Although how m6A mediates inflammatory responses in the context of MDD is still unknown, m6A has been found to mediate the inflammatory response in the context of brain diseases such as brain stroke. Using an ischemic stroke model in mice, researchers identified an increase in m6A methylation in pathways vital to the inflammatory response, including tumour necrosis factor (TNF), Toll-like receptors (TLR), and NFkB [60].
References
- Desrosiers, R.; Friderici, K.; Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA 1974, 71, 3971–3975.
- Boccaletto, P.; Stefaniak, F.; Ray, A.; Cappannini, A.; Mukherjee, S.; Purta, E.; Kurkowska, M.; Shirvanizadeh, N.; Destefanis, E.; Groza, P.; et al. MODOMICS: A database of RNA modification pathways. 2021 update. Nucleic Acids Res. 2022, 50, D231–D235.
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206.
- Yang, C.; Hu, Y.; Zhou, B.; Bao, Y.; Li, Z.; Gong, C.; Yang, H.; Wang, S.; Xiao, Y. The role of m6A modification in physiology and disease. Cell Death Dis. 2020, 11, 960.
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012, 149, 1635–1646.
- Linder, B.; Grozhik, A.V.; Olarerin-George, A.O.; Meydan, C.; Mason, C.E.; Jaffrey, S.R. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 2015, 12, 767–772.
- He, P.C.; He, C. m6A RNA methylation: From mechanisms to therapeutic potential. EMBO J. 2021, 40, e105977.
- Yue, Y.; Liu, J.; Cui, X.; Cao, J.; Luo, G.; Zhang, Z.; Cheng, T.; Gao, M.; Shu, X.; Ma, H.; et al. VIRMA mediates preferential m6A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018, 4, 10.
- Patil, D.P.; Chen, C.K.; Pickering, B.F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S.R. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016, 537, 369–373.
- Knuckles, P.; Lence, T.; Haussmann, I.U.; Jacob, D.; Kreim, N.; Carl, S.H.; Masiello, I.; Hares, T.; Villasenor, R.; Hess, D.; et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d. Genes Dev. 2018, 32, 415–429.
- Wen, J.; Lv, R.; Ma, H.; Shen, H.; He, C.; Wang, J.; Jiao, F.; Liu, H.; Yang, P.; Tan, L.; et al. Zc3h13 Regulates Nuclear RNA m6A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol. Cell 2018, 69, 1028–1038.e6.
- Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 2014, 15, 293–306.
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.S.; Hao, Y.J.; Sun, B.F.; Sun, H.Y.; Li, A.; Ping, X.L.; Lai, W.Y.; et al. Nuclear m6A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 2016, 61, 507–519.
- Roundtree, I.A.; Luo, G.Z.; Zhang, Z.; Wang, X.; Zhou, T.; Cui, Y.; Sha, J.; Huang, X.; Guerrero, L.; Xie, P.; et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. eLife 2017, 6, e31311.
- Mao, Y.; Dong, L.; Liu, X.M.; Guo, J.; Ma, H.; Shen, B.; Qian, S.B. m6A in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat. Commun. 2019, 10, 5332.
- Kretschmer, J.; Rao, H.; Hackert, P.; Sloan, K.E.; Hobartner, C.; Bohnsack, M.T. The m6A reader protein YTHDC2 interacts with the small ribosomal subunit and the 5′-3′ exoribonuclease XRN1. RNA 2018, 24, 1339–1350.
- Du, H.; Zhao, Y.; He, J.; Zhang, Y.; Xi, H.; Liu, M.; Ma, J.; Wu, L. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 2016, 7, 12626.
- Shi, H.; Wang, X.; Lu, Z.; Zhao, B.S.; Ma, H.; Hsu, P.J.; Liu, C.; He, C. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017, 27, 315–328.
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 2015, 161, 1388–1399.
- Zaccara, S.; Jaffrey, S.R. A Unified Model for the Function of YTHDF Proteins in Regulating m6A-Modified mRNA. Cell 2020, 181, 1582–1595.e18.
- Hazra, D.; Chapat, C.; Graille, M. m6A mRNA Destiny: Chained to the rhYTHm by the YTH-Containing Proteins. Genes 2019, 10, 49.
- Hsu, P.J.; Shi, H.; Zhu, A.C.; Lu, Z.; Miller, N.; Edens, B.M.; Ma, Y.C.; He, C. The RNA-binding protein FMRP facilitates the nuclear export of N6-methyladenosine-containing mRNAs. J. Biol. Chem. 2019, 294, 19889–19895.
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518, 560–564.
- Alarcon, C.R.; Goodarzi, H.; Lee, H.; Liu, X.; Tavazoie, S.; Tavazoie, S.F. HNRNPA2B1 Is a Mediator of m6A-Dependent Nuclear RNA Processing Events. Cell 2015, 162, 1299–1308.
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.S.; Mesquita, A.; Liu, C.; Yuan, C.L.; et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018, 20, 285–295.
- Chang, M.; Lv, H.; Zhang, W.; Ma, C.; He, X.; Zhao, S.; Zhang, Z.W.; Zeng, Y.X.; Song, S.; Niu, Y.; et al. Region-specific RNA m6A methylation represents a new layer of control in the gene regulatory network in the mouse brain. Open Biol. 2017, 7.
- Xiong, X.; Hou, L.; Park, Y.P.; Molinie, B.; Gregory, R.I.; Kellis, M.; Consortium, G.T.; Gregory, R.I.; Kellis, M. Genetic drivers of m6A methylation in human brain, lung, heart and muscle. Nat. Genet. 2021, 53, 1156–1165.
- Xu, H.; Dzhashiashvili, Y.; Shah, A.; Kunjamma, R.B.; Weng, Y.L.; Elbaz, B.; Fei, Q.; Jones, J.S.; Li, Y.I.; Zhuang, X.; et al. m6A mRNA Methylation Is Essential for Oligodendrocyte Maturation and CNS Myelination. Neuron 2020, 105, 293–309.e5.
- Wu, R.; Li, A.; Sun, B.; Sun, J.G.; Zhang, J.; Zhang, T.; Chen, Y.; Xiao, Y.; Gao, Y.; Zhang, Q.; et al. A novel m6A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 2019, 29, 23–41.
- Merkurjev, D.; Hong, W.T.; Iida, K.; Oomoto, I.; Goldie, B.J.; Yamaguti, H.; Ohara, T.; Kawaguchi, S.Y.; Hirano, T.; Martin, K.C.; et al. Synaptic N6-methyladenosine (m6A) epitranscriptome reveals functional partitioning of localized transcripts. Nat. Neurosci. 2018, 21, 1004–1014.
- Flamand, M.N.; Meyer, K.D. m6A and YTHDF proteins contribute to the localization of select neuronal mRNAs. Nucleic Acids. Res. 2022, 1, 13–14.
- Madugalle, S.U.; Meyer, K.; Wang, D.O.; Bredy, T.W. RNA N6-Methyladenosine and the Regulation of RNA Localization and Function in the Brain. Trends Neurosci. 2020, 43, 1011–1023.
- Shi, H.; Zhang, X.; Weng, Y.L.; Lu, Z.; Liu, Y.; Lu, Z.; Li, J.; Hao, P.; Zhang, Y.; Zhang, F.; et al. m6A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature 2018, 563, 249–253.
- Yu, J.; Chen, M.; Huang, H.; Zhu, J.J.; Song, H.; Zhu, J.J.; Park, J.; Ji, S.J. Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Res. 2018, 46, 1412–1423.
- Yu, J.; She, Y.; Yang, L.; Zhuang, M.; Han, P.; Liu, J.; Lin, X.; Wang, N.; Chen, M.; Jiang, C.; et al. The m 6 A Readers YTHDF1 and YTHDF2 Synergistically Control Cerebellar Parallel Fiber Growth by Regulating Local Translation of the Key Wnt5a Signaling Components in Axons. Adv. Sci. 2021, 8, e2101329.
- Zhang, Z.; Wang, M.; Xie, D.; Huang, Z.; Zhang, L.; Yang, Y.; Ma, D.; Li, W.; Zhou, Q.; Yang, Y.G.; et al. METTL3-mediated N6-methyladenosine mRNA modification enhances long-term memory consolidation. Cell Res. 2018, 28, 1050–1061.
- Widagdo, J.; Zhao, Q.Y.; Kempen, M.J.; Tan, M.C.; Ratnu, V.S.; Wei, W.; Leighton, L.; Spadaro, P.A.; Edson, J.; Anggono, V.; et al. Experience-Dependent Accumulation of N6-Methyladenosine in the Prefrontal Cortex Is Associated with Memory Processes in Mice. J. Neurosci. 2016, 36, 6771–6777.
- Walters, B.J.; Mercaldo, V.; Gillon, C.J.; Yip, M.; Neve, R.L.; Boyce, F.M.; Frankland, P.W.; Josselyn, S.A. The Role of The RNA Demethylase FTO (Fat Mass and Obesity-Associated) and mRNA Methylation in Hippocampal Memory Formation. Neuropsychopharmacology 2017, 42, 1502–1510.
- Engel, M.; Eggert, C.; Kaplick, P.M.; Eder, M.; Roh, S.; Tietze, L.; Namendorf, C.; Arloth, J.; Weber, P.; Rex-Haffner, M.; et al. The Role of m6A/m-RNA Methylation in Stress Response Regulation. Neuron 2018, 99, 389–403.e9.
- Spychalaid, A.; Rü, U. FTO affects hippocampal function by regulation of BDNF processing. PLoS ONE 2019, 14, e0211937.
- Han, M.; Liu, Z.; Xu, Y.; Liu, X.; Wang, D.; Li, F.; Wang, Y.; Bi, J. Abnormality of m6A mRNA Methylation Is Involved in Alzheimer’s Disease. Front. Neurosci. 2020, 14, 98.
- Li, H.; Ren, Y.; Mao, K.; Hua, F.; Yang, Y.; Wei, N.; Yue, C.; Li, D.; Zhang, H. FTO is involved in Alzheimer’s disease by targeting TSC1-mTOR-Tau signaling. Biochem. Biophys. Res. Commun. 2018, 498, 234–239.
- Shafik, A.M.; Zhang, F.; Guo, Z.; Dai, Q.; Pajdzik, K.; Li, Y.; Kang, Y.; Yao, B.; Wu, H.; He, C.; et al. N6-methyladenosine dynamics in neurodevelopment and aging, and its potential role in Alzheimer’s disease. Genome Biol. 2021, 22, 1–19.
- Chen, Y.; Chen, Y.; Song, Y. Regulatory mechanism of FTO in Parkinson’s disease cell model. FASEB J. 2021, 35.
- Pupak, A.; Singh, A.; Sancho-Balsells, A.; Alcalá-Vida, R.; Espina, M.; Giralt, A.; Martí, E.; Ørom, U.A.V.; Ginés, S.; Brito, V. Altered m6A RNA methylation contributes to hippocampal memory deficits in Huntington’s disease mice. Cell. Mol. Life Sci. CMLS 2022, 79, 416.
- Liu, Y.; Zhang, H. RNA m6A Modification Changes in Postmortem Nucleus Accumbens of Subjects with Alcohol Use Disorder: A Pilot Study. Genes 2022, 13, 958.
- Reis, A.L.M.; Hammond, J.M.; Stevanovski, I.; Arnold, J.C.; McGregor, I.S.; Deveson, I.W.; Gururajan, A. Sex-specific transcriptomic and epitranscriptomic signatures of PTSD-like fear acquisition. iScience 2022, 25, 104861.
- Zhang, N.; Ding, C.; Zuo, Y.; Peng, Y.; Zuo, L. N6-methyladenosine and Neurological Diseases. Mol. Neurobiol. 2022, 59, 1925–1937.
- Zhang, R.; Zhang, Y.; Guo, F.; Li, S.; Cui, H. RNA N6-Methyladenosine Modifications and Its Roles in Alzheimer’s Disease. Front. Cell. Neurosci. 2022, 16, 820378.
- Duman, R.S.; Aghajanian, G.K.; Sanacora, G.; Krystal, J.H. Synaptic plasticity and depression: New insights from stress and rapid-acting antidepressants. Nat. Med. 2016, 22, 238–249.
- Takeuchi, T.; Duszkiewicz, A.J.; Morris, R.G. The synaptic plasticity and memory hypothesis: Encoding, storage and persistence. Philos. Trans. R. Soc. B 2014, 369, 20130288.
- Li, L.; Zang, L.; Zhang, F.; Chen, J.; Shen, H.; Shu, L.; Liang, F.; Feng, C.; Chen, D.; Tao, H.; et al. Fat mass and obesity-associated (FTO) protein regulates adult neurogenesis. Hum. Mol. Genet. 2017, 26, 2398–2411.
- Cao, Y.; Zhuang, Y.; Chen, J.; Xu, W.; Shou, Y.; Huang, X.; Shu, Q.; Li, X. Dynamic effects of Fto in regulating the proliferation and differentiation of adult neural stem cells of mice. Hum. Mol. Genet 2020, 29, 727–735.
- Sun, L.; Ma, L.; Zhang, H.; Cao, Y.; Wang, C.; Hou, N.; Huang, N.; von Deneen, K.M.; Zhao, C.; Shi, Y.; et al. FTO deficiency reduces anxiety- and depression-like behaviors in mice via alterations in gut microbiota. Theranostics 2019, 9, 721–733.
- Chen, J.; Zhang, Y.J.Y.C.; Huang, C.; Shen, H.; Sun, B.; Cheng, X.; Zhang, Y.J.Y.C.; Yang, Y.G.Y.; Shu, Q.; Yang, Y.G.Y.; et al. m6A Regulates Neurogenesis and Neuronal Development by Modulating Histone Methyltransferase Ezh2. Genom. Proteom. Bioinform. 2019, 17, 154–168.
- Li, Q.; Wen, S.; Ye, W.; Zhao, S.; Liu, X. The potential roles of m6A modification in regulating the inflammatory response in microglia. J. Neuroinflammation 2021, 18, 149.
- Gu, X.; Zhang, Y.; Li, D.; Cai, H.; Cai, L.; Xu, Q. N6-methyladenosine demethylase FTO promotes M1 and M2 macrophage activation. Cell. Signal. 2020, 69, 109553.
- Liu, Y.; Liu, Z.; Tang, H.; Shen, Y.; Gong, Z.; Xie, N.; Zhang, X.; Wang, W.; Kong, W.; Zhou, Y.; et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 facilitates M1 macrophage polarization through the methylation of STAT1 mRNA. Am. J. Physiol. Cell Physiol. 2019, 317, C762–C775.
- Yu, R.; Li, Q.; Feng, Z.; Cai, L.; Xu, Q. m6A Reader YTHDF2 Regulates LPS-Induced Inflammatory Response. Int. J. Mol. Sci. 2019, 20, 1323.
- Chokkalla, A.K.; Mehta, S.L.; Kim, T.; Chelluboina, B.; Kim, J.; Vemuganti, R. Transient Focal Ischemia Significantly Alters the m6A Epitranscriptomic Tagging of RNAs in the Brain. Stroke 2019, 50, 2912–2921.
More
Information
Subjects:
Neurosciences
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.0K
Revisions:
2 times
(View History)
Update Date:
16 Jun 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




