Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Stefania Nobili | -- | 3298 | 2023-06-13 02:52:38 | | | |
| 2 | Conner Chen | + 2 word(s) | 3300 | 2023-06-15 05:06:16 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Rejali, L.; Seifollahi Asl, R.; Sanjabi, F.; Fatemi, N.; Asadzadeh Aghdaei, H.; Saeedi Niasar, M.; Ketabi Moghadam, P.; Nazemalhosseini Mojarad, E.; Mini, E.; Nobili, S. Colorectal Cancer Management Based on Staging Systems. Encyclopedia. Available online: https://encyclopedia.pub/entry/45473 (accessed on 12 January 2026).
Rejali L, Seifollahi Asl R, Sanjabi F, Fatemi N, Asadzadeh Aghdaei H, Saeedi Niasar M, et al. Colorectal Cancer Management Based on Staging Systems. Encyclopedia. Available at: https://encyclopedia.pub/entry/45473. Accessed January 12, 2026.
Rejali, Leili, Romina Seifollahi Asl, Fatemeh Sanjabi, Nayeralsadat Fatemi, Hamid Asadzadeh Aghdaei, Mahsa Saeedi Niasar, Pardis Ketabi Moghadam, Ehsan Nazemalhosseini Mojarad, Enrico Mini, Stefania Nobili. "Colorectal Cancer Management Based on Staging Systems" Encyclopedia, https://encyclopedia.pub/entry/45473 (accessed January 12, 2026).
Rejali, L., Seifollahi Asl, R., Sanjabi, F., Fatemi, N., Asadzadeh Aghdaei, H., Saeedi Niasar, M., Ketabi Moghadam, P., Nazemalhosseini Mojarad, E., Mini, E., & Nobili, S. (2023, June 13). Colorectal Cancer Management Based on Staging Systems. In Encyclopedia. https://encyclopedia.pub/entry/45473
Rejali, Leili, et al. "Colorectal Cancer Management Based on Staging Systems." Encyclopedia. Web. 13 June, 2023.
Copy Citation
Colorectal cancer (CRC) is the second cause of cancer-related deaths in both sexes globally and presents different clinical outcomes that are described by a range of genomic and epigenomic alterations. Despite the advancements in CRC screening plans and treatment strategies, the prognosis of CRC is dismal. In the last two decades, molecular biomarkers predictive of prognosis have been identified in CRC, although biomarkers predictive of treatment response are only available for specific biological drugs used in stage IV CRC. The CRC staging system is based on the TNM (T—primary tumor, N—regional lymph nodes’ status, and M—distant metastases) staging system.
colorectal cancer
cancer risk
consensus molecular subtype
1. TNM Staging, a Pathological Evaluation of CRC
Colorectal cancer (CRC) is a molecularly heterogeneous malignancy with various clinical outcomes. This characteristic makes the CRC classification more challenging [1]. The management of CRC usually still basically relies on the tumor location, and the stage of disease is defined according to the American Joint Committee on Cancer (AJCC), with TNM staging (tumor, nodes, metastasis) as the main predictive factor of prognosis [2][3][4]. The TNM staging provides information about the prognosis of the patient’s disease as a post-surgical stratification based on the pathologist’s evaluation of the resected tumor, assists in decision making for the adjuvant treatment, if needed, and confirms the presence/absence of metastasis at the time of diagnosis. The main failing point of TNM classification is the inability to discriminate between “good” and “poor” cancer prognosis within the same stage. Inadequate prediction of the exact outcome of patients is concerning, particularly in stages II and III, because nearly 20% of patients defined in stage II may still die due to recurrence [5]. Patients detected within the same TNM stage may also experience different outcomes in relation to various genotypic and phenotypic differences that exist in CRC patients [6]. Accordingly, unnecessary treatment or over-treatment may occur through inaccurate discrimination of the disease. The TNM system, widely used in cancer staging, places considerable emphasis on the lymph node status. This aspect has been the subject of significant controversies, given the imprecise nature of the system. Consequently, researchers are actively exploring alternative or supplementary features that could enhance the precision of cancer staging [5]. Therefore, refinements of biological parameters to improve the stratification of patients for tumor treatment or surveillance strategies are under investigation.
2. Molecular Staging, Genetic and Epigenetic Characteristics of CRC
The molecular subtyping application of CRC as a heterogeneous disease is potentially beneficial [7]. Molecular classifications based on genetic and epigenetic characteristics in CRC patients rely on mutations, microsatellite instability (MSI), CpG island methylator phenotype (CIMP), chromosomal instability (CIN), copy-number deviations (SCNA), and significant pathways that affect CRC initiation and progression, such as WNT and MYC, which are employed for CRC stratification [3][8].
Differences in tumor biology are reflected by specific supervised expression signatures [6]. Several expression-based assays (e.g., Oncotype DX Colon, ColoPrint, and ColDx/GeneFX) with prognostic value for CRC are commercially available, but none of these signatures are yet validated and recommended for clinical use [9][10].
Developments in genomic, transcriptomic, and big-data technologies enable investigators to explore the molecular characteristics of tumors and define their clinical relevance. The integrative omics analyses reveal the potential of incorporating different biological levels in which the transcriptome may play a valuable role [10]. These items are discussed in more detail below.
- (i)
-
CRC development from benign to malignant lesions, induced by some key driver genes, acquires a series of mutations over time. Among the key driver genes that play role in carcinogenesis, the adenomatous polyposis coli (APC), accompanied by its mutations, regulates growth advantages in epithelial cells and results in the formation of a small adenoma. Later, the generation of mutations in KRAS and BRAF provides a second round of expansion for cells, involving transformation to a large adenoma. Finally, the occurrence of PIK3CA, SMAD4, and p53 mutations develops a malignant tumor that has the potential for invasion and metastasis. It remains unclear which mutations of these key driver genes are involved in the metastasis of CRC [11].
- (ii)
-
MSI is caused by mutations in DNA mismatch repair genes (MLH1, MSH2, MSH6, and PMS2) and EPCAM. The occurrence of mutations in MLH1 or MSH2 genes leads to an increased risk (70–80%) of developing cancer, while mutations in the MSH6 or PMS2 genes have a comparatively lower risk (25–60%) of cancer development [12]. Nearly 15–20% of primary CRCs have the MSI phenotype, whereas the remainders are microsatellite stable (MSS). MSI tumors demonstrate multiple single nucleotide variants (SNVs) and insertion/deletions (indels). The five microsatellite markers were used as a standard panel to access the MSI status of cancer, including BAT26 and BAT25 as mononucleotide repeats, plus three dinucleotide repeats (D2S123, D5S346, and D17S250), as defined by the Bethesda Guidelines [13]. A subset of tumors with unstable loci in ≥30% markers are defined as “microsatellite high” (MSI-H), a subset of tumors with 10–29% unstable loci are classified as “microsatellite low” (MSI-L), and “microsatellite stable” (MSS) tumors are marked with no unstable markers [12]. In a study conducted by Mori et al. [14], a comprehensive genomic screening of microsatellite coding regions was performed, revealing the presence of mutations in nine loci (TGF-βR2, Bax, MSH3, ActRIIB, SEC63, AIM2, NADH–ubiquinone oxidoreductase, COBLL1, and EBP1) in over 20% of tumors. It has been observed that the presence of MSI-H in CRC is associated with superior anti-tumor immune response, inhibition of tumor cell growth, and an improved prognosis when compared to patients displaying MSI-L or MSS status. MSI status is a conceivable predictor for the treatment decision strategy method in MSI-H and MSI-L tumors. Furthermore, MSI tumors are more frequently located in the proximal colon, and present as more poorly differentiated cancers [15]. MSI is rarely found in polyps, except in Lynch syndrome, due to germline mutations in one of the MMR genes. MSI is the hallmark of HNPCC or Lynch syndrome and occurs in >95% of HNPCC cases [12].
- (iii)
-
CIN nominated as the suppressor pathway phenotype is observed in 70–85% of CRC tumors and is often considered equal to MSS status [16]. CIN is induced by the occasional gain or loss of the whole chromosome during mitosis. Accordingly, CIN tumors are often aneuploid with structural or numerical aberrations. Furthermore, numerous significant events contribute to the development of the CIN, such as encompassing mutations in some oncogenes, such as APC, KRAS, TP53, CTNNB1, and PIK3CA, and loss of heterozygosity (LOH) chromosome in 18q with some of the tumor suppressor genes, such as SMAD2, SMAD4, and DCC, in the location. Traditionally, CRC develops through the adenoma to carcinoma pathway. APC gene mutation and inactivation are early events that are followed by the activation of KRAS as an oncogene due to mutation appearance in the adenomatous stage, followed by the inactivation of TP53 on chromosome 17p and the deletion of chromosome 18q, leading to metastatic carcinoma [17]. APC mutations are present at the preliminary stages of neoplasia and are majorly linked with the classic tubular adenoma pathway and CIN cancers [18].
- (iv)
-
CIMP is an epigenetic event that has been observed to precede the onset of cancer. The process involves an increase in methylation levels within the promoter region, which can lead to the silencing of tumor-suppressor genes. Conversely, global hypomethylation has been linked to genomic instability and chromosomal abnormalities. In CRC, epigenetic instability manifests as hypermethylation of CpG islands, often in tandem with global DNA hypomethylation. In CIMP-positive CRC, promoter regions of tumor suppressor genes are frequently hypermethylated, resulting in loss-of-function in these genes.
CIMP was described by Toyota et al. [19] as a tumorigenesis pathway in 1999. Furthermore, CIMP tumors coincide to a considerable extent with MSI. CIMP is characterized by high promoter methylation of various genes, such as p16, MINT clones, THBS, and MLH1, and is correlated with some significant clinical, pathological, and molecular features, such as female sex, old age, right-sided tumors, high MSI, and BRAF V600E mutations [20][21]. CIMP is a subclassification that is determined by the integration of genetic and epigenetic instability, and it is further divided into two categories: CIMP-low and CIMP-high. Analysis of DNA methylation profiles has demonstrated that roughly 20% of CRCs fall into the CIMP category [12].
3. Supervised Gene Expression Profiles for Prognosis Prediction of Early Stages CRC
Gene expression profiles (GEP) aim to predict the likelihood of tumor recurrence after surgery in the early stages of CRC and could also provide predictive information about the benefit of pharmacological treatment [22]. The commercially available gene expression profile assays include the following (Table 1):
-
The Oncotype DX colon cancer assay (Genomic Health, Redwood City, CA, USA, http://www.genomichealth.com (accessed on 27 February 2023) predicts recurrence in stage II colon cancer patients after surgical resection [23][24]. Oncotype DX includes a 12-gene expression assay (7 cancer-related—BGN, C-MYC, FAP, GADD45B, INHBA, Ki-67, MYBL2—and 5 reference genes—ATP5E, GPX1, PGK1, UBB, VDAC2 [25]—based on reverse transcriptase-polymerase chain reaction (RT-PCR). It has been tested on archival formalin-fixed and paraffin-embedded (FFPE) tumor tissue specimens in the QUASAR trial [10].
-
Coloprint (Agendia, Amsterdam, The Netherlands, http://www.agendia.com (accessed on 27 February 2023) is a test based on an 18-mRNA signature that predicts CRC relapse in the early stages of the disease. Furthermore, Coloprint uses whole-genome expression data analysis and has been validated in several independent cohorts. In addition, ColoPrint can predict the development of distant metastasis in stage II CRC patients and facilitates treatment strategy decisions for patients who may be safely managed without chemotherapy [26].
-
A further prognostic gene expression microarray-based assay, ColDx, commercially administered as GeneFx Colon, was developed by Almac (Almac Group Ltd., Craigavon, UK) [27]. It is based on a 634-gene signature panel [28] and performed on FFPE tumor samples [29]. This assay differentiates stage II tumors into low- and high-risk for disease recurrence [10].
-
CologuideEX (Oslo University Hospital, Oslo, Norway) stratifies CRC patients into high- and low-recurrence risk groups based on molecular markers. It contains a small panel of 13 genes (PIGR, CXCL13, MMP3, TUBA1B, SESN1, AZGP1, KLK6, EPHA7, SEMA3A, DSC3, CXCL10, ENPP3, and BNIP3) optimally combined for robust risk stratification of CRC stage II patients using a consecutive Norwegian test series [30] by Affymetrix exon-based microarrays (Affymetrix, Santa Clara, CA, USA). The test was performed on fresh-frozen samples [31].
-
The OncoDefender-CRC test investigates the expression profiles of 5 genes (BMI1, ETV6, H3F3B, RPS10, and VEGFA) on archival FFPE tumor tissue by RT-PCR using the custom 384-well TaqMan low-density arrays. This dichotomous 5-gene prognostic signature is able to differentiate between patients with lymph node-negative, invasive CRCs at “high risk” from those at “low risk” of recurrence within 3 years after curative resection [32].
-
The ColoGuidePro was also created at Oslo University Hospital (Norway) to predict good or poor prognosis in stages II and III of CRC patients. This assay includes a panel of seven genes, and it is based on Affymetrix exon-based microarrays to determine the expression signature of fresh-frozen tissue samples [33].
Despite the availability of these platforms, their use in the clinic is not currently recommended by international guidelines due to unclear clinical utility for risk stratification and a lack of strong validation in predicting treatment benefits. In contrast, there are contradictions among gene expression-based CRC classifications that need to be resolved to correlate cancer cell phenotypic features with clinical behavior and guide targeted treatments.
Table 1. Available gene expression assays predictive of prognosis in early-stage CRC.
| Assay | Company | Gene Signature | Specimen Used | Technique for GEP | Actual Status | Refs. |
|---|---|---|---|---|---|---|
| ColoPrint colon cancer recurrence assay | Agendia, Inc. | 18-gene expression profile | Fresh tissue or fresh, frozen tissue | Agendia customized whole-genome high-density microarrays | Not FDA approved | [34][35][36] |
| GeneFx Colon | Licensed by Precision Therapeutics (originally developed by Almac Diagnostics as the ColDX assay) | 634 probe-set signatures | Formalin-fixed paraffin-embedded tissue | Amplified product hybridized to Almac Colorectal Cancer DSA on Affymetrix 7G scanner | Not FDA approved | [28] |
| OncoDefender-CRC (colon and rectal cancer) | Everist Genomics | 5 genes | Formalin-fixed paraffin-embedded tissue | TaqMan Array | Not FDA approved | [32] |
| Oncotype DX colon cancer assay | Genomic Health, Inc. | 12 genes (7 prognostics and 5 reference genes) | Formalin-fixed paraffin-embedded tissue | Reverse transcription-quantitative PCR (RT-qPCR) | Not FDA approved | [23][24][25] |
| ColoGuideEx | Oslo University Hospital, Norway | 13 genes | Fresh, frozen biopsies | Exon-based microarray | Unknown status | [30] |
| ColoGuidePro (Derived from ColoGuideEx) |
Oslo University Hospital, Norway | 7 genes | Fresh, frozen biopsies | Exon-based microarray | Unknown status | [33] |
4. The Consensus Molecular Subtyping as a Transcriptome-Based Staging
In recent years, several unsupervised gene expression-based classifications have been obtained due to the advancements in sequencing methods. Despite the availability of such classifications, their clinical utility has still not been recognized due to technical problems (e.g., the lack of standard methodology, different data processing, and diverse gene expression values, and practical reasons). In 2015, Guinney et al. integrated previously available classifications to obtain a distinct classification of CRC, and, thus, tried to eliminate the discrepancies between previous subtyping systems [37]. The CRC subtyping consortium (CRCSC) normalized and integrated data from 6 previously available CRC subtyping classifications for a total of approximately 4000 primary tumors from 18 CRC datasets, by obtaining 4 distinct subtypes of CRC (CMS1–4) characterized by different prognoses, i.e., the “consensus molecular subtype (CMS)” [37] (Table 2).
Table 2. The Consensus molecular subtype characteristics.
| CMS1 | CMS2 | CMS3 | CMS4 | Unclassified/Mixed | |
|---|---|---|---|---|---|
| Distribution | 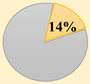 |
 |
 |
 |
 |
| Biological features | Immune subtype MSI Tumors BRAFV600E mutation Hypermutated CIMP  DNA damage repair genes  |
CIN  Epithelial differentiation Tumor suppressor genes  TP53 mutation |
KRAS mutation 30% hypermutated CIMP  |
SCNA  EMT  |
|
| Pathways activation |
MAPK RTK JAK-STAT |
WNT MYC |
MAPK RTK Epithelial and metabolism signatures |
TGFβ signaling Angiogenesis Matrix remodeling Stromal infiltration Complement inflammatory system |
|
| MicroRNA | MiR-17–92  |
Let 7-miR family  |
miR-200 family associated with EMT regulation  |
||
| Cell line | Undifferentiated | Colon-like | Colon-like | Undifferentiated | |
| Prognostic value | Good prognosis in early stage  Cytotoxic lymphocytes genes overexpression Poor prognosis after recurrence |
Stage 2 tumors:  Highest OS |
Stage 2 tumors:  Lowest OS |
Poorer prognosis | Adequate nintedanib therapy response |
| Predict value | Bevacizumab may be more efficient than cetuximab | Sensitivity to oxaliplatin  Addition of bevacizumab to capecitabine-based chemotherapy is beneficial |
Addition of bevacizumab to capecitabine-based chemotherapy is beneficial  |
Stage III resistant to anti-EGFR therapy/no benefit from adjuvant therapy IRI-based regimen is highly beneficial Cetuximab may be more effective than Bevacizumab |
|
| Polyps and adenomas | Same in both sexes Mostly right colon Mostly HP and SSA polyps Immune and stromal infiltration Activation of JAK-STAT and MAPK signaling BRAFV600E mutation |
Same in both sexes Mostly left colon Significant enrichment for WNT and MYC pathways and E2F transcription factors/G2/M checkpoint/ mitotic spindle assembly/PI3K/ mTOR genes Most of the FAP, LS, and sporadic polyps Most of AP polyps |
Chang et al. study: a small proportion of polyps in Komor et al. study: metabolism-associated Most prevalent (73%) Low risk of developing CRC Enrichment in haem, fatty acid, and sugar metabolism genes |
Mesenchymal and stromal signatures TGFβ activation Mostly in SSA and HP polyps Low frequency |
|
| Immunoscore | In non-pedunculated T1 CRC: I-low | In non-pedunculated T1 CRC: I-high |
|||
| Immunotherapy and Immune Subtypes | C1 wound healing IS (46%) C2 IFN-g dominant IS (53%) Other IS rare The most likely to profit from immunotherapy |
C1 wound healing IS (91%) C2 IFN-g dominant IS (8%) Other IS rare Immunity in KRAS mutant CMS2 is more suppressed |
C1 wound healing IS (77–78%) C2 IFN-g dominant (11–13%) C3 inflammatory IS (7%) C4 lymphocyte-depleted IS (4%) |
C1 wound healing IS (77–78%) C2 IFN-g dominant (11–13%) C3 inflammatory IS (6%) TGF-β dominant phenotype (2.3%) exclusively for CMS4 |
|
| Gut microbiome | Fusobacteria  Bacteroidetes  Fusobacterium Hwasookii  Porphyromonas Gingivalis  Fusobacterium nucleatum  Parvimonas micra  Peptostreptococcus stomatis  Firmicutes  Proteobacteria  |
Selenomas  Prevotella  Bacteroides  |
Selenomas  Prevotella  |
For further information on Table 2 content refer to 10.3390/cancers15102746
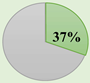
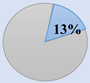
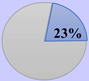
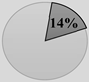
CRCSC was established to assess the core subtypes’ fundamental molecular features of CRC among the previously available gene expression-based classifications and to merge all the accessible data sources, such as mutations, methylation status, copy-number variations, microRNAs, and proteomics, to examine whether CMS can be extensively utilized in clinical practice approach [37]. Then, they determined the biological and molecular features of each subtype and ultimately assessed the prognostic and clinical association of the CMS subtypes. The CMS stratification named as a “gold standard” consists of CMS1 (immune subtype, 14%), CMS2 (canonical subtype, 37%), CMS3 (metabolic subtype, 13%), and CMS4 (mesenchymal subtype, 23%). Heterogeneous samples with mixed characteristics are classified as mixed or indeterminate samples (14%) [37].
In terms of biological features, considering the genomic aberrations, CMS1 comprised the more significant number of MSI tumors that demonstrated the hypermethylation status. They were also hyper-mutated samples with a low prevalence of somatic copy-number alterations. Likewise, they had over-expression of proteins involved in DNA damage repair. CMS1 is the immune subtype with high expression levels of genes involved in the immune response. In CMS1, an enhanced expression of genes involved in the diffuse immune infiltration, comprising TH1 and cytotoxic T cells, accompanied the immune invasion pathways. This subtype is widely hypermethylated and has a low prevalence of SCNAs [38].
CMS2 or “canonical subtype” was characterized by epithelial differentiation and has the highest distribution of tumors with high copy number counts in oncogenes and low copy number counts in tumor suppressor genes. Marked activation of WNT and MYC signaling, typical of CRC carcinogenesis was also present [37].
CMS3 tumors were defined as CIN tumors with fewer SCNAs and higher CIMP status. In addition, almost 30% of CMS3 tumors were hyper-mutated that overlapped with other MSI phenotypes [37]. The CMS3 or “metabolic subtype” indicates epithelial features and metabolic deregulation.
Ultimately, CMS4, or the “mesenchymal subtype”, comprises mesenchymal-like tumors with high stromal infiltration in combination with normal cells [39]. A significant up-regulation of genes involved in epithelial–mesenchymal transition (EMT) and of signatures related to the TGβ signaling activation, angiogenesis, matrix remodeling pathways, and the complement inflammatory system has been observed in CMS4 [37].
CMS4 relates to poorer patient prognosis and poor response to anti-EGFR drugs and routine chemotherapy regimens. Additionally, CMS4 is correlated with worse disease-free and overall survival, with the highest tendency to develop distant metastasis among other subtypes [40].
Overall, CMS2–4 have revealed an increased level of CIN [37].
The integrative analysis of mutations and copy-number variations based on The Cancer Genome Atlas (TCGA) data showed that BRAF mutations are frequently found in CMS1 and related to the MSI phenotype. KRAS mutations are frequently seen in CMS3. Additionally, the receptor tyrosine kinase (RTK) and mitogen-activated protein kinase (MAPK) pathways are generally activated in CMS1 and CMS3. However, none of these genetic aberrations exclusively belong to specific CMS subtypes. The observed heterogeneity in CMS status among CRCs with commonly accepted driver events underscores the significant variability in the biological behavior of such tumors. Additionally, this highlights the poor correlation between genotype and phenotype in CRC [37].
In a molecular analysis of the AGITG MAX clinical trial, there was no significant difference in the proportion of RAS mutations across CMS groups, but the incidence of BRAF V600E mutation was diagnosed with the highest proportion in CMS1 (34%) compared to CMS2 and CMS4 (each <2%). MSI was not common in this CMS sub-study and was found to be 7% in both CMS1 and CMS3. Finally, CMS1 included a high percentage (39%) of CIMP-high tumors in comparison with CMS2 (3%) and CMS4 (6%) [41].
In association with the clinical variables, several findings have been revealed. The CMS1 group is generally right-sided, presents mainly in women with a greater histopathological grade, and patients in this subtype showed poor survival after recurrence. In contrast, CMS2 subtypes are mostly left-sided, with a higher survival rate after relapse. CMS4 tumors are mainly related to AJCC stages III and IV, and patients with CMS4 CRC display poorer overall and relapse-free survival. Consequently, the poor prognosis after relapse is relevant for patients with MSI and BRAF mutations [37]. Therefore, it has been suggested that during the time of transition from the adenoma stage to the advanced carcinoma phase, cells change from CMS1–3 to CMS4 [42]. Most peritoneal metastases have been observed in CMS4 due to colorectal peritoneal carcinomatosis and adverse histopathological features. These observations comprised a high stroma level in both primary and metastatic tumors, inadequate differentiation grade, and increased tumor budding in primary tumors [43].
Individuals with inflammatory bowel diseases (IBD) are at an increased risk for CRC development. The signaling pathways involved in colitis-associated cancer are believed to be similar to those observed in sporadic CRCs but with a distinct sequence of events. Recently, the molecular signature of these cancers has been identified in a study that shed light on the lack of CMS2 tumors among IBD-CRCs, which were, instead, skewed toward CMS4 [44].
Generally, it is believed that the CMSs of CRC may better inform clinicians about prognosis, treatment response, and potential novel therapeutic strategies [37].
References
- Vogelstein, B.; Fearon, E.R.; Hamilton, S.R.; Kern, S.E.; Preisinger, A.C.; Leppert, M.; Nakamura, Y.; White, R.; Smits, A.M.; Bos, J.L. Genetic alterations during colorectal-tumor development. N. Engl. J. Med. 1988, 319, 525–532.
- Weiser, M.R. AJCC 8th Edition: Colorectal Cancer. Ann. Surg. Oncol. 2018, 25, 1454–1455.
- Purcell, R.V.; Schmeier, S.; Lau, Y.C.; Pearson, J.F.; Frizelle, F.A. Molecular subtyping improves prognostication of Stage 2 colorectal cancer. BMC Cancer 2019, 19, 1155.
- Rosen, R.D.; Sapra, A. TNM Classification. In StatPearls; StatPearls Publishing LLC: Treasure Island, FL, USA, 2022.
- Lea, D.; Håland, S.; Hagland, H.R.; Søreide, K. Accuracy of TNM staging in colorectal cancer: A review of current culprits, the modern role of morphology and stepping-stones for improvements in the molecular era. Scand. J. Gastroenterol. 2014, 49, 1153–1163.
- Wang, W.; Kandimalla, R.; Huang, H.; Zhu, L.; Li, Y.; Gao, F.; Goel, A.; Wang, X. Molecular subtyping of colorectal cancer: Recent progress, new challenges and emerging opportunities. Semin. Cancer Biol. 2019, 55, 37–52.
- Soldevilla, B.; Carretero-Puche, C.; Gomez-Lopez, G.; Al-Shahrour, F.; Riesco, M.C.; Gil-Calderon, B.; Alvarez-Vallina, L.; Espinosa-Olarte, P.; Gomez-Esteves, G.; Rubio-Cuesta, B.; et al. The correlation between immune subtypes and consensus molecular subtypes in colorectal cancer identifies novel tumour microenvironment profiles, with prognostic and therapeutic implications. Eur. J. Cancer 2019, 123, 118–129.
- Vivas, C.S.; Sanz-Pamplona, R.; Grasselli, J.; Mulet-Margalef, N.; Soler, R.S. Are Gene Signatures Ready for Use in the Selection of Patients for Adjuvant Treatment? Curr. Color. Cancer Rep. 2016, 12, 18–26.
- Benson, A.B. 2023 NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) Colon Cancer Version 3.2022–January 25, 2023. J. Natl. Compr. Cancer Netw. 2023. Available online: https://www.nccn.org/guidelines/guidelines-detail?category=1&id=1428 (accessed on 27 February 2023).
- Qian, Y.; Daza, J.; Itzel, T.; Betge, J.; Zhan, T.; Marmé, F.; Teufel, A. Prognostic cancer gene expression signatures: Current status and challenges. Cells 2021, 10, 648.
- Huang, D.; Sun, W.; Zhou, Y.; Li, P.; Chen, F.; Chen, H.; Xia, D.; Xu, E.; Lai, M.; Wu, Y. Mutations of key driver genes in colorectal cancer progression and metastasis. Cancer Metastasis Rev. 2018, 37, 173–187.
- Malki, A.; ElRuz, R.A.; Gupta, I.; Allouch, A.; Vranic, S.; Al Moustafa, A.E. Molecular Mechanisms of Colon Cancer Progression and Metastasis: Recent Insights and Advancements. Int. J. Mol. Sci. 2020, 22, 130.
- Boland, C.R.; Thibodeau, S.N.; Hamilton, S.R.; Sidransky, D.; Eshleman, J.R.; Burt, R.W.; Meltzer, S.J.; Rodriguez-Bigas, M.A.; Fodde, R.; Ranzani, G.N.; et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: Development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998, 58, 5248–5257.
- Mori, Y.; Yin, J.; Rashid, A.; Leggett, B.A.; Young, J.; Simms, L.; Kuehl, P.M.; Langenberg, P.; Meltzer, S.J.; Stine, O.C. Instabilotyping: Comprehensive identification of frameshift mutations caused by coding region microsatellite instability. Cancer Res. 2001, 61, 6046–6049.
- Kang, S.; Na, Y.; Joung, S.Y.; Lee, S.I.; Oh, S.C.; Min, B.W. The significance of microsatellite instability in colorectal cancer after controlling for clinicopathological factors. Medicine. 2018, 97, e0019.
- Requena, D.O.; Garcia-Buitrago, M. Molecular Insights Into Colorectal Carcinoma. Arch. Med. Res. 2020, 51, 839–844.
- Ahmad, R.; Singh, J.K.; Wunnava, A.; Al-Obeed, O.; Abdulla, M.; Srivastava, S.K. Emerging trends in colorectal cancer: Dysregulated signaling pathways (Review). Int. J. Mol. Med. 2021, 47, 1.
- Zhang, Z.H.; Jhaveri, D.J.; Marshall, V.M.; Bauer, D.C.; Edson, J.; Narayanan, R.K.; Robinson, G.J.; Lundberg, A.E.; Bartlett, P.F.; Wray, N.R.; et al. A Comparative Study of Techniques for Differential Expression Analysis on RNA-Seq Data. PLoS ONE 2014, 9, e103207.
- Toyota, M.; Ahuja, N.; Ohe-Toyota, M.; Herman, J.G.; Baylin, S.B.; Issa, J.-P.J. CpG island methylator phenotype in colorectal cancer. Proc. Natl. Acad. Sci. USA 1999, 96, 8681–8686.
- Zong, L.; Abe, M.; Ji, J.; Zhu, W.G.; Yu, D. Tracking the Correlation Between CpG Island Methylator Phenotype and Other Molecular Features and Clinicopathological Features in Human Colorectal Cancers: A Systematic Review and Meta-Analysis. Clin. Transl. Gastroenterol. 2016, 7, e151.
- Advani, S.M.; Advani, P.; DeSantis, S.M.; Brown, D.; VonVille, H.M.; Lam, M.; Loree, J.M.; Mehrvarz Sarshekeh, A.; Bressler, J.; Lopez, D.S.; et al. Clinical, Pathological, and Molecular Characteristics of CpG Island Methylator Phenotype in Colorectal Cancer: A Systematic Review and Meta-analysis. Transl. Oncol. 2018, 11, 1188–1201.
- Russell, M.C.; Chang, G.J. Molecular profiling for stage II colon cancer. Color. Cancer 2012, 1, 479–490.
- Marisa, L.; de Reyniès, A.; Duval, A.; Selves, J.; Gaub, M.P.; Vescovo, L.; Etienne-Grimaldi, M.C.; Schiappa, R.; Guenot, D.; Ayadi, M.; et al. Gene expression classification of colon cancer into molecular subtypes: Characterization, validation, and prognostic value. PLoS Med. 2013, 10, e1001453.
- Clark-Langone, K.M.; Sangli, C.; Krishnakumar, J.; Watson, D. Translating tumor biology into personalized treatment planning: Analytical performance characteristics of the Oncotype DX®Colon Cancer Assay. BMC Cancer 2010, 10, 691.
- Yau, T.O. Precision treatment in colorectal cancer: Now and the future. JGH Open 2019, 3, 361–369.
- Maak, M.; Simon, I.; Nitsche, U.; Roepman, P.; Snel, M.; Glas, A.M.; Schuster, T.; Keller, G.; Zeestraten, E.; Goossens, I. Independent validation of a prognostic genomic signature (ColoPrint) for patients with stage II colon cancer. Ann. Surg. 2013, 257, 1053–1058.
- Sharif, S.; O’Connell, M.J. Gene signatures in stage II colon cancer: A clinical review. Curr. Color. Cancer Rep. 2012, 8, 225–231.
- Fang, S.H.; Efron, J.E.; Berho, M.E.; Wexner, S.D. Dilemma of stage II colon cancer and decision making for adjuvant chemotherapy. J. Am. Coll. Surg. 2014, 219, 1056–1069.
- Graham, D.M.; Coyle, V.M.; Kennedy, R.D.; Wilson, R.H. Molecular subtypes and personalized therapy in metastatic colorectal cancer. Curr. Color. Cancer Rep. 2016, 12, 141–150.
- Ågesen, T.H.; Sveen, A.; Merok, M.A.; Lind, G.E.; Nesbakken, A.; Skotheim, R.I.; Lothe, R.A. ColoGuideEx: A robust gene classifier specific for stage II colorectal cancer prognosis. Gut 2012, 61, 1560–1567.
- Lopez, N.E.; Weiss, A.C.; Robles, J.; Fanta, P.; Ramamoorthy, S.L. A systematic review of clinically available gene expression profiling assays for stage II colorectal cancer: Initial steps toward genetic staging. Am. J. Surg. 2016, 212, 700–714.
- Lenehan, P.F.; Boardman, L.A.; Riegert-Johnson, D.; De Petris, G.; Fry, D.W.; Ohrnberger, J.; Heyman, E.R.; Gerard, B.; Almal, A.A.; Worzel, W.P. Generation and external validation of a tumor-derived 5-gene prognostic signature for recurrence of lymph node-negative, invasive colorectal carcinoma. Cancer 2012, 118, 5234–5244.
- Sveen, A.; Ågesen, T.H.; Nesbakken, A.; Meling, G.I.; Rognum, T.O.; Liestøl, K.; Skotheim, R.I.; Lothe, R.A. ColoGuidePro: A Prognostic 7-Gene Expression Signature for Stage III Colorectal Cancer PatientsPrognostic Gene Expression Classifier of Colorectal Cancer. Clin. Cancer Res. 2012, 18, 6001–6010.
- Salazar, R.; Roepman, P.; Capella, G.; Moreno, V.; Simon, I.; Dreezen, C.; Lopez-Doriga, A.; Santos, C.; Marijnen, C.; Westerga, J. Gene expression signature to improve prognosis prediction of stage II and III colorectal cancer. J. Clin. Oncol. 2011, 29, 17–24.
- Tan, I.B.; Tan, P. An 18-gene signature (ColoPrint®) for colon cancer prognosis. Nat. Rev. Clin. Oncol. 2011, 8, 131–133.
- Kopetz, S.; Tabernero, J.; Rosenberg, R.; Jiang, Z.-Q.; Moreno, V.; Bachleitner-Hofmann, T.; Lanza, G.; Stork-Sloots, L.; Maru, D.; Simon, I. Genomic classifier ColoPrint predicts recurrence in stage II colorectal cancer patients more accurately than clinical factors. Oncol. 2015, 20, 127–133.
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356.
- Llosa, N.J.; Cruise, M.; Tam, A.; Wicks, E.C.; Hechenbleikner, E.M.; Taube, J.M.; Blosser, R.L.; Fan, H.; Wang, H.; Luber, B.S. The Vigorous Immune Microenvironment of Microsatellite Instable Colon Cancer Is Balanced by Multiple Counter-Inhibitory CheckpointsImmune Checkpoints in Human Colorectal Cancer. Cancer Discov. 2015, 5, 43–51.
- Anitei, M.-G.; Zeitoun, G.; Mlecnik, B.; Marliot, F.; Haicheur, N.; Todosi, A.-M.; Kirilovsky, A.; Lagorce, C.; Bindea, G.; Ferariu, D. Prognostic and predictive values of the immunoscore in patients with rectal cancer. Clin. Cancer Res. 2014, 20, 1891–1899.
- Eide, P.W.; Bruun, J.; Lothe, R.A.; Sveen, A. CMScaller: An R package for consensus molecular subtyping of colorectal cancer pre-clinical models. Sci. Rep. 2017, 7, 16618.
- Mooi, J.K.; Wirapati, P.; Asher, R.; Lee, C.K.; Savas, P.; Price, T.J.; Townsend, A.; Hardingham, J.; Buchanan, D.; Williams, D.; et al. The prognostic impact of consensus molecular subtypes (CMS) and its predictive effects for bevacizumab benefit in metastatic colorectal cancer: Molecular analysis of the AGITG MAX clinical trial. Ann. Oncol. 2018, 29, 2240–2246.
- Haasnoot, K.J.C.; Backes, Y.; Moons, L.M.G.; Kranenburg, O.; Trinh, A.; Vermeulen, L.; Noë, M.; Tuynman, J.B.; van Lent, A.U.G.; van Ginneken, R.; et al. Associations of non-pedunculated T1 colorectal adenocarcinoma outcome with consensus molecular subtypes, immunoscore, and microsatellite status: A multicenter case-cohort study. Mod. Pathol. 2020, 33, 2626–2636.
- Ubink, I.; van Eden, W.J.; Snaebjornsson, P.; Kok, N.F.M.; van Kuik, J.; van Grevenstein, W.M.U.; Laclé, M.M.; Sanders, J.; Fijneman, R.J.A.; Elias, S.G.; et al. Histopathological and molecular classification of colorectal cancer and corresponding peritoneal metastases. Br. J. Surg. 2018, 105, e204–e211.
- Rajamäki, K.; Taira, A.; Katainen, R.; Välimäki, N.; Kuosmanen, A.; Plaketti, R.M.; Seppälä, T.T.; Ahtiainen, M.; Wirta, E.V.; Vartiainen, E.; et al. Genetic and Epigenetic Characteristics of Inflammatory Bowel Disease-Associated Colorectal Cancer. Gastroenterology 2021, 161, 592–607.
More
Information
Subjects:
Oncology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
590
Revisions:
2 times
(View History)
Update Date:
15 Jun 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




