
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Guodong Zhu | -- | 2647 | 2023-06-06 15:51:22 | | | |
| 2 | Fanny Huang | Meta information modification | 2647 | 2023-06-07 05:20:32 | | |
Video Upload Options
Kidney tumors comprise a broad spectrum of different histopathological entities, with more than 0.4 million newly diagnosed cases each year, mostly in middle-aged and older men. Based on the description of the 2022 World Health Organization (WHO) classification of renal cell carcinoma (RCC), some new categories of tumor types have been added according to their specific molecular typing. However, studies on these types of RCC are still superficial, many types of these RCC currently lack accurate diagnostic standards in the clinic, and treatment protocols are largely consistent with the treatment guidelines for clear cell RCC (ccRCC), which might result in worse treatment outcomes for patients with these types of molecularly defined RCC.
1. Introduction
| Molecularly Defined Renal Cell Carcinoma Types | TFE3-Rearranged Renal Cell Carcinomas | TFEB-Altered Renal Cell Carcinomas | Elongin C (ELOC, Formerly TCEB1)-Mutated Renal Cell Carcinoma | Fumarate Hydratase-Deficient Renal Cell Carcinoma | Succinate Dehydrogenase-Deficient Renal Cell Carcinoma | ALK-Rearranged Renal Cell Carcinomas | SMARCB1-Deficient Renal Medullary Carcinoma |
|---|---|---|---|---|---|---|---|
| Mutated genes | Transcription factor binding to IGHM enhancer 3 (TFE3) | Transcription factor EB (TFEB) | Elongin C (ELOC) | Fumarate hydratase (FH) gene | Succinate dehydrogenase (SDH) | Anaplastic lymphoma kinase (ALK) | Subfamily B member 1 (SMARCB1) |
| Location of genes | Xp11.23 | 6p21 | 8q21.11 | 1q43 | SDHA: 5p15 SDHB: lp35-p36.1 SDHC: 1q21 SDHD: 11q23 |
2p23 | 22q11.2 |
| Prevalence age | Childhood | Childhood | Middle and old age | Adult | All ages | Childhood | Teenage |
| Clinical Syndromes | None | None | None | Hereditary leiomyomatosis and renal cell carcinoma (HLRCC) | SDH-deficient tumor syndrome | None | Rhabdoid tumor predisposition syndrome; familial schwannomatosis syndrome |
| Chaperone genes | ASPL, PRCC, SFPQ, CLTC, PARP14, RBM10, NONO, MED15 | MALAT1, CLTC, KHDRBS2, CADM2 | None | None | None | VCL, TPM3, EML4, STRN, HOOK1 | None |
| Mode of inheritance | Dominant inheritance | Dominant inheritance | Dominant inheritance | Dominant inheritance | Dominant inheritance | Dominant inheritance | Dominant inheritance |
| Morphological characteristics | Transparent eosinophils; papillary architecture and psammoma bodies under the microscope | TFEB-translocated RCC: the biphasic growth pattern consisting of large and small tumor cells; smaller cells around the basement membrane-like structures; extensive hyalinization; papillary architecture; clear cell morphology. TFEB-amplified RCC: above pattern was less common |
A clear cellular morphology under the microscope; thick fibromuscular bands; branching glandular vesicular; tubular structures | The papillary type or solid, tubulocystic, sieve-like type; abundant eosinophilic granulocytes, perinuclear halo | Cuboidal tumor cells, nested or tubular growth pattern. Characteristic morphology: the presence of vesicles or flocculent inclusions in the cytoplasm |
ALK-rearranged RCC with VCL as a fusion gene: sickle-cell trait; eosinophilic granulocytic stroma; cytoplasmic lumen. Other ALK-rearranged RCC: similar to PRCC; consist of abundant intracellular and extracellular mucins; eosinophilic granuloplasm |
At a high grade at the time of detection; infiltrative growth; sieve or reticular appearance |
| Ancillary test (IHC, FISH) | Positive: PAX8 (100%); TFE3 (95%); CD10 (89%); achromatase (82%). Negative: cytokeratin 7 (CK7); carbonic anhydrase 9 (CA9); GATA3 |
Positive: histone K; Melan-A TFEB-amplified RCC: diffusely or patchily positive when tested for TFEB levels |
Positive: CK7; ELOC; CA9; CD10; ELOC in the nucleus. | Positive: PAX8; succinate dehydrogenase B abnormal succinate semicarbonate (2SC)S-(2-succino)-cysteine. Negative: FH; CK7; TFE3 |
Positive: PAX8; epithelial membrane antigen (EMA). Negative: SDHB; CK7; CD117; histone K; TFE3; HMB45. SDHA-deficient RCC showed negativity for SDHA |
Positive: PAX7; CK10; AMACR; CD3; cytokeratin; ALK. Negative: carbonic anhydrase IX; TFE45; histone enzyme K; Melan A; HMB45 |
Negative: SMARCB1 |
| Oncological behavior and prognosis | May develop metastases within 20–30 years after diagnosis | TFEB-amplified RCC had higher tumor aggressiveness than TFEB-rearranged tumors. The 5-year survival rate for TFEB-amplified RCC was 48% |
Has an aggressive oncological behavior | Have highly staged or distant metastases when diagnosed | Most cases are low grade and have a good prognosis with a low probability of metastasis | ALK-rearranged RCC with VCL as a fusion gene: no recurrence or distant metastasis. Other ALK-rearranged RCC: more aggressive clinical course |
Often found at an advanced stage or with distant metastases; highly aggressive nature of the tumor. Average overall survival: 6–8 months |
2. TFE3-Rearranged Renal Cell Carcinomas
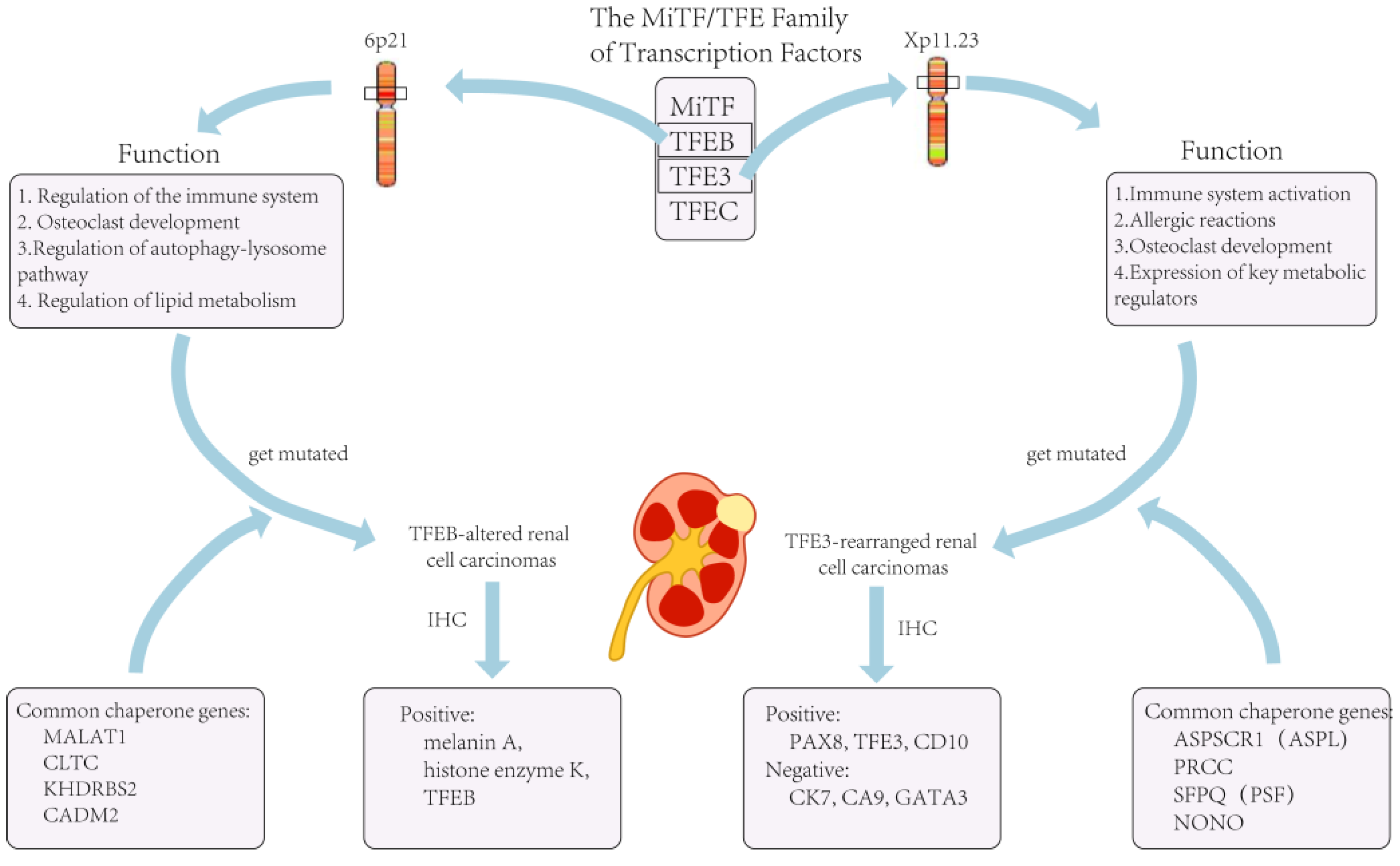
3. TFEB-Altered Renal Cell Carcinomas
References
- Capitanio, U.; Montorsi, F. Renal cancer. Lancet 2016, 10021, 894–906.
- Dibajnia, P.; Cardenas, L.M.; Lalani, A.A. The emerging landscape of neo/adjuvant immunotherapy in renal cell carcinoma. Hum. Vaccin. Immunother. 2023, 1, 2178217.
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 6, 394–424.
- Motzer, R.J.; Jonasch, E.; Agarwal, N.; Alva, A.; Baine, M.; Beckermann, K.; Carlo, M.I.; Choueiri, T.K.; Costello, B.A.; Derweesh, I.H.; et al. Kidney Cancer, Version 3.2022, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2022, 1, 71–90.
- Moch, H.; Amin, M.B.; Berney, D.M.; Compérat, E.M.; Gill, A.J.; Hartmann, A.; Menon, S.; Raspollini, M.R.; Rubin, M.A.; Srigley, J.R.; et al. The 2022 World Health Organization Classification of Tumours of the Urinary System and Male Genital Organs-Part A: Renal, Penile, and Testicular Tumours. Eur. Urol. 2022, 5, 458–468.
- Gray, R.E.; Harris, G.T. Renal Cell Carcinoma: Diagnosis and Management. Am. Fam. Physician 2019, 3, 179–184.
- Padala, S.A.; Barsouk, A.; Thandra, K.C.; Saginala, K.; Mohammed, A.; Vakiti, A.; Rawla, P.; Barsouk, A. Epidemiology of Renal Cell Carcinoma. World J. Oncol. 2020, 3, 79–87.
- Yong, C.; Stewart, G.D.; Frezza, C. Oncometabolites in renal cancer. Nat. Rev. Nephrol. 2020, 3, 156–172.
- Navani, V.; Heng., D.Y.C. Treatment Selection in First-line Metastatic Renal Cell Carcinoma-The Contemporary Treatment Paradigm in the Age of Combination Therapy: A Review. JAMA Oncol. 2022, 8, 292–299.
- Sandoval, J.; Esteller, M. Cancer epigenomics: Beyond genomics. Curr. Opin. Genet. Dev. 2012, 1, 50–55.
- Mitchell, T.J.; Turajlic, S.; Rowan, A.; Nicol, D.; Farmery, J.H.R.; O’Brien, T.; Martincorena, I.; Tarpey, P.; Angelopoulos, N.; Yates, L.R.; et al. TRACERx Renal Consortium. Timing the Landmark Events in the Evolution of Clear Cell Renal Cell Cancer: TRACERx Renal. Cell 2018, 3, 611–623.e17.
- Xing, T.; He, H. Epigenomics of clear cell renal cell carcinoma: Mechanisms and potential use in molecular pathology. Chin. J. Cancer Res. 2016, 1, 80–91.
- Pastore, N.; Vainshtein, A.; Klisch, T.J.; Armani, A.; Huynh, T.; Herz, N.J.; Polishchuk, E.V.; Sandri, M.; Ballabio, A. TFE3 Regulates whole-body energy metabolism in cooperation with TFEB. EMBO Mol. Med. 2017, 5, 605–621.
- Argani, P. Translocation carcinomas of the kidney. Genes. Chromosomes Cancer 2022, 5, 219–227.
- Caliò, A.; Segala, D.; Munari, E.; Brunelli, M.; Martignoni, G. MiT Family Translocation Renal Cell Carcinoma: From the Early Descriptions to the Current Knowledge. Cancers 2019, 8, 1110.
- Sukov, W.R.; Hodge, J.C.; Lohse, C.M.; Leibovich, B.C.; Thompson, R.H.; Pearce, K.E.; Wiktor, A.E.; Cheville, J.C. TFE3 rearrangements in adult renal cell carcinoma: Clinical and pathologic features with outcome in a large series of consecutively treated patients. Am. J. Surg. Pathol. 2012, 5, 663–670.
- Akgul, M.; Williamson, S.R.; Ertoy, D.; Argani, P.; Gupta, S.; Caliò, A.; Reuter, V.; Tickoo, S.; Al-Ahmadie, H.A.; Netto, G.J.; et al. Diagnostic approach in TFE3-rearranged renal cell carcinoma: A multi-institutional international survey. J. Clin. Pathol. 2021, 5, 291–299.
- Kmeid, M.; Akgul, M. TFE3 Rearrangement and Expression in Renal Cell Carcinoma. Int. J. Surg. Pathol. 2022, 1, 10668969221108517.
- Caliò, A.; Marletta, S.; Brunelli, M.; Pedron, S.; Portillo, S.C.; Segala, D.; Bariani, E.; Gobbo, S.; Netto, G.; Martignoni, G. TFE3 and TFEB-rearranged renal cell carcinomas: An immunohistochemical panel to differentiate from common renal cell neoplasms. Virchows Arch. 2022, 6, 877–891.
- Argani, P.; Zhong, M.; Reuter, V.E.; Fallon, J.T.; Epstein, J.I.; Netto, G.J.; Antonescu, C.R. TFE3-Fusion Variant Analysis Defines Specific Clinicopathologic Associations Among Xp11 Translocation Cancers. Am. J. Surg. Pathol. 2016, 6, 723–737.
- Argani, P. MiT family translocation renal cell carcinoma. Semin. Diagn. Pathol. 2015, 2, 103–113.
- Tretiakova, M.S. Chameleon TFE3-translocation RCC and How Gene Partners Can Change Morphology: Accurate Diagnosis Using Contemporary Modalities. Adv. Anat. Pathol. 2022, 3, 131–140.
- Green, W.M.; Yonescu, R.; Morsberger, L.; Morris, K.; Netto, G.J.; Epstein, J.I.; Illei, P.B.; Allaf, M.; Ladanyi, M.; Griffin, C.A.; et al. Utilization of a TFE3 break-apart FISH assay in a renal tumor consultation service. Am. J. Surg. Pathol. 2013, 8, 1150–1163.
- Lee, H.J.; Shin, D.H.; Kim, S.Y.; Hwang, C.S.; Lee, J.H.; Park, W.Y.; Choi, K.U.; Kim, J.Y.; Lee, C.H.; Sol, M.Y.; et al. TFE3 translocation and protein expression in renal cell carcinoma are correlated with poor prognosis. Histopathology 2018, 5, 758–766.
- Wang, X.M.; Zhang, Y.; Mannan, R.; Skala, S.L.; Rangaswamy, R.; Chinnaiyan, A.; Su, F.; Cao, X.; Zelenka-Wang, S.; McMurry, L.; et al. TRIM63 is a sensitive and specific biomarker for MiT family aberration-associated renal cell carcinoma. Mod. Pathol. 2021, 8, 1596–1607.
- Aldera, A.P.; Ramburan, A.; John, J. TFE3-rearranged renal cell carcinoma with osseous metaplasia and indolent behaviour. Urol. Case Rep. 2022, 42, 102041.
- Gupta, S.; Argani, P.; Jungbluth, A.A.; Chen, Y.B.; Tickoo, S.K.; Fine, S.W.; Gopalan, A.; Al-Ahmadie, H.A.; Sirintrapun, S.J.; Sanchez, A.; et al. TFEB Expression Profiling in Renal Cell Carcinomas: Clinicopathologic Correlations. Am. J. Surg. Pathol. 2019, 11, 1445–1461.
- Argani, P.; Reuter, V.E.; Zhang, L.; Sung, Y.S.; Ning, Y.; Epstein, J.I.; Netto, G.J.; Antonescu, C.R. TFEB-amplified Renal Cell Carcinomas: An Aggressive Molecular Subset Demonstrating Variable Melanocytic Marker Expression and Morphologic Heterogeneity. Am. J. Surg. Pathol. 2016, 11, 1484–1495.
- Wyvekens, N.; Rechsteiner, M.; Fritz, C.; Wagner, U.; Tchinda, J.; Wenzel, C.; Kuithan, F.; Horn, L.C.; Moch, H. Histological and molecular characterization of TFEB-rearranged renal cell carcinomas. Virchows Arch. 2019, 5, 625–631.
- Williamson, S.R.; Grignon, D.J.; Cheng, L.; Favazza, L.; Gondim, D.D.; Carskadon, S.; Gupta, N.S.; Chitale, D.A.; Kalyana-Sundaram, S.; Palanisamy, N. Renal Cell Carcinoma with Chromosome 6p Amplification Including the TFEB Gene: A Novel Mechanism of Tumor Pathogenesis? Am. J. Surg. Pathol. 2017, 3, 287–298.
- Williamson, S.R.; Eble, J.N.; Palanisamy, N. Sclerosing TFEB-rearrangement renal cell carcinoma: A recurring histologic pattern. Hum. Pathol. 2017, 62, 175–179.
- Gupta, S.; Johnson, S.H.; Vasmatzis, G.; Porath, B.; Rustin, J.G.; Rao, P.; Costello, B.A.; Leibovich, B.C.; Thompson, R.H.; Cheville, J.C.; et al. TFEB-VEGFA (6p21.1) co-amplified renal cell carcinoma: A distinct entity with potential implications for clinical management. Mod. Pathol. 2017, 7, 998–1012.




