Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ademola Hammed | -- | 3913 | 2023-05-30 16:28:50 | | | |
| 2 | Jessie Wu | Meta information modification | 3913 | 2023-05-31 05:37:17 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Adeniyi, A.; Bello, I.; Mukaila, T.; Sarker, N.C.; Hammed, A. Ammonia Classification and Biological Ammonia Production. Encyclopedia. Available online: https://encyclopedia.pub/entry/45020 (accessed on 07 February 2026).
Adeniyi A, Bello I, Mukaila T, Sarker NC, Hammed A. Ammonia Classification and Biological Ammonia Production. Encyclopedia. Available at: https://encyclopedia.pub/entry/45020. Accessed February 07, 2026.
Adeniyi, Adewale, Ibrahim Bello, Taofeek Mukaila, Niloy Chandra Sarker, Ademola Hammed. "Ammonia Classification and Biological Ammonia Production" Encyclopedia, https://encyclopedia.pub/entry/45020 (accessed February 07, 2026).
Adeniyi, A., Bello, I., Mukaila, T., Sarker, N.C., & Hammed, A. (2023, May 30). Ammonia Classification and Biological Ammonia Production. In Encyclopedia. https://encyclopedia.pub/entry/45020
Adeniyi, Adewale, et al. "Ammonia Classification and Biological Ammonia Production." Encyclopedia. Web. 30 May, 2023.
Copy Citation
Ammonia, a compound with the chemical formula NH3, is composed of two of the most ubiquitous elements on Earth—nitrogen and hydrogen. Ammonia is colorless and characterized by its pungent odor. Ammonia has a wide range of industrial and agricultural applications due to its unique properties
biological ammonia
bioprocessing
bioengineering
fermentation
enzyme immobilization
1. Ammonia Classification
Ammonia production systems can be classified into three categories based on the carbon emissions from the production processes but not the type of ammonia being produced: brown (or grey) ammonia, blue ammonia, and green ammonia.
1.1. Brown (or Grey) Ammonia
The Haber-Bosch process is the conventional method for ammonia production. It is responsible for more than 60% of the ammonia produced globally. Due to its high energy requirement and significant contribution to CO2 emissions, the resulting ammonia from the Haber-Bosch process is termed brown ammonia. The Haber-Bosch process is the reaction of nitrogen (N2) and hydrogen (H2) in the presence of an iron catalyst and other oxide promoters such as K2O, Al2O, and CaO. The reaction runs at around 400–600 °C for efficient catalysis and up to 200–400 atmospheres of gas pressure to enhance entropy, an energy-hungry reaction that sucks up about 1% of global energy production and is thermodynamically exothermic [1].
N2(g) + 3H2(g) ⇌ 2NH3(g) ∆Ho = −92 kJ
While the nitrogen in the reaction above is extracted from the air, the hydrogen comes from natural gas (methane), oil, or coal through industrial processes that release CO2. Steam methane reforming is the most commonly used method to generate hydrogen, generating CO2 emissions. The current form of the Haber-Bosch process begins by generating hydrogen from fossil-fuel feedstocks, usually coal or oil. A reformer converts the feedstocks into a mixture of gases (syngas), which includes hydrogen. Thereafter, a carbon monoxide shift converter mixes water and the carbon monoxide from the preformed syngas to form carbon dioxide (CO2) and more hydrogen. The final steps involve the separation of hydrogen from ammonia synthesis by acid gas removal. At various steps of the process, CO2 is released. For every molecule of natural gas (methane) used, three molecules of CO2 are generated, and 1.6 tons of CO2 is emitted per ton of ammonia produced from the most efficient ammonia production plants [2][3].
CH4 + H2O + 4N2 ↝ 8NH3 + 3CO2
1.2. Blue Ammonia
Efforts by engineers across the world to make ammonia production less energy consuming and sustainable gave rise to the concept of blue ammonia. Blue ammonia, like brown ammonia, is produced from hydrocarbon feedstocks, but carbon capture utilization and storage (CCUS) technologies are integrated into ammonia production plants to sequester the resulting CO2. Of all the CCUS technologies known, amine absorption technology is the most widely used and commercially available [4]. Amines (or alkanolamines) are organic compounds with a basic nitrogen atom. They can be used to separate CO2 from the gas stream during ammonia production through the exothermic reaction of CO2 with an amine. Another CCUS technology is based on the principle that CO2 from any gas mixture (syngas) can be separated by cooling and condensation [5]. The technology, termed cryogenic separation, facilitates the direct production of liquid CO2, which can be transported. Although the amount of energy required for cooling in the process is relatively high and water must be removed to prevent cooling of the blocks by gas flow, the use of membranes in the gas separation process is promising [6]. Some of the membranes known to decompose CO2 are palladium membranes, polymeric membranes, and zeolites [7].
Another notable CCUS technology uses an adsorption device, rotary concentrator, on solids. The solid materials used for the adsorption include activated carbons, activated aluminum oxide (Al2O3), clays, zeolites, and silicon dioxide (SiO2). A modified version of this technology is pressurized swing adsorption (PSA), in which the gas mixture flows in the direction of the packed bed of the adsorbent at high pressure until the concentration of the desired gas to be separated reaches equilibrium [8]. The captured CO2 can be stored by several methods and used for a variety of production processes, including increased oil recovery, coal bed methane extraction, and deep ocean injection, among others [3][9]. In the long run, CCUS blue ammonia production technology will not be beneficial as high energy is still being used to drive the process, and the lack of CCUS infrastructure as well as transportation of CO2 poses yet another challenge.
1.3. Green Ammonia
The ammonia production process targeted at reducing or completely removing carbon dioxide emissions birthed the concept of green ammonia. To achieve zero carbon emissions during ammonia production, renewable feedstocks coupled with reduced energy usage are harnessed. At present, the most desirable but expensive green ammonia production method generates hydrogen from water electrolysis powered by solar, wind, hydroelectric, or geothermal energy [10]. This approach is also known as electrochemical ammonia synthesis (EAS). Electrolytes used for the EAS are diverse; they include solid electrolytes such as polymer electrolyte membrane (PEM) and anion exchange membrane (AEM), chlorine salts, melt hydroxides, and acidic electrolytes in liquid form [11]. Electrolysis in the latter is mostly done by the deposition of ammonium salts in solution to cause rapid changes in the pH of the solution. The low solubility of nitrogen often hinders electrolysis in its solutions. Thus, gas diffusion in electrodes is required for high efficiency and production rates [3].
In the EAS method, electrocatalysts commonly used based on the physical state and pH of the electrolyte include precious metals, metal nitrides, and metal oxides [12]. Transition metal-free catalysts such as black phosphorus and nitrogen-doped carbons are also known catalysts. The use of these catalysts minimizes the loss of nitrogen and improves process efficiency for high ammonia synthesis [13]. One of two reactors could be used to conduct the electrolysis: hydrogen generation reactors or nitrogen reduction reactors. The latter is preferred for low-temperature applications and gives more yield in downstream ammonia synthesis. Higher ammonia production efficiency can, however, be achieved in the hydrogen generation reactor by adding ZrO2 to the ruthenium catalyst. Similarly, reducing the number of protons on the catalyst surface by using high-pH electrolytes has also been shown to solve the underproduction problem in the hydrogen generation reactor [14]. The source of nitrogen is also crucial in the EAS method. The moisture content of the air used as the nitrogen source is an important parameter that affects the ammonia conversion rate. Using high-purity nitrogen from the air with reduced moisture will significantly increase ammonia synthesis [15].
Albeit the innovation of EAS to produce ammonia in an environmentally friendly manner, energy consumption is still unacceptably high due to the high current density utilized for hydrogen production, and the process occurs at a low capacity (10−9 to 10−11 mol cm−2 s−1) [16]. The water electrolyzer used in EAS requires a continuous supply of high-purity, pretreated water for its operation. Consequently, nine tons of water are consumed for every ton of hydrogen produced, and for the production of an amount of ammonia by EAS through water electrolysis, approximately double the amount of water is required, deepening the worldwide water crisis [10].
2. Biological Ammonia Production
Biological approaches are considered eco-friendly as they are natural processes that do not produce any harmful by-products. There are several approaches for biological ammonia production, including nitrogen fixation, nitrification, nitrate/nitrite reduction, urea hydrolysis, metabolic engineering of microorganisms, and in vitro ruminal microbial fermentation of protein biomass, but the most reported methods are biological nitrogen fixation (BNF) and metabolic engineering of microorganisms. Biological ammonia production by rumen bacteria fermentation of protein biomass, as experimented on in this research, is a relatively new approach and has shown the potential to complement ammonia bioproduction.
2.1. Biological Nitrogen Fixation by Nitrogenase
Biological nitrogen fixation (BNF) is a natural process that converts atmospheric molecular nitrogen (N2) to ammonia (NH3). BNF, an ATP-dependent reduction reaction catalyzed by the nitrogenase enzyme, is responsible for approximately half of the bioavailable nitrogen that supports all life forms [17]. Relative to the Haber-Bosch process, which requires high temperature and pressure conditions to break down molecular nitrogen, nitrogen-fixing microorganisms produce ammonia at ambient temperature and pressure. Nitrogen-fixing microbes are robust and have been explored to produce biofertilizers in commercial quantities [18][19]. Researchers are actively making attempts to mimic the natural process of BNF by isolating nitrogen-fixing bacteria (Figure 1) and nitrogenase for synthetic ammonia production. The major challenge with this research effort is that nitrogenase catalysis is highly energy dependent, making its reaction rate slower than most enzymes in nature [20].
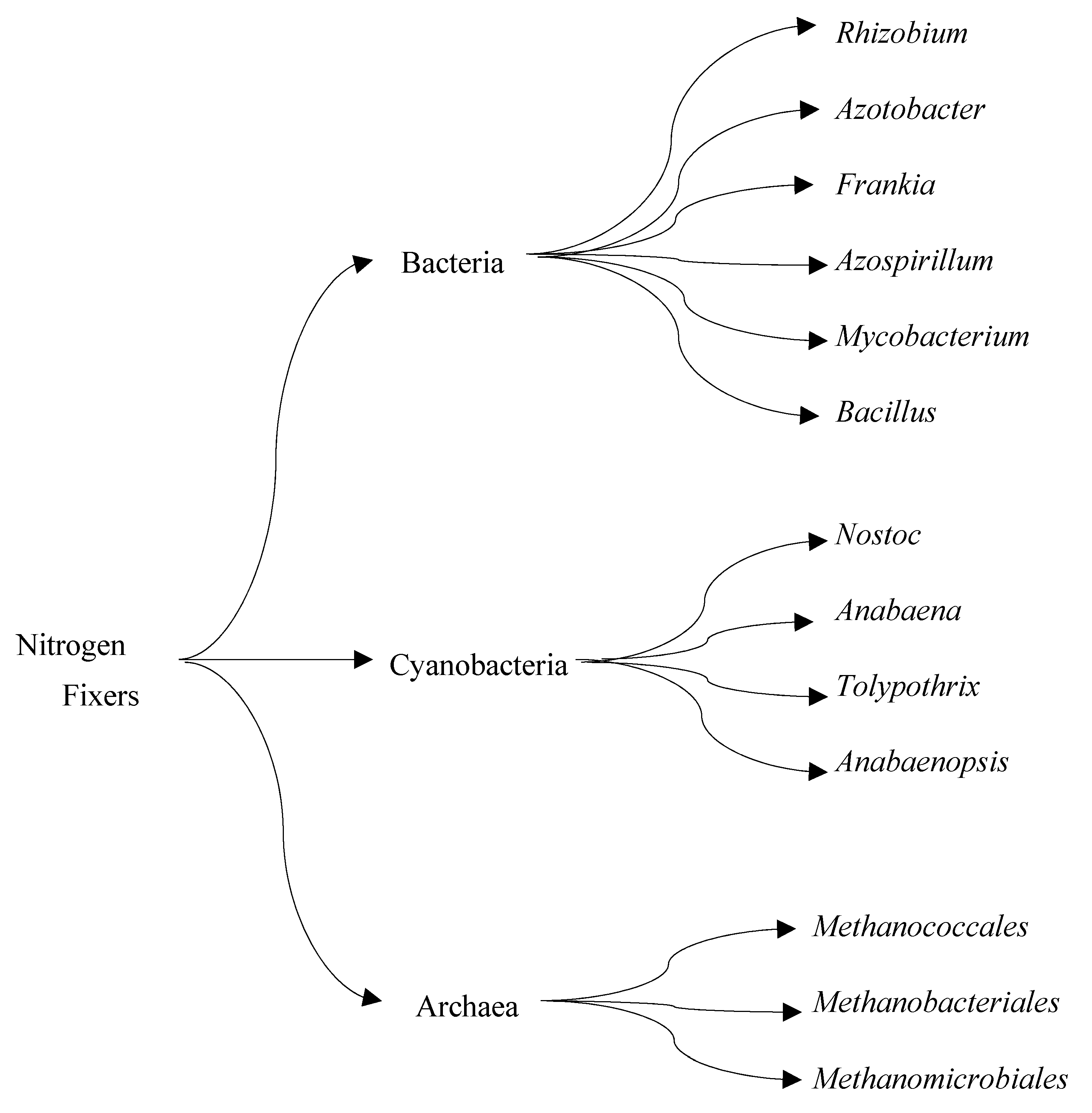
Figure 1. Three groups of nitrogen-fixing microorganisms.
The main microorganisms that possess nitrogenase and carry out nitrogen fixation are the genus Rhizobia, which colonizes the root of legumes, and species in the genera Azotobacter and Klebsiella that can fix nitrogen without parasitizing plant roots. The latter group is the main focus of research on synthetic BNF [21][22]. Nitrogenase requires up to eight molecules of ATP to produce a molecule of ammonia in an anoxic condition. Although the reaction mechanism of nitrogenase is unclear due to its multiple interrelated subunits, scientists have attempted to construct a heterologous expression system for Klebsiella nitrogenase subunits in E. coli [23]. Similarly, heterologous expression of the Klebsiella nitrogenase gene cluster has been constructed in E. coli and yeast to understand the mechanism by which nitrogenase functions without oxygen as well as to increase its activity [24][25]. Various studies have also investigated how nitrogen-fixing bacteria can function under aerobic conditions without inactivating nitrogenase. Such research involves the use of polysaccharide membranes to protect nitrogenase from oxygen exposure [25][26].
For industrial applications of nitrogen-fixing bacteria, some biotechnology companies have engineered Enterobacter sp. lacking glutamine due to low expression of the transcription factor GlnR to increase intracellular glutamine and, consequently, synthesize ammonia in the presence of nitrogenase [27]. Steady nutrient supply through BNF has also been successful with non-leguminous crop plants such as corn. The use of anaerobic microflora is also a known strategy for ammonia bioproduction by BNF, and a plethora of methodologies for ammonia recovery have been established. A notable one is the evaporation of solution following fermentation and pH increases [28][29][30].
2.2. Cell and Metabolic Engineering for Ammonia Production
Various biomasses, including food waste, microbial biomass, and protein-rich crop residues, can be fermented by engineered microorganisms whose metabolisms are well understood for ammonia bioproduction. In a metabolic engineering study on the conversion of protein wastes into biofuels and ammonia using microbes, the codY gene (a transcriptional regulator), in Bacillus subtilis was knocked out. The codY gene regulates the activity of several other genes involved in different processes, such as producing branched-chain amino acids (ilvABHCD and leuABCD), removing amino groups from other molecules (ybgE, ald, yhdC, appBC, and dppBC), and inhibiting the expression of genes that cause protein breakdown and uptake (yhdG, appBC, and dppBC). In bacteria, proteins are encoded for amino acid biosynthesis by the ilv-leu operon. The deletion of the codY gene removed regulatory constraints on this operon, causing a significant increase in the production and uptake of branched-chain amino acids (BCAA) due to the derepression of the ilv-leu operon and subsequent upregulation of genes responsible for BCAA synthesis.
In addition to the deletion of the codY gene, the BkdB gene in Bacillus subtilis was also knocked out. BkdB is a lipoamide acyltransferase enzyme that helps in the biosynthesis of branched-chain fatty acids by converting branched-chain keto acids into their acyl-CoA derivatives. This conversion inhibits the production of biofuels and ammonia. The BkdB gene knockout had a significant impact on the production of branched-chain fatty acids in Bacillus subtilis. Obstruction of production resulted in increased availability of metabolic precursors for the production of biofuels and ammonia. To completely transform B. subtilis to favor ammonia synthesis, an alcohol dehydrogenase gene, leuDH, and two-keto-acid decarboxylase were overexpressed. LeuDH is an alcohol dehydrogenase gene that plays an important role in the conversion of amino acids to alpha-keto acids, while two-keto-acid decarboxylase is an enzyme that catalyzes the decarboxylation of alpha-keto acids, which are important metabolic intermediates in amino acid biosynthesis. Overexpression of LeuDH increased the rate of amino-acid nitrogen reflux, which helped to increase the efficiency of protein conversion. Similarly, overexpressing two-keto-acid decarboxylase led to the increased availability of metabolic precursors such as alpha-ketoisocaproate (KIC) and alpha ketoglutarate (AKG) for the production of ammonia. The resulting final strain of B. subtilis was employed in the fermentation of protein biomass obtained from E. coli cells. This process produced ammonia with a theoretical yield of about 50% [31].
A similar study on ammonia production from amino acid-based biomass-like sources using engineered E. coli has been reported [32]. Since E.coli assimilates ammonia intracellularly [33], the two genes involved in the ammonia assimilation pathway, glnA and gdhA which are both glutamine assimilation genes, were knocked out to enhance ammonia production. glnA encodes for enzyme glutamine synthetase (GS) and catalyzes the conversion of glutamate and ammonia to glutamine, while gdhA encodes for the enzyme glutamate dehydrogenase (GDH) and catalyzes the reversible conversion of glutamate and ammonia to alpha-ketoglutarate. The deletion of glnA promotes the extracellular leaching of ammonia, while the deletion of gdhA increases ammonia flux to produce more glutamate, a known precursor of ammonia. In the research, deleting the two genes redirected the nitrogen assimilation pathways in E. coli toward ammonia production, resulting in a peak titer yield of 458 mg/L, equivalent to an overall yield of 47.8% [32].
Further studies on the metabolic engineering of E. coli for ammonia production converted different food wastes, including soy sauce cake, mirin cake, and tomato peel, to ammonia. Using metabolic profiling to assess the correlation between substances in the media (amino acids, sugars, and organic acids) and ammonia production, glucose was implicated as an inhibitor of ammonia production. When glucose was added to the amino acid-containing medium at different concentrations, a negative correlation with ammonia production was obtained. Thus, E. coli was engineered to hinder the inhibitory effect of glucose by knocking out the transporter gene, ptsG, and the phosphotransferase system, which transports glucose and other sugars. Briefly, the polymerase chain reaction (PCR) technique was used to amplify and copy specific fragments of genes that encoded resistance to pts’G-Kim and glnA-Km (amplified from pKD13) using primers ptsGF and ptsGR. The amplified DNA fragments were then transferred into E. coli cells through electroporation. Following the transfer, E. coli cells were grown on LB agar containing specific antibiotics—ampicillin and kanamycin. This allowed only the cells that had taken up the amplified DNA fragments to survive and grow, while the others died off. By repeating this process with different combinations of DNA fragments and antibiotics, more varieties of E. coli strains with different genetic modifications, such as AptsG and AglnA, were created. To ensure that the modified DNA fragments had been inserted into the correct location in the E. coli genome, PCR was used to amplify and sequence the insertion region using insertion-checking primers. The resulting E. coli strain succeeded in producing ammonia in a glucose-containing amino acid medium, with up to 73% yield [34]. In the studies described above, ammonia was, however, produced intracellularly. As a result, theproduced ammonia can still be used up by these microbes for growth [33]. Therefore, a system that can produce ammonia extracellularly without impeding microbial growth may improve productivity.
Studies on yeast for extracellular ammonia production have been attempted. Prominent among such studies is the use of yeast cell surface engineering (YCSE) systems to avoid ammonia toxicity and assimilation. In YCSE, the protein to be converted to ammonia is displayed on the cell surface, usually by the attachment of a secretory signal to the N-terminus of the target protein and a signal sequence, an α-agglutin containing a glycosylphosphatidylinositol anchor, on its C-terminus. Briefly, the plasmid for yeast cell surface display of L-amino acid oxidase was constructed by synthesizing and inserting the codon-optimized sequence of HcLAAO (L-amino acid oxidase) into pULDl, resulting in a plasmid named pULDl-HcLAAO. A strep-tag negative control plasmid called pULDl-s was also constructed. The yeast strain Saccharomyces cerevisiae BY4741/sedlA was utilized to display HcLAAO on the cell surface. The constructed plasmid was then introduced into the yeast strain. Yeast cells were then transformed, grown in a synthetic dextrose medium and cultured in SDC buffer at pH 7.0. Using this approach, up to106 target proteins could be displayed on the yeast cell surface, which are then used as biocatalysts for enzyme immobilization [33][35][36].
Ammonia production from soybean residues has been successful with the YCSE technique [37]. Amino acid catabolic enzymes that produce ammonia from amino acid precursors, such as ammonia lyases, have attracted interest for their efficiency in being displayed on the yeast cell surface because their catalysis does not require cofactors, unlike nitrogenases. With yeast cells displaying glutamine ammonia-lyases, ammonia was produced from glutamine solution, reaching a titer of up to 3.34 g/L and an efficiency of 83.2% [37]. The limitation of this approach is that only glutamine, of the 20 amino acids, can be utilized. Interestingly, L-amino acid oxidase with a broad substrate specificity can be displayed for ammonia production from several amino acids [38][39]. These are lab-scale studies that may be difficult to transition to an industrial scale for eco-friendly biological ammonia production. Table 1 shows a summary of the metabolic engineering route for biological ammonia production.
Table 1. Summary of metabolic engineering approaches for biological ammonia production.
| Approach | Description | Host | Substrates | Ref |
|---|---|---|---|---|
| Gene knockout | Deletion of CodY gene which regulates genes:
|
Bacillus subtilis | Amino acid | [31] |
| Gene knockout | Deletion of gene BkdB which helps in the biosynthesis of branched chain fatty acids | Bacillus subtilis | Amino acid | [31] |
| Gene overexpression | Over expression of proteins leuDH, and two-keto-acid decarboxylase which respectively converts amino acids to important metabolic intermediates and increases the availability of metabolic precursors for ammonia production | Bacillus subtilis | Amino acid | [31] |
| Gene knockout | Deletion of genes glnA and gdhA which aids ammonia assimilation | Eschericia coli | Amino acid | [32] |
| Gene knockout | Deletion of ptsG (glucose transporter gene) and deletion of phosphoenol pyruvate (glucose transporter) | Eschericia coli | Soybean residue and food waste | [34] |
| Cell surface engineering | HcLAAO (L-amino acid oxidase) display on yeast cell surface by gene insertion | Yeast cells | Amino acids from soybean residue | [33] |
| Cell surface engineering | Glutaminase gene (Ybas) display on yeast cell surface by gene insertion. | Yeast cells | Soybean residue and glutamine | [37]. |
2.3. Ammonia from Wastewater Treatment Plants
Microbial fuel cell technology can be used to produce ammonia in wastewater treatment plants through a process called ammonia oxidation [40][41]. Ammonia oxidation involves the use of specialized bacteria that are capable of oxidizing ammonia to produce electrons, which can then be used to generate electricity. In a typical microbial fuel cell system for ammonia production, the wastewater is first pumped into an anaerobic anode chamber. The anaerobic environment allows the bacteria to break down organic matter in the wastewater, releasing electrons in the process. The bacteria responsible for ammonia oxidation are then introduced into the anode chamber. These bacteria are able to use the electrons produced by the organic matter breakdown to oxidize ammonia in the wastewater [42]. Consequently, the wastewater is cleaned up, and ammonium is removed and converted into harmless gaseous N2 [43].
Ammonia can also be generated in wastewater treatment plants through ammonification. Ammonification, the breakdown of food waste, human waste, and other nitrogen-containing biological materials present in wastewater, converts the nitrogen-containing organic matter into ammonia [44][45]. Following the removal of large solids and debris from influent wastewater [46], the resulting wastewater is made to undergo a series of treatment processes to reduce nutrient levels, including a specialized approach called biological nutrient removal [47]. This process employs specific anaerobic bacteria such as Clostridium perfringens, Peptostreptococcus, Actinomyces meyeri, Bifidobacterium species, Propionibacterium, Bacterioides, and Fusobacterium [48] to break down organic matter and convert nitrogen compounds into ammonia. The ammonia produced is then further transformed into nitrate and nitrite ions through a process called nitrification [49][50]. Although there have been numerous studies conducted on ammonia production from wastewater treatment plants [43][51][52][53][54][55][56][57][58][59][60], most of the processes involved are highly energy-intensive and economically non-viable [43].
2.4. Hyper Ammonia-Producing Bacteria Route
The digestive compartment of ruminant animals, the rumen, is a biorefinery for ammonia production. Ruminal microorganisms can break down plant materials containing carbohydrates and proteins in their feeds for energy. The products of protein degradation, including peptides and amino acids, are metabolized to protein and/or ammonia. The microbial protein thus formed is required for animal products, but the ammonia is absorbed from the rumen, metabolized, and excreted in the urine. This is an inefficient use of dietary proteins with devastating consequences for the environment through environmental nitrogen pollution [61].
Several studies in the animal sciences have sought strategies to promote microbial protein synthesis and regulate ammonia production. These studies revealed the identity of a certain group of bacteria whose rate of ammonia production is much higher than can be used up by the ruminal microbes for other functions, including microbial protein synthesis [62][63][64]. This group of bacteria, known as the hyper-ammonia-producing bacteria (HAB), can effectively convert dietary protein to surplus ammonia [65][66]. This type of natural ammonia is produced when the digestive systems of humans and animals undergo a biochemical reaction leading to the breakdown of nitrogen-containing amines (NH2) in proteins into ammonia or the ionic form (ammonium). It is referred to as biological ammonia.
The first step towards the degradation of amino acids is deamination, which is the removal of an amine group to convert it to ammonia. It has been reported that amino acid deamination in the rumen produces more ammonia than can be utilized by the bacteria [67]. Deamination may occur through oxidation, reduction, hydrolysis, or the removal of elements. It helps to free the carbon skeleton by removing the amine group from the amino acid. Furthermore, deamination could be carried out on either a single amino acid, pairs of amino acids as in the case of the Stickland reaction, or a combination of amino acids and a non-nitrogenous compound, all resulting in ammonia and keto-acids as major products [68].
The next biochemical reaction is called ammonification, which is the second stage of mineralization [69]. Useful energy can also be derived metabolically by bacteria and related microorganisms through ammonification. Ammonium (NH4+) is thus produced by microorganisms, and if in excess, it is excreted into the environment as nutrients for uptake by plants or as feedstock for further nitrification [69]. HABs have been implicated in converting ~50% of ruminal dietary protein to ammonia [70][71][72].
HABs are found in cattle rumen or swine manure stored in the pit [73][74][75]. Additionally, HABs thrive in the rumen of the hay-fed cattle compared to grain-fed cattle [62] because the pH of hay-fed cattle rumen environment is relatively neutral, thus providing a favorable condition for their growth compared to the slightly acidic pH (<6.0) observed in grain-fed cattle [76]. HABs are capable of producing up to 40 mM (0.6812 mg/L) of ammonia in peptone-amino acid medium, depending on energy and carbon source [74][77]. HABs can operate in both anaerobic and aerobic environments, but anaerobic-HABs are more prominent and of major concern because they convert a large percentage of dietary protein in the rumen to ammonia [78]. Although HABs are detrimental to ruminant metabolism due to excess ammonia generation causing toxicity to rumen microbes and hyperammonemia in farm animals [78], they can be harnessed as a sustainable source for large-scale ammonia production with low energy requirements and zero emissions.
There are several strains of hyper-ammonia-producing bacteria (HAB) with different biological ammonia-production capacities. Selenomonas ruminantium, Peptostreptococcus elsdenii, and Bacteroides ruminicola are HAB strains that are capable of producing at least 1 µM of biological ammonia on a lab scale through deamination. S. ruminantium catabolizes cysteine hydrolysate, while P. elsdenii breaks down casein hydrolysate and specific amino acids (L-serine, L-threonine, and L-cysteine) to produce biological ammonia [79][80]. Depending on HAB strain and environmental conditions, it is also possible to produce much higher concentrations of biological ammonia (>24 mM) [77].
References
- Paschkewitz, T.M. Ammonia Production at Ambient Temperature and Pressure. Ph.D. Thesis, University of Iowa, Iowa City, IA, USA, 2012.
- Boerner, L.K. Industrial Ammonia Production Emits More CO2 than Any Other Chemical-Making Reaction. Chemists Want to Change That. Chem. Eng. News 2019, 97, 1–9.
- Yüzbaşıoğlu, A.E.; Tatarhan, A.H.; Gezerman, A.O. Decarbonization in Ammonia Production, New Technological Methods in Industrial Scale Ammonia Production and Critical Evaluations. Heliyon 2021, 7, e08257.
- Hussin, F.; Aroua, M.K. Recent Trends in the Development of Adsorption Technologies for Carbon Dioxide Capture: A Brief Literature and Patent Reviews (2014–2018). J. Clean. Prod. 2020, 253, 119707.
- Abdelkareem, M.A.; Lootah, M.A.; Sayed, E.T.; Wilberforce, T.; Alawadhi, H.; Yousef, B.A.A.; Olabi, A.G. Fuel Cells for Carbon Capture Applications. Sci. Total Environ. 2021, 769, 144243.
- Kamble, A.R.; Patel, C.M.; Murthy, Z.V.P. A Review on the Recent Advances in Mixed Matrix Membranes for Gas Separation Processes. Renew. Sustain. Energy Rev. 2021, 145, 111062.
- Mutch, G.A.; Qu, L.; Triantafyllou, G.; Xing, W.; Fontaine, M.L.; Metcalfe, I.S. Supported Molten-Salt Membranes for Carbon Dioxide Permeation. J. Mater. Chem. A 2019, 7, 12951–12973.
- Siqueira, R.M.; Freitas, G.R.; Peixoto, H.R.; Nascimento, J.F.D.; Musse, A.P.S.; Torres, A.E.B.; Azevedo, D.C.S.; Bastos-Neto, M. Carbon Dioxide Capture by Pressure Swing Adsorption. Energy Procedia 2017, 114, 2182–2192.
- Bello, I.; Rasaq, N.; Adeniyi, A.; Hammed, A. Enzyme Aided Processing of Oil. Int. J. Halal Res. 2021, 3, 60–72.
- Ghavam, S.; Vahdati, M.; Wilson, I.G.; Styring, P. Sustainable Ammonia Production Processes. Front. Energy Res. 2021, 9, 580808.
- Miller, H.A.; Bouzek, K.; Hnat, J.; Loos, S.; Bernäcker, C.I.; Weißgärber, T.; Röntzsch, L.; Meier-Haack, J. Green Hydrogen from Anion Exchange Membrane Water Electrolysis: A Review of Recent Developments in Critical Materials and Operating Conditions. Sustain. Energy Fuels 2020, 4, 2114–2133.
- Yang, J.; Weng, W.; Xiao, W. Electrochemical Synthesis of Ammonia in Molten Salts. J. Energy Chem. 2020, 43, 195–207.
- Zhang, L.; Ji, X.; Ren, X.; Ma, Y.; Shi, X.; Tian, Z.; Asiri, A.M.; Chen, L.; Tang, B.; Sun, X. Electrochemical Ammonia Synthesis via Nitrogen Reduction Reaction on a MoS2 Catalyst: Theoretical and Experimental Studies. Adv. Mater. 2018, 30, 1800191.
- Casallas, C.; Dincer, I. Assessment of an Integrated Solar Hydrogen System for Electrochemical Synthesis of Ammonia. Int. J. Hydrogen Energy 2017, 42, 21495–21500.
- Giddey, S.; Badwal, S.P.S.; Kulkarni, A. Review of Electrochemical Ammonia Production Technologies and Materials. Int. J. Hydrogen Energy 2013, 38, 14576–14594.
- Frattini, D.; Cinti, G.; Bidini, G.; Desideri, U.; Cioffi, R.; Jannelli, E. A System Approach in Energy Evaluation of Different Renewable Energies Sources Integration in Ammonia Production Plants. Renew. Energy 2016, 99, 472–482.
- Boyd, E.S.; Peters, J.W. New Insights into the Evolutionary History of Biological Nitrogen Fixation. Front. Microbiol. 2013, 4, 201.
- Swarnalakshmi, K.; Yadav, V.; Murugeasn, S.; Dhar, D. Biofertilizers for Higher Pulse Production in India: Scope, Accessibility and Challenges. Indian J. Agron. 2016, 61, 173–181.
- Khosro, M.; Yousef, S. Bacterial Biofertilizers for Sustainable Crop Production: A Review. J. Agric. Bological Sci. 2012, 7, 307–316.
- Rapson, T.D.; Wood, C.C. Analysis of the Ammonia Production Rates by Nitrogenase. Catalysts 2022, 12, 844.
- Lindström, K.; Mousavi, S.A. Effectiveness of Nitrogen Fixation in Rhizobia. Microb. Biotechnol. 2020, 13, 1314–1335.
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; Van Der Putten, W.H. Going Back to the Roots: The Microbial Ecology of the Rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799.
- Temme, K.; Zhao, D.; Voigt, C.A. Refactoring the Nitrogen Fixation Gene Cluster from Klebsiella Oxytoca. Proc. Natl. Acad. Sci. USA 2012, 109, 7085–7090.
- López-Torrejón, G.; Burén, S.; Veldhuizen, M.; Rubio, L.M. Biosynthesis of Cofactor-Activatable Iron-Only Nitrogenase in Saccharomyces Cerevisiae. Microb. Biotechnol. 2021, 14, 1073–1083.
- Takimoto, R.; Tatemichi, Y.; Aoki, W.; Kosaka, Y.; Minakuchi, H.; Ueda, M.; Kuroda, K. A Critical Role of an Oxygen-Responsive Gene for Aerobic Nitrogenase Activity in Azotobacter Vinelandii and Its Application to Escherichia coli. Sci. Rep. 2022, 12, 4182.
- Wang, D.; Xu, A.; Elmerich, C.; Ma, L.Z. Biofilm Formation Enables Free-Living Nitrogen-Fixing Rhizobacteria to Fix Nitrogen under Aerobic Conditions. ISME J. 2017, 11, 1602–1613.
- Bhatti, M.; Feng, P.C.C.; Pitkin, J. Methods and Compositions for Improving Plant Health. U.S. Patent 8,754,011, 17 June 2014.
- Yenigün, O.; Demirel, B. Ammonia Inhibition in Anaerobic Digestion: A Review. Process Biochem. 2013, 48, 901–911.
- Whelan, M.J.; Everitt, T.; Villa, R. A Mass Transfer Model of Ammonia Volatilisation from Anaerobic Digestate. Waste Manag. 2010, 30, 1808–1812.
- Walker, M.; Iyer, K.; Heaven, S.; Banks, C.J. Ammonia Removal in Anaerobic Digestion by Biogas Stripping: An Evaluation of Process Alternatives Using a First Order Rate Model Based on Experimental Findings. Chem. Eng. J. 2011, 178, 138–145.
- Choi, K.; Wernick, D.G.; Tat, C.A.; Liao, J.C. Consolidated Conversion of Protein Waste into Biofuels and Ammonia Using Bacillus Subtilis. Metab. Eng. 2014, 23, 53–61.
- Mikami, Y.; Yoneda, H.; Tatsukami, Y.; Aoki, W.; Ueda, M. Ammonia Production from Amino Acid—Based Biomass—like Sources by Engineered Escherichia coli. AMB Express 2017, 7, 83.
- Watanabe, Y.; Aoki, W.; Ueda, M. Improved Ammonia Production from Soybean Residues by Cell Surface-Displayed l-Amino Acid Oxidase on Yeast. Biosci. Biotechnol. Biochem. 2021, 85, 972–980.
- Tatemichi, Y.; Kuroda, K.; Nakahara, T.; Ueda, M. Efficient Ammonia Production from Food by—Products by Engineered Escherichia coli. AMB Express 2020, 10, 150.
- Ueda, M. Establishment of Cell Surface Engineering and Its Development. Biosci. Biotechnol. Biochem. 2016, 80, 1243–1253.
- Kuroda, K.; Ueda, M. Cell Surface Engineering of Yeast for Applications in White Biotechnology. Biotechnol. Lett. 2011, 33, 1–9.
- Watanabe, Y.; Kuroda, K.; Tatemichi, Y.; Nakahara, T.; Aoki, W.; Ueda, M. Construction of Engineered Yeast Producing Ammonia from Glutamine and Soybean Residues (Okara). AMB Express 2020, 10, 70.
- Lu, P.; Ma, D.; Chen, Y.; Guo, Y.; Chen, G.Q.; Deng, H.; Shi, Y. L-Glutamine Provides Acid Resistance for Escherichia coli through Enzymatic Release of Ammonia. Cell Res. 2013, 23, 635–644.
- Bloess, S.; Beuel, T.; Krüger, T.; Sewald, N.; Dierks, T.; Fischer von Mollard, G. Expression, Characterization, and Site-Specific Covalent Immobilization of an L-Amino Acid Oxidase from the Fungus Hebeloma Cylindrosporum. Appl. Microbiol. Biotechnol. 2019, 103, 2229–2241.
- Fenner, K.; Men, Y. Comment on “Role of Ammonia Oxidation in Organic Micropollutant Transformation during Wastewater Treatment”: Overlooked Evidence to the Contrary. Environ. Sci. Technol. 2021, 55, 12128–12129.
- Wang, S.; Peng, Y.; Ma, B.; Wang, S.; Zhu, G. Anaerobic Ammonium Oxidation in Traditional Municipal Wastewater Treatment Plants with Low-Strength Ammonium Loading: Widespread but Overlooked. Water Res. 2015, 84, 66–75.
- Kumar, R.; Singh, L.; Zularisam, A.W. Exoelectrogens: Recent Advances in Molecular Drivers Involved in Extracellular Electron Transfer and Strategies Used to Improve It for Microbial Fuel Cell Applications. Renew. Sustain. Energy Rev. 2016, 56, 1322–1336.
- Ye, Y.; Ngo, H.H.; Guo, W.; Liu, Y.; Chang, S.W.; Nguyen, D.D.; Liang, H.; Wang, J. A Critical Review on Ammonium Recovery from Wastewater for Sustainable Wastewater Management. Bioresour. Technol. 2018, 268, 749–758.
- Sánchez-Monedero, M.A.; Roig, A.; Paredes, C.; Bernal, M.P. Nitrogen Transformation during Organic Waste Composting by the Rutgers System and Its Effects on PH, EC and Maturity of the Composting Mixtures. Bioresour. Technol. 2001, 78, 301–308.
- Wu, K.; Yau, Y.; Sze, E.T.-P. Application of Anaerobic Bacterial Ammonification Pretreatment to Microalgal Food Waste Leachate Cultivation and Biofuel Production. Mar. Pollut. Bull. 2020, 153, 111007.
- Bello, I.A. Challenges in Textile Wastewater and Current Palliative Methods: An Overview. IIUM Eng. J. 2017, 18, 71–78.
- Palatsi, J.; Ripoll, F.; Benzal, A.; Pijuan, M.; Romero-Güiza, M.S. Enhancement of Biological Nutrient Removal Process with Advanced Process Control Tools in Full-Scale Wastewater Treatment Plant. Water Res. 2021, 200, 117212.
- Cyprowski, M.; Stobnicka-Kupiec, A.; Lawniczek-Walczyk, A.; Bakal-Kijek, A.; Golofit-Szymczak, M.; Górny, R.L. Anaerobic Bacteria in Wastewater Treatment Plant. Int. Arch. Occup. Environ. Health 2018, 91, 571–579.
- Haynes, R.J. Nitrification. In Mineral Nitrogen in the Plant-Soil System; Academic Press: Cambridge, MA, USA, 1986; pp. 127–165.
- Van Kessel, M.A.; Speth, D.R.; Albertsen, M.; Nielsen, P.H.; Op den Camp, H.J.; Kartal, B.; Jetten, M.S.; Lücker, S. Complete Nitrification by a Single Microorganism. Nature 2015, 528, 555–559.
- Foley, J.; De Haas, D.; Hartley, K.; Lant, P. Comprehensive Life Cycle Inventories of Alternative Wastewater Treatment Systems. Water Res. 2010, 44, 1654–1666.
- Iskander, S.M.; Brazil, B.; Novak, J.T.; He, Z. Resource Recovery from Landfill Leachate Using Bioelectrochemical Systems: Opportunities, Challenges, and Perspectives. Bioresour. Technol. 2016, 201, 347–354.
- Huang, J.; Kankanamge, N.R.; Chow, C.; Welsh, D.T.; Li, T.; Teasdale, P.R. Removing Ammonium from Water and Wastewater Using Cost-Effective Adsorbents: A Review. J. Environ. Sci. 2018, 63, 174–197.
- Barbera, E.; Bertucco, A.; Kumar, S. Nutrients Recovery and Recycling in Algae Processing for Biofuels Production. Renew. Sustain. Energy Rev. 2018, 90, 28–42.
- He, C.; Wang, K.; Yang, Y.; Amaniampong, P.N.; Wang, J.-Y. Effective Nitrogen Removal and Recovery from Dewatered Sewage Sludge Using a Novel Integrated System of Accelerated Hydrothermal Deamination and Air Stripping. Environ. Sci. Technol. 2015, 49, 6872–6880.
- Huang, H.; Yang, J.; Li, D. Recovery and Removal of Ammonia–Nitrogen and Phosphate from Swine Wastewater by Internal Recycling of Struvite Chlorination Product. Bioresour. Technol. 2014, 172, 253–259.
- Husnain, T.; Mi, B.; Riffat, R. A Combined Forward Osmosis and Membrane Distillation System for Sidestream Treatment. J. Water Resour. Prot. 2015, 7, 1111.
- Katehis, D.; Diyamandoglu, V.; Fillos, J. Stripping and Recovery of Ammonia from Centrate of Anaerobically Digested Biosolids at Elevated Temperatures. Water Environ. Res. 1998, 70, 231–240.
- Kim, T.; An, J.; Jang, J.K.; Chang, I.S. Coupling of Anaerobic Digester and Microbial Fuel Cell for COD Removal and Ammonia Recovery. Bioresour. Technol. 2015, 195, 217–222.
- Kuntke, P.; Śmiech, K.; Bruning, H.; Zeeman, G.; Saakes, M.; Sleutels, T.; Hamelers, H.V.M.; Buisman, C.J.N. Ammonium Recovery and Energy Production from Urine by a Microbial Fuel Cell. Water Res. 2012, 46, 2627–2636.
- Puniya, A.K.; Singh, R.; Kamra, D.N. Rumen Microbiology: From Evolution to Revolution. In Rumen Microbiology: From Evolution to Revolution; Springer: New Delhi, India, 2015; pp. 1–379.
- Rychlik, J.L.; Russell, J.B. Mathematical Estimations of Hyper-Ammonia Producing Ruminal Bacteria and Evidence for Bacterial Antagonism That Decreases Ruminal Ammonia Production. FEMS Microbiol. Ecol. 2000, 32, 121–128.
- Pengpeng, W.; Tan, Z. Ammonia Assimilation in Rumen Bacteria: A Review. Anim. Biotechnol. 2013, 24, 107–128.
- Rychlik, J.L.; Russell, J.B. Bacteriocin-like Activity of Butyrivibrio Fibrisolvens JL5 and Its Effect on Other Ruminal Bacteria and Ammonia Production. Appl. Environ. Microbiol. 2002, 68, 1040–1046.
- Bach, A.; Calsamiglia, S.; Stern, M.D. Nitrogen Metabolism in the Rumen. J. Dairy Sci. 2005, 88, E9–E21.
- Van Soest, P.J. Nitrogen Metabolism. In Nutritional Ecology of the Ruminant; Cornell University Press: Ithaca, NY, USA, 1994.
- Rychlik, J.L.; Lavera, R.; Russell, J.B. Amino Acid Deamination by Ruminal Megasphaera elsdenii Strains. Curr. Microbiol. 2002, 45, 340–345.
- Doelle, H.W. Nitrogen Metabolism as an Energy Source for Anaerobic Microorganisms (Clostridium). In Bacterial Metabolism; Elsevier: Amsterdam, The Netherlands, 1969; pp. 402–422.
- Strock, J.S. Ammonification. In Encyclopedia of Ecology, Five-Volume Set; Elsevier Inc.: Amsterdam, The Netherlands, 2008; pp. 162–165.
- Nolan, J.V. Quantitative Models of Nitrogen Metabolism in Sheep. In Digenstion and Metabolism in the Ruminant; Oxford University Press: Oxford, UK, 1975.
- Valente, T.N.P.; da Silva Lima, E.; dos Santos, W.B.R.; Cesário, A.S.; Tavares, C.J.; de Freitas, M.A.M. Ruminal Microorganism Consideration and Protein Used in the Metabolism of the Ruminants: A Review. Afr. J. Microbiol. Res. 2016, 10, 456–464.
- Adeniyi, A.; Bello, I.; Mukaila, T.; Monono, E.; Hammed, A. Developing Rumen Mimicry Process for Biological Ammonia Synthesis. Bioprocess Biosyst. Eng. 2023, 1–10.
- Nagaraja, T.G. Microbiology of the Rumen. In Rumenology; Springer: Berlin/Heidelberg, Germany, 2016; pp. 39–61.
- Whitehead, T.R.; Cotta, M.A. Isolation and Identification of Hyper-Ammonia Producing Bacteria from Swine Manure Storage Pits. Curr. Microbiol. 2004, 48, 20–26.
- Adeniyi, A.; Bello, I.; Mukaila, T.; Hammed, A. A Review of Microbial Molecular Profiling during Biomass Valorization. Biotechnol. Bioprocess Eng. 2022, 27, 515–532.
- Slyter, L.L. Influence of Acidosis on Rumen Function. J. Anim. Sci. 1976, 43, 910–929.
- Chen, G.; Russell, J.B. More Monensin-Sensitive, Ammonia-Producing Bacteria from the Rumen. Appl. Environ. Microbiol. 1989, 55, 1052–1057.
- McDonald, I.W. The Absorption of Ammonia from the Rumen of the Sheep. Biochem. J. 1948, 42, 584.
- Bryant, M.P. The Characteristics of Strains of Selenomonas Isolated from Bovine Rumen Contents. J. Bacteriol. 1956, 72, 162–167.
- Bryant, M.P.; Small, N. The Anaerobic Monotrichous Butyric Acid-Producing Curved Rod-Shaped Bacteria of the Rumen. J. Bacteriol. 1956, 72, 16–21.
More
Information
Subjects:
Biotechnology & Applied Microbiology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
3.0K
Revisions:
2 times
(View History)
Update Date:
31 May 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




