Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Dong, H.; Yang, X.; Shi, J.; Xiao, C.; Zhang, Y. Advantages and Prospects for In Vitro PHA Synthesis. Encyclopedia. Available online: https://encyclopedia.pub/entry/44970 (accessed on 07 February 2026).
Dong H, Yang X, Shi J, Xiao C, Zhang Y. Advantages and Prospects for In Vitro PHA Synthesis. Encyclopedia. Available at: https://encyclopedia.pub/entry/44970. Accessed February 07, 2026.
Dong, Huaming, Xue Yang, Jingjing Shi, Chunqiao Xiao, Yanfei Zhang. "Advantages and Prospects for In Vitro PHA Synthesis" Encyclopedia, https://encyclopedia.pub/entry/44970 (accessed February 07, 2026).
Dong, H., Yang, X., Shi, J., Xiao, C., & Zhang, Y. (2023, May 29). Advantages and Prospects for In Vitro PHA Synthesis. In Encyclopedia. https://encyclopedia.pub/entry/44970
Dong, Huaming, et al. "Advantages and Prospects for In Vitro PHA Synthesis." Encyclopedia. Web. 29 May, 2023.
Copy Citation
Polyhydroxyalkanoates (PHAs) are a series of structurally diverse storage polyesters that are accumulated by various bacterial species and stored intracellularly in the form of granules. They primarily act as carbon and energy storage compounds to sustain cell survival during starvation.
cell-free synthesis system
Polyhydroxyalkanoate (PHA)
PHA synthesis pathway
1. Overview of Polyhydroxyalkanoates (PHAs)
Polyhydroxyalkanoates (PHAs) are a series of structurally diverse storage polyesters that are accumulated by various bacterial species and stored intracellularly in the form of granules [1]. They primarily act as carbon and energy storage compounds to sustain cell survival during starvation [2]. In 1926, Lemoigne [3] first discovered these biological polymers. Since then, PHAs have attracted significant commercial and research interest in the green polymer market due to their desirable properties such as biocompatibility, biodegradability, and diversified chemical structures. PHAs have been recognized as a green alternative to conventional petroleum-derived plastics [4].
Based on the number of carbon atoms present in the (R)-HA monomer unit, PHA polymers can be broadly classified into three types: short-chain-length PHAs (scl-PHA, C3-C5), medium-chain-length PHAs (mcl-PHA, C6-C14) and long-chain-length PHAs (lcl-PHA, containing more than 14 carbon atoms) [5]. Many bacteria can produce scl-PHA, including the model species Cupriavidus necator (also known as Ralstonia eutropha) [6], while mcl-PHA producers are mainly found in Pseudomonas sp. [7][8][9]. The monomer composition of PHA polymers defines most of the physical–chemical properties of the material, which is a determining factor in its engineering application, for example, mcl-PHA and its copolymers are suitable for a range of biomedical applications requiring flexible biological materials [7]. Additionally, PHA can be synthesized in different forms depending on the bacterial species and the substrate provided. These include homopolymers that are composed of only one type of monomer [10], random copolymers [11] that contain two or more different monomers, and block copolymers [12] that are made up of at least two homopolymers connected by covalent bonds.
2. In Vitro PHA Synthesis
2.1. Exploration of PHA Synthetase
Cell-free synthetic biology, in conjunction with the prototyping design, may offer a promising approach for PHA synthesis. In the 1970s, research into the in vitro synthesis of PHA using purified enzymes [13] was carried out, primarily aimed at exploring the activity of PhaC to optimize the PHA synthesis capability of the strain. Since then, this approach has been widely adopted by researchers. Qi, et al. [14] purified the type II PHA synthases PhaC1 and PhaC2 from Pseudomonas aeruginosa and determined their kinetic parameters for PHA synthesis. A molar mass of 9.8 × 106 g/mol of PHB was obtained when using (R,S)-3-hydroxybutyryl-CoA as a substrate. Additionally, PhaC activity can be inhibited by large amounts of CoA. The reaction pathway catalyzed by PhaC from acyl-CoA to PHA is a single step and provides direct and clear guidance for subsequent work with the in vitro synthesis of PHA.
Subsequently, other researchers developed cell-free synthesis methods that improved the polymer yield. Jossek and Steinbüchel [15] established an in vitro PHB three-enzyme biosynthetic system. In this system, the release of CoA was coupled with acetate, which was reused as an acetyl group donor catalyzed by acetyl-CoA synthase to generate 3-hydroxybutyrate. This method made the in vitro synthesis of PHB independent of the costly consumption of CoA. In subsequent research, Han, et al. [16] discovered a method for synthesizing non-natural PHA using a chemical-enzyme approach. Their system consisted of an organic phase of hexane and a buffered aqueous phase. In the aqueous phase, propionyl-CoA transferase, with pan-substrate activity, catalyzed the CoA transfer from acetyl-CoA to form a CoA-activated precursor for a PHA monomer with the release of acetate. During polymerization, CoA was released into the aqueous reaction phase and reacetylated in the intermediate phase. Ultimately, the maximum titer of PHB achieved was 1.2 g/L, which is comparable to that of natural producers [17]. Unconventional PHA cell-free synthesis schemes have also been reported, such as the use of class II and III PHA synthases to catalyze PHA surface coatings on hydrophobic carriers [18]. Additional reports related to cell-free PHA synthesis are summarized in Table 1.
Table 1. Summary for PHA synthesis exploration in vitro.
| Substrates | Enzymes | Source of Enzymes | Products | Titer (mg/mL) | Conversion Rate (%) | Average M (g/mol) | Ref. |
|---|---|---|---|---|---|---|---|
| (R,S)-3HD-CoA | PhaC1, PhaC2 | P. aeruginosa | P(3HD) | 0.068 | 12 | 9.8 × 106 | [14] |
| (R)-LA and (R)-3HB, acetyl-CoA | PhaC1SG, PhaC2SG LPE, CoA-transferase, and acetyl-CoA-synthetase | Pseudomonas sp. SG4502 and Pseudomonas sp. 61-3 |
P(LA-co-3HB) | 0.77 | / | 2.7 × 105 | [19] |
| D-(-)-3-hydroxybutyric acid | acetyl-CoA-synthetase, PCT, and PhaECcv |
C. vinosum and C. propionicum | PHB | 2.75 | 73 | 1–2 × 106 | [15] |
| (R)-3-Hydroxyoctanoyl-CoA | PhaC1PP and PhaECAv |
P. putida and Allochromatium vinosum | PHB | / | / | / | [18] |
| HB−NAC and HB−CoA | PhaECAv | A. vinosum | PHB | / | / | 5.84–7.97 × 104 | [20] |
| β-butyrolactone | FadB1x, PhaCDa, and PhaJAc |
A. caviae, D. acidovorans, and P. putida KT2440 | PHB | 1.1575 | 27 | 1.2 × 105 | [21] |
| thiophenyl (R)-3-hydroxybutyrate | PhaC | R. eutropha | PHB | 0.36 | 47 | 1.6 × 106 | [22] |
| (R)-2HB, AcETG, (R)-3HB | PHA synthase I, PHA synthase II, and PHA synthase IV PCT (propionate CoA transferase) | R. eutropha, Pseudomonas sp. SG4502, Synechocystis sp. PCC6803, Bacillus sp. INT005, and C. propionicum JCM1430 | P(2HB), PHB | 1.2 | / | 1.00 × 105 | [16][17] |
| Glucose, NADPH, acetoacetyl-CoA | PHA synthase, acetoacetyl-CoA reductase, and glucose dehydrogenase | A. eutrophus | PHB | / | / | 10 × 106 | [23] |
| crotonic anhydride | PhaJ and PhaC | A. caviae FA440 and R. eutropha H16 | PHB | / | 58 | 6.4 × 106 | [24] |
| Acetate, CoA | ACS, PhaA, PhaB, and PhaC | E. coli JM109 and R. eutropha H I6 | PHB | 1.12 | 45.5 | 6.64 × 106 | [25] |
Note: M indicates molar mass.
2.2. In Vitro Multienzyme Systems for PHA Synthesis
Flexible and controllable multienzyme cascade systems have been employed to test entire metabolic pathways [26][27][28][29]. It is reassuring that the rapid growth of synthetic biology has largely contributed to the development of controllable cell-free synthesis systems, the ability to guide substrates toward desired products, and the effective combination of facilities that can freely assemble heterologous enzymes [30]. In many ways, the cell-free synthesis method is a promising strategy for PHA production.
In vitro multienzyme systems for PHA synthesis have been investigated by several research groups. In 1998, Jossek and Steinbüchel [15] established an extracellular PHB tri-enzyme system with acetate and 3HB as substrates, which could be prepared on a semi-preparative scale. Five years later, Satoh, Tajima, Tannai and Munekata [25] developed an improved new enzyme-catalyzed PHB synthesis system using five enzymes including β-ketothiolase (PhaA), NADPH-dependent acetoacetyl-CoA reductase (PhaB), PhaC, acetyl-CoA synthetase (ACS), and glucose dehydrogenase (GDH). In this system, acetyl-CoA was synthesized from acetate, CoA, and ATP through ACS, and then, two acetyl-CoA molecules were condensed by PhaA to synthesize acetyl-CoA, which was transformed into (R)-3-hydroxybutanyl-CoA (3HB-CoA) by PhaB. Finally, 3HB-CoA was polymerized and transformed into PHB by PhaC, with a titer of 1.12 mg/mL. Subsequently, cell-free PHA (mainly PHB) synthesis systems based on PhaA, PhaB, and PhaC were combined into multiple enzyme reaction modules. For instance, Opgenorth, et al. [31] designed a PBG (pentose–bifido–glycolysis) cycle that effectively converted glucose into acetyl-CoA and demonstrated its utility in PHB production. In a recent study, Li, et al. [32] developed an in vitro system that coupled plant cystoid membranes and five enzymes to synthesize PHB and regenerate NADPH. Furthermore, Zhang, et al. [33] demonstrated PHB production from methanol by utilizing the artificial synthetic acetyl-CoA (SACA) pathway. It is noteworthy that although the previous studies highlighted the usefulness of the acetyl-CoA synthesis, they did not focus on enhancing PHA yield. Nonetheless, the considerable yield and rate data collected in these studies pave the way for the future of in vitro PHA synthesis.
3. Advantages and Prospects for In Vitro PHA Synthesis
3.1. Advantages of In Vitro PHA Synthesis
Cell-free methods have enormous potential in accelerating the design–build–test (DBT) cycle by avoiding resource conflicts between cell growth and product synthesis [34]. Cell-free synthetic technology is a promising approach that can complement in vivo metabolic engineering [35]. For instance, by mixing the substrates and enzymes, the efficiency of the biosynthesis pathway can be conveniently optimized in vitro [36][37]. Moreover, cell-free catalysis offers significant advantages over the bottlenecks encountered in PHA synthesis, as outlined below:
3.1.1. The Limitation of Cell Volume Is Eliminated in Cell-Free Catalysis
Intracellular crowding can be exacerbated by the accumulation of PHA particles, leading to the termination of PHA production [38]. Despite attempts to alter cell morphology [39][40], the constraint of cell volume remains a limitation. On the other hand, a cell-free system offers an advantage by allowing for the easy separation of PHA from the system. This eliminates the spatial constraint and enables the system to maintain its continuous productivity as long as possible [31].
3.1.2. Real-Time Monitoring of Product Concentration Can Be Achieved
The real-time detection of intracellular PHA content remains a significant challenge, often requiring post-fermentation analysis due to the lack of effective methods. Despite attempts to develop biosensors for this purpose [41][42], technical challenges, such as response specificity and accuracy in complex cellular systems, continue to pose significant obstacles. In contrast, Opgenorth, Korman and Bowie [31] successfully achieved real-time monitoring of PHB content in an in vitro system by measuring the absorbance value at 600 nm, which guided glucose replenishment and resulted in a high conversion rate of 93.6% [31]. Presently, achieving yields or conversions close to the theoretical maximum is already common in in vitro synthetic systems [30][43][44].
3.1.3. Significant Reduction in Product Separation Costs Can Be Achieved
The production cost of PHA is significantly impacted by the energy consumption required for cell crushing and the cost of solvents used for extraction [45][46]. This energy-intensive and environmentally unfriendly production process poses a major challenge for PHA production. In contrast, in vitro PHA synthesis, with simple composition and minimal interference from metabolites, offers a promising alternative. This method only requires centrifugation to obtain PHA products with high purity and low cost [47]. Therefore, in vitro PHA synthesis holds immense potential for the development of sustainable and cost-effective PHA production processes.
3.1.4. Compatible with New Pathways and Heterologous Enzymes
Introducing new pathways and heterologous enzymes into cells is a technically challenging process due to the complex regulatory system inherent to cells. Compatibility issues often arise, resulting in suboptimal results for artificial pathways with high theoretical yields. Conversely, in vitro systems offer a highly compatible and straightforward approach to pathway development, making them a preferred choice for many researchers. Moreover, PHA, being a classical acetyl-CoA derivative, is frequently chosen as a product for new pathway development [33][48]. Therefore, in vitro PHA synthesis presents a promising avenue for the development of novel and efficient biopolymer production processes with improved compatibility and technical feasibility.
3.1.5. The Monomer Composition Is Better Controlled
The customization of the monomer composition of intracellular PHAs presents a significant challenge that hinders the study of PHA material properties. However, in vitro PHA synthesis provides a promising solution to this challenge, allowing for better control over the polymerization process and monomer composition by combining specific substrates, enzymes, and polymerases in a cell-free catalytic system. Therefore, in vitro PHA synthesis offers a unique opportunity for the production of tailored biopolymers with specific and customizable properties, facilitating the study of PHA material properties and expanding their potential applications.
3.2. Challenges with In Vitro PHA Synthesis
Despite the advantages of cell-free methods in overcoming cell growth constraints [49], many challenges remain to be solved before PHA products can be commercially available on a large scale.
3.2.1. Insufficient Diversity in Monomer Composition
Scl-PHA is primarily synthesized from acetyl-CoA through the PhaCAB pathway, whereas mcl/lcl-PHA synthesis typically involves either the fatty acid β-oxidation or fatty acid synthesis pathway (Figure 1). However, the implementation of the fatty acid synthesis and degradation pathway in vitro is a challenging task due to the requirement for complex coenzymes or acyl-carrying proteins. Although there have been numerous reports on the in vitro synthesis of PHB using acetyl-CoA as the sole precursor [31], progress in the in vitro synthesis of mcl-PHA has been limited. Additionally, the inhibitory effect of CoA on PhaC [14] poses another challenge that needs to be addressed in the in vitro synthesis of PHA.
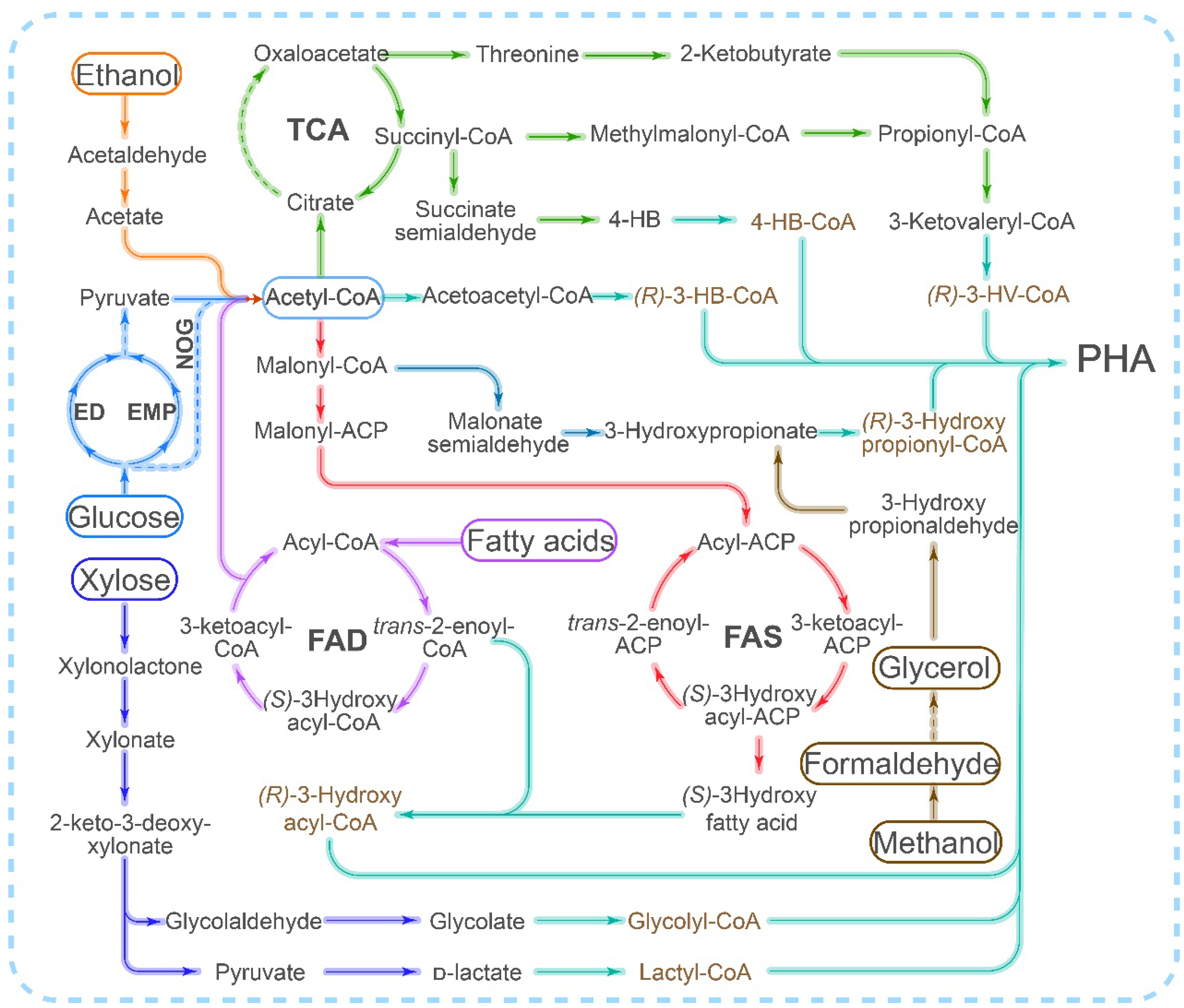
Figure 1. Different pathways for PHA synthesis utilized by different carbon sources. ED: Entner–Doudoroff pathway; EMP: Embden–Meyerhof–Parnas Pathway (glycolytic pathway); FAD: fatty acid degradation pathway (beta-oxidation pathway); FAS: fatty acid synthesis pathway; TCA: tricarboxylic acid cycle; NOG: nonoxidative glycolysis pathway.
3.2.2. Insufficient Enzyme Activity and Stability
Numerous in vitro multistep pathways have been developed to enhance product yield from low-cost substrates, using either crude extract or purified enzymes (Figure 2). However, the apparent activity and stability of enzymes in cell-free systems are often lower than those in live cells. To tackle this challenge, various potential solutions are worth exploring. For instance, employing thermostable enzymes [50] or improving enzyme stability through protein engineering [51][52], or multienzyme immobilization technology [53], could potentially address the issue of activity and stability. Furthermore, optimizing enzyme production efficiency and developing cost-effective production methods will play a crucial role in determining the commercial viability of in vitro PHA synthesis systems.
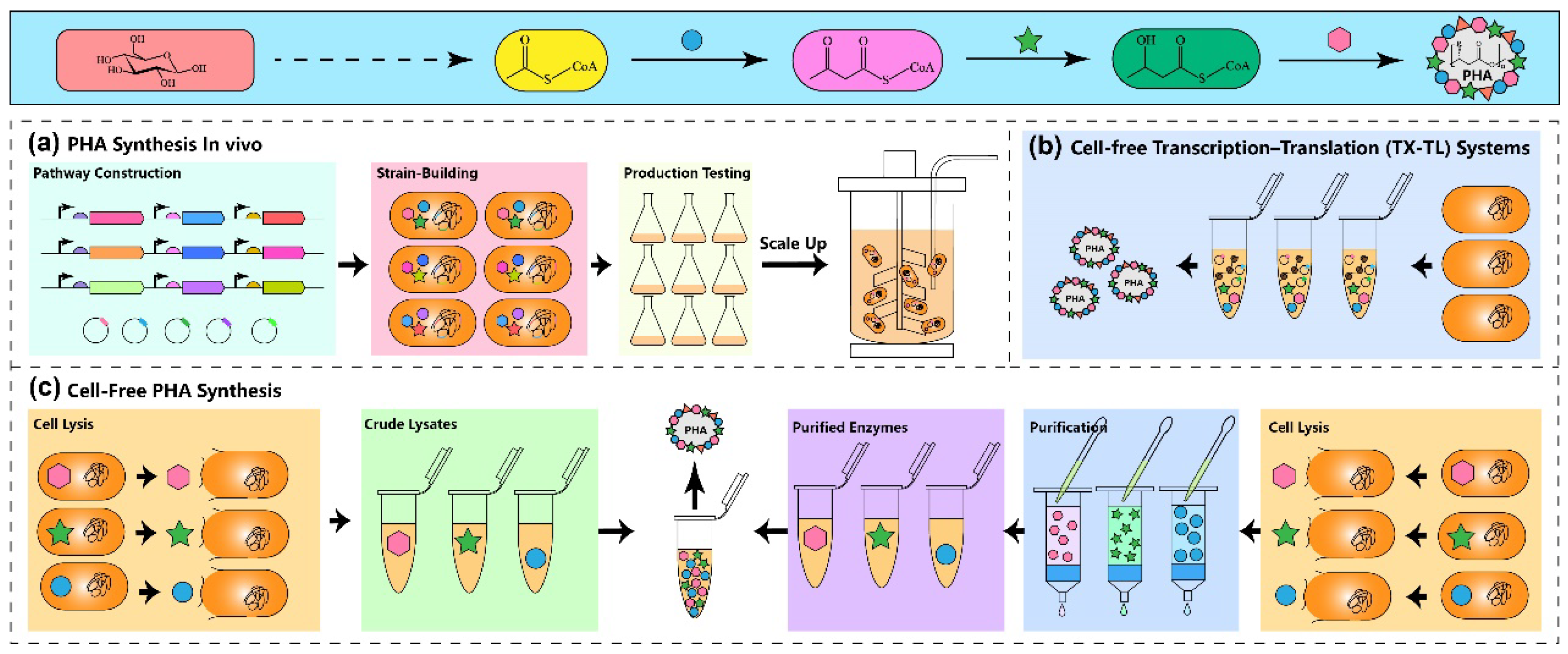
Figure 2. The patterns of PHA production in vivo and in vitro. (a) Traditional in vivo PHA production involves strain construction, metabolic pathways engineering, fermentation optimization, and process scaling up. (b) PHA synthesis using a transcription–translation system. In this system, the cell crude extract contains plasmid DNA and the enzymes required for PHA synthesis are first transcribed and translated in the cell crude extract to form the PHA synthesis pathway, which can synthesize PHA when the substrate is available. (c) Traditional cell-free PHA synthesis. The cell crude extracts were directly used as the reaction system (left), or mixing purified enzymes to form a cell-free system for PHA synthesis (right). In the figure, the blue circle indicates PhaA, the green star indicates PhaB, and the pink hexagon indicates PhaC.
3.2.3. Dependence on Energy and Cofactors
Cofactors such as ATP, NAD(P)+, CoA, and FAD2+ play an indispensable role in cell-free PHA biosynthesis [54]. However, in industrial biocatalysis, these cofactors can be expensive supplements. For instance, the cost of NADH is approximately $260 g−1 [55]. To enhance economic feasibility, researchers are striving to regenerate and reuse cofactors [56][57] or reduce the use of expensive ones [58][59][60][61]. Artificial analogs can substitute for expensive cofactors [62] and can be designed to be more stable than their natural counterparts while still being recognized by enzymes. The CoA cycle is crucial for PHA synthesis, but its dependence on chemicals for cofactor repair in vitro hinders CoA from being stable for extended periods [63]. More comprehensive reviews of cofactor and ATP regeneration systems have been reported [54][64][65].
3.3. Feasible Solutions (What to Work On)
The in vitro synthesis of PHA holds promising prospects, emphasizing the need to actively explore methods to tackle the challenges outlined above. Here, several feasible solutions that have the potential to reduce the cost of in vitro PHA synthesis have been compiled or designed. It is anticipated that these efforts will lead to the enhancement of cost-effectiveness in the entire process of in vitro PHA production and the facilitation of the mitigation of future market challenges.
3.3.1. Utilization of Unconventional Substrates
Currently, the utilization of cell-free synthetic systems is limited due to the high cost of raw materials. To compete with petroleum-based products, it is essential to reduce the material cost. One promising approach is to introduce nonconventional substrates, such as CO2, CH4, and lignocellulose, into the cell-free synthetic system. This can be achieved by incorporating a degrading absorption module [66][67] that enables the utilization of nonconventional substrates. However, the introduction of these substrates may alter the composition of the PHA monomer, resulting in PHA products with varying properties. This can pose additional challenges and complexity, but it also presents an opportunity to expand monomer diversity and create new materials with unique properties.
3.3.2. Low-Cost Acquisition and Stability Modification of Enzymes
The use of purified enzymes in cell-free biotechnology is a costly and time-consuming process. A viable solution to this issue is whole-cell catalysis, which involves the use of unpurified cell extracts and lower-quality enzymes, making it a more accessible and affordable option [30]. This approach involves expressing the required enzymes in the cells and then breaking the cell walls to release them into the buffer system. However, it is important to note that developing low-cost purification methods is a more promising strategy. To improve the stability of enzymes in the PHA in vitro synthesis pathways, several techniques such as directed evolution, enzyme immobilization, and screening of natural thermostable enzymes can be employed in conjunction with developing low-cost enzyme purification technology to achieve efficient PHA production. Immobilized cell-free systems immobilize enzymes and cofactors on or within a carrier material, resulting in a series of biochemical reactions with reduced or prevented enzyme migration [68]. Compared to free or soluble enzymes, immobilized enzymes possess higher stability, reusability, and simplified recovery and purification processes [69]. Moreover, by developing efficient protein expression and secretion systems [70], it is possible to circumvent the energy and materials consumption and enzyme activity damage associated with cell lysis and protein purification. The continuous optimization of cell surface display technology [71][72] and the immobilization of multiple cells [73] can facilitate the creation of multienzyme systems that reduce PHA production costs through improved activity, stability, and reusability of enzymes.
3.3.3. Cofactor Regeneration
In cell-free systems, the use of cofactors can be costly. Consequently, many efforts have been made in developing strategies for balancing and regenerating cofactors. One approach is to integrate coenzyme reactions, while another is to utilize light and electrical energy for regeneration. Additionally, using inexpensive substrate cofactors to regenerate spent cofactors and designing cofactor-free pathway systems are also viable options. For example, the regeneration of NAD(P)+ can be accomplished through the use of NAD(P)H oxidase (Nox), which is formed by hydration and oxidizes NAD(P)H to NAD(P)+ [31]. NAD+ can be recovered from NADH and less expensive substrates, such as glucose, by using coupled reactions that involve glucose dehydrogenase [74]. Regenerating ATP can be achieved through the use of polyphosphate kinase (Ppk) [56], which accepts inexpensive polyphosphates and synthesize ATP from ADP or AMP in ATP-dependent reactions [67][75]. Clean energy sources such as light and electricity are recommended due to their cost-effectiveness. Li, Wei, Zhang, Liu, You and Zhu [32] utilized light by incorporating thylakoid membranes from spinach leaves into the system to provide ATP and NADP+ for the PHA synthesis pathway. Additionally, CoA can be released from the condensation and polymerization catalyzed by PhaA and PhaC in the PHB synthesis pathway and then reused in acetyl-CoA synthesis [25].
3.3.4. Modeling In Silico Provides Quantitative Guidance
Computational modeling offers a quantitative framework for pathway validation and optimization by predicting intracellular metabolite concentrations and reaction fluxes [76]. In cell-free processes, changes in reaction rates and intermediate concentrations can be attributed to enzymatic activities. Kinetic pathway modeling can facilitate the adjustment of enzyme, substrate, and cofactor concentrations, contributing to the rational understanding of cell-free systems [77][78]. For instance, Rollin, et al. [79] fitted a nonlinear kinetic model to experimental data, achieving a complete conversion of glucose and xylose to H2 and CO2 in a cell-free system. Through the analysis of enzymes with the greatest control coefficient for the desired reaction flux, the optimized enzyme usage led to a production rate of 32 mmol H2·L−1·h−1. Horvath et al. developed a genome-scale cell-free protein synthesis (CFPS) kinetic model to predict protein production pathways [80].
Moreover, genome-scale constraint-based models, such as stoichiometric metabolic network models, enzyme-constrained models [81], and thermodynamically constrained models [82][83], have been applied to guide pathway selection and to perform thermodynamic driving force and enzyme cost analyses for products and biomass synthesis. These models have broad applicability and have been applied to, or are expected to serve in, the in vitro PHA synthesis process [84]. Meanwhile, genome-scale kinetic modeling [85], or large-scale whole-cell models [86], have provided promising visions for rational product synthesis. The rapid development of artificial intelligence in parameter mining also provides greater opportunities for the deep integration of experimentation and simulation [87].
3.3.5. Exploring the Modular PHA Synthesis
The synthesis pathway of PHA exhibits significant flexibility and modularity (as shown in Figure 3). Modular PHA synthesis can be achieved by utilizing the modular assembly of multienzyme systems through cell-free metabolic pathways to construct complete PHA synthesis pathways. Various substrates are metabolized through pathways such as central carbon metabolism, β-oxidation, and fatty acid synthesis (FAS) to provide precursors for PHA production [88]. Scl-PHA synthesis begins with acetyl-CoA as the precursor, which is polymerized by PhaA, PhaB, and PhaC. For mcl- and lcl-PHA synthesis, various intermediate products of the fatty acid degradation and synthesis pathways can also serve as precursors. When fatty acid substrates undergo β-oxidation degradation, reducing the activity of FadB and FadA can redirect intermediate metabolites toward mcl-PHA synthesis [88]. Non-fatty acid carbon sources, such as glucose, xylose, glycerol, ethanol, and one-carbon metabolites, require a connection between the substrate assimilation pathways and PHA monomer precursor synthesis pathways, such as acetyl-, glycolyl-, lactyl-, and various hydroxylated fatty acyl-CoAs [89][90]. The modular assembly approach is particularly advantageous because it can efficiently incorporate new reactions and pathway modules, resulting in the rapid formation of novel synthesis routes. This approach holds great promise for the development of new production modes that can surpass original yield limitations, ultimately leading to reduced costs associated with PHA production.
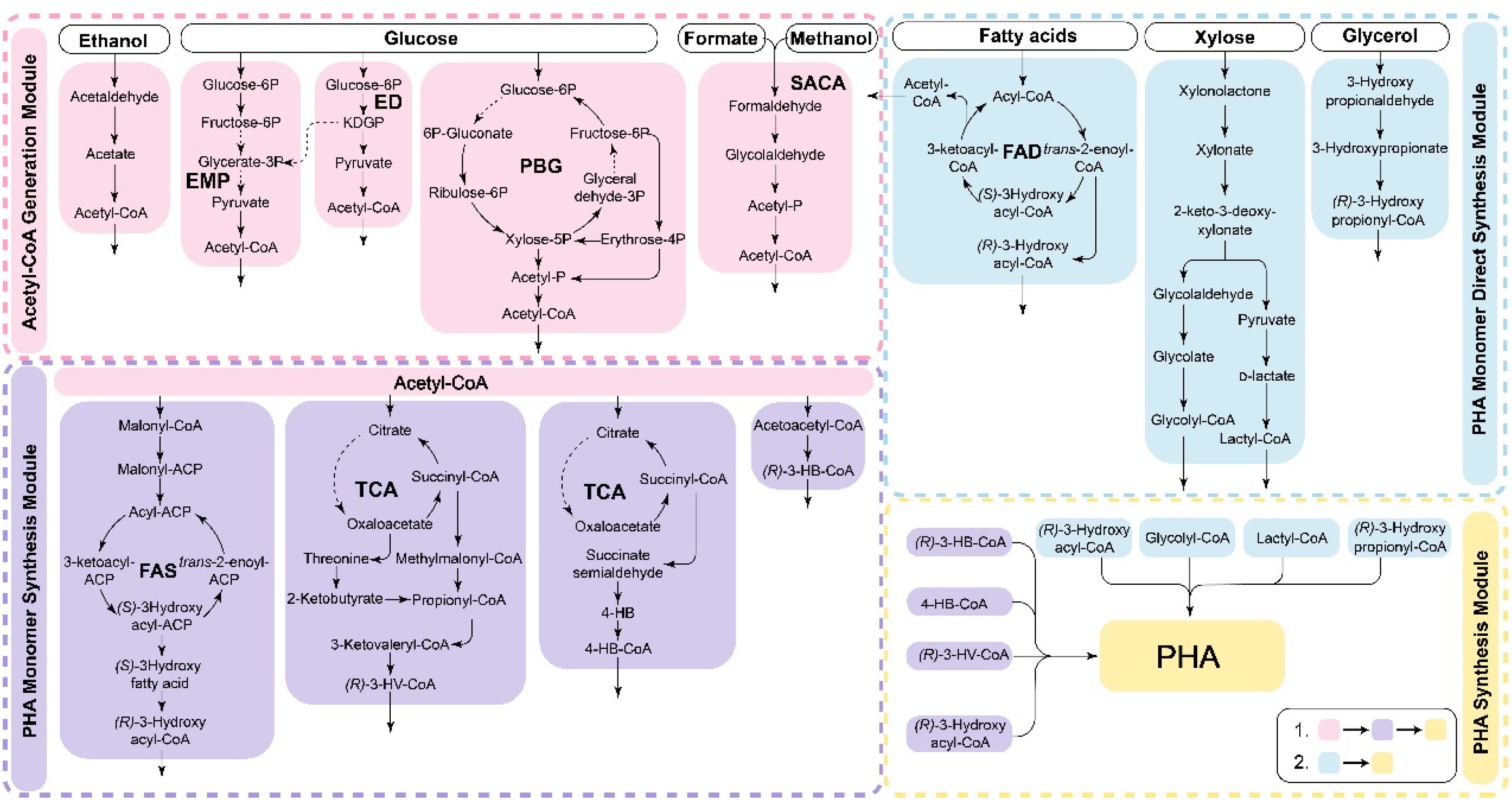
Figure 3. The in vitro modular assembly of synthetic PHA. The different metabolic modules are assembled and coupled to synthesize PHA using different substrates. Cell-free PHA synthesis can be achieved in vitro by assembling various metabolic modules and then coupling the PHA synthesis pathway to achieve the utilization of different substrates. The process typically involves generating acetyl-CoA modules and then combining them with the acetyl-CoA utilization pathway to synthesize the precursors of PHA. The enzymes related to central metabolism exhibit high activity, leading to a high rate of acetyl-CoA production and ensuring a sufficient substrate supply for the subsequent PHA synthesis pathway. While the utilization of fatty acids can directly lead to the synthesis of mcl-PHA, non-fatty acid substrates usually need to be converted into acetyl-CoA before participating in the synthesis of PHA. PBG: pentose, bifido, and glycolysis pathway [31]; SACA: synthetic acetyl-CoA pathway [48].
References
- Jendrossek, D. Polyhydroxyalkanoate granules are complex subcellular organelles (Carbonosomes). J. Bacteriol. 2009, 191, 3195–3202.
- Koller, M.; Mukherjee, A.; Obruca, S.; Zinn, M. Polyhydroxyalkanoates (PHA): Microbial synthesis of natural polyesters. In Microbial Production of High-Value Products; Rehm, B.H.A., Wibowo, D., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 185–236.
- Lemoigne, M. Études sur l’autolyse microbienne origine de l’acide β-oxybutyrique formé par autolyse. Ann. Inst. Pasteur 1927, 41, 148.
- Acharjee, S.A.; Bharali, P.; Gogoi, B.; Sorhie, V.; Walling, B.; Alemtoshi. PHA-based bioplastic: A potential alternative to address microplastic pollution. Water Air Soil Pollut. 2022, 234, 21.
- Li, Z.; Yang, J.; Loh, X.J. Polyhydroxyalkanoates: Opening doors for a sustainable future. NPG Asia Mater. 2016, 8, e265.
- Mravec, F.; Obruca, S.; Krzyzanek, V.; Sedlacek, P.; Hrubanova, K.; Samek, O.; Kucera, D.; Benesova, P.; Nebesarova, J. Accumulation of PHA granules in Cupriavidus necator as seen by confocal fluorescence microscopy. FEMS Microbiol. Lett. 2016, 363, fnw094.
- Rai, R.; Keshavarz, T.; Roether, J.A.; Boccaccini, A.R.; Roy, I. Medium chain length polyhydroxyalkanoates, promising new biomedical materials for the future. Mater. Sci. Eng. R Rep. 2011, 72, 29–47.
- Xu, Z.; Xu, M.; Cai, C.; Chen, S.; Jin, M. Microbial polyhydroxyalkanoate production from lignin by Pseudomonas putida NX-1. Bioresour. Technol. 2021, 319, 124210.
- Wang, X.; Lin, L.; Dong, J.; Ling, J.; Wang, W.; Wang, H.; Zhang, Z.; Yu, X. Simultaneous Improvements of pseudomonas cell growth and polyhydroxyalkanoate production from a pignin derivative for lignin-consolidated bioprocessing. Appl. Environ. Microbiol. 2018, 84, e01469-18.
- Gao, J.; Vo, M.T.; Ramsay, J.A.; Ramsay, B.A. Overproduction of MCL-PHA with high 3-hydroxydecanoate Content. Biotechnol. Bioeng. 2018, 115, 390–400.
- Wang, Y.; Chung, A.; Chen, G.-Q. Synthesis of Medium-Chain-Length polyhydroxyalkanoate homopolymers, random copolymers, and block copolymers by an engineered strain of Pseudomonas entomophila. Adv. Healthc. Mater. 2017, 6, 1601017.
- Pederson, E.N.; McChalicher, C.W.J.; Srienc, F. Bacterial synthesis of PHA block copolymers. Biomacromolecules 2006, 7, 1904–1911.
- Fukui, T.; Yoshimoto, A.; Matsumoto, M.; Hosokawa, S.; Saito, T.; Nishikawa, H.; Tomita, K. Enzymatic synthesis of poly-β-hydroxybutyrate inZoogloea ramigera. Arch. Microbiol. 1976, 110, 149–156.
- Qi, Q.; Steinbüchel, A.; Rehm, B.H.A. In vitro synthesis of poly(3-hydroxydecanoate): Purification and enzymatic characterization of type II polyhydroxyalkanoate synthases PhaC1 and PhaC2 from Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 2000, 54, 37–43.
- Jossek, R.; Steinbüchel, A. In vitro synthesis of poly(3-hydroxybutyric acid) by using an enzymatic coenzyme A recycling system. FEMS Microbiol. Lett. 1998, 168, 319–324.
- Han, X.; Satoh, Y.; Satoh, T.; Matsumoto, K.i.; Kakuchi, T.; Taguchi, S.; Dairi, T.; Munekata, M.; Tajima, K. Chemo-enzymatic synthesis of polyhydroxyalkanoate (PHA) incorporating 2-hydroxybutyrate by wild-type class I PHA synthase from Ralstonia eutropha. Appl. Microbiol. Biotechnol. 2011, 92, 509–517.
- Han, X.; Satoh, Y.; Tajima, K.; Matsushima, T.; Munekata, M. Chemo-enzymatic synthesis of polyhydroxyalkanoate by an improved two-phase reaction system (TPRS). J. Biosci. Bioeng. 2009, 108, 517–523.
- Sato, S.; Minato, M.; Kikkawa, Y.; Abe, H.; Tsuge, T. In vitro synthesis of polyhydroxyalkanoate catalyzed by class II and III PHA synthases: A useful technique for surface coatings of a hydrophobic support with PHA. J. Chem. Technol. Biotechnol. 2010, 85, 779–782.
- Tajima, K.; Han, X.; Satoh, Y.; Ishii, A.; Araki, Y.; Munekata, M.; Taguchi, S. In vitro synthesis of polyhydroxyalkanoate (PHA) incorporating lactate (LA) with a block sequence by using a newly engineered thermostable PHA synthase from Pseudomonas sp. SG4502 with acquired LA-polymerizing activity. Appl. Microbiol. Biotechnol. 2012, 94, 365–376.
- Lawrence, A.G.; Choi, J.; Rha, C.; Stubbe, J.; Sinskey, A.J. In vitro analysis of the chain termination reaction in the synthesis of poly-(R)-β-hydroxybutyrate by the class III Synthase from Allochromatium vinosum. Biomacromolecules 2005, 6, 2113–2119.
- Tomizawa, S.; Sato, S.; Lan, J.C.-W.; Nakamura, Y.; Abe, H.; Tsuge, T. In vitro evidence of chain transfer to tetraethylene glycols in enzymatic polymerization of polyhydroxyalkanoate. Appl. Microbiol. Biotechnol. 2013, 97, 4821–4829.
- Tajima, K.; Satoh, Y.; Nakazawa, K.; Tannai, H.; Erata, T.; Munekata, M.; Kamachi, M.; Lenz, R.W. Chemoenzymatic synthesis of poly(3-hydroxybutyrate) in a water-organic solvent two-phase system. Macromolecules 2004, 37, 4544–4546.
- Gerngross, T.U.; Martin, D.P. Enzyme-catalyzed synthesis of poly: Formation of macroscopic granules in vitro. Proc. Natl. Acad. Sci. USA 1995, 92, 6279–6283.
- Tomizawa, S.; Yoshioka, M.; Ushimaru, K.; Tsuge, T. Preparative synthesis of poly monomer for enzymatic cell-free polymerization. Polym. J. 2012, 44, 982–985.
- Satoh, Y.; Tajima, K.; Tannai, H.; Munekata, M. Enzyme-catalyzed poly(3-hydroxybutyrate) synthesis from acetate with CoA recycling and NADPH regeneration in vitro. J. Biosci. Bioeng. 2003, 95, 335–341.
- Cherubini, F. The biorefinery concept: Using biomass instead of oil for producing energy and chemicals. Energy Convers. Manag. 2010, 51, 1412–1421.
- Schmidt-Dannert, C.; Lopez-Gallego, F. A roadmap for biocatalysis—Functional and spatial orchestration of enzyme cascades. Microb. Biotechnol. 2016, 9, 601–609.
- Taniguchi, H.; Okano, K.; Honda, K. Modules for in vitro metabolic engineering: Pathway assembly for bio-based production of value-added chemicals. Synth. Syst. Biotechnol. 2017, 2, 65–74.
- Wilding, K.M.; Schinn, S.-M.; Long, E.A.; Bundy, B.C. The emerging impact of cell-free chemical biosynthesis. Curr. Opin. Biotechnol. 2018, 53, 115–121.
- You, C.; Zhang, Y.H.P. Cell-free biosystems for biomanufacturing. In Future Trends in Biotechnology; Zhong, J.-J., Ed.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 89–119.
- Opgenorth, P.H.; Korman, T.P.; Bowie, J.U. A synthetic biochemistry module for production of bio-based chemicals from glucose. Nat. Chem. Biol. 2016, 12, 393–395.
- Li, F.; Wei, X.; Zhang, L.; Liu, C.; You, C.; Zhu, Z. Installing a green engine to drive an enzyme cascade: A light-powered in vitro biosystem for poly(3-hydroxybutyrate) synthesis. Angew. Chem. Int. Ed. 2022, 61, e202111054.
- Zhang, J.; Liu, D.; Liu, Y.; Chu, H.; Bai, J.; Cheng, J.; Zhao, H.; Fu, S.; Liu, H.; Fu, Y.; et al. Hybrid synthesis of polyhydroxybutyrate bioplastics from carbon dioxide. Green Chem. 2023, 25, 3247–3255.
- Grubbe, W.S.; Rasor, B.J.; Krüger, A.; Jewett, M.C.; Karim, A.S. Cell-free styrene biosynthesis at high titers. bioRxiv 2020.
- Karim, A.S.; Dudley, Q.M.; Juminaga, A.; Yuan, Y.; Crowe, S.A.; Heggestad, J.T.; Garg, S.; Abdalla, T.; Grubbe, W.S.; Rasor, B.J.; et al. In vitro prototyping and rapid optimization of biosynthetic enzymes for cell design. Nat. Chem. Biol. 2020, 16, 912–919.
- Guo, W.; Sheng, J.; Feng, X. Mini-review: In vitro metabolic engineering for biomanufacturing of high-value products. Comput. Struct. Biotechnol. J. 2017, 15, 161–167.
- Zhang, Y.; Meng, Q.; Ma, H.; Liu, Y.; Cao, G.; Zhang, X.; Zheng, P.; Sun, J.; Zhang, D.; Jiang, W.; et al. Determination of key enzymes for threonine synthesis through in vitro metabolic pathway analysis. Microb. Cell Factories 2015, 14, 86.
- van den Berg, J.; Boersma, A.J.; Poolman, B. Microorganisms maintain crowding homeostasis. Nat. Rev. Microbiol. 2017, 15, 309–318.
- Wu, H.; Chen, J.; Chen, G.-Q. Engineering the growth pattern and cell morphology for enhanced PHB production by Escherichia coli. Appl. Microbiol. Biotechnol. 2016, 100, 9907–9916.
- Tan, D.; Wu, Q.; Chen, J.-C.; Chen, G.-Q. Engineering Halomonas TD01 for the low-cost production of polyhydroxyalkanoates. Metab. Eng. 2014, 26, 34–47.
- Poltronieri, P. Polyhydroxyalkanoate production in biofermentor monitored through biosensor application. Int. J. Biosens. Bioelectron. 2018, 4, 235–240.
- Wang, W.; Wei, Q.; Zhang, J.; Zhang, M.; Wang, C.; Qu, R.; Wang, Y.; Yang, G.; Wang, J. A ratiometric fluorescent biosensor reveals dynamic regulation of long-chain fatty acyl-coa esters metabolism. Angew. Chem. Int. Ed. 2021, 60, 13996–14004.
- Mao, Y.; Yuan, Q.; Yang, X.; Liu, P.; Cheng, Y.; Luo, J.; Liu, H.; Yao, Y.; Sun, H.; Cai, T.; et al. Non-natural aldol reactions enable the design and construction of novel one-carbon assimilation pathways in vitro. Front. Microbiol. 2021, 12, 677596.
- Bogorad, I.W.; Lin, T.-S.; Liao, J.C. Synthetic non-oxidative glycolysis enables complete carbon conservation. Nature 2013, 502, 693–697.
- Madkour, M.H.; Heinrich, D.; Alghamdi, M.A.; Shabbaj, I.I.; Steinbüchel, A. PHA recovery from biomass. Biomacromolecules 2013, 14, 2963–2972.
- Ong, S.Y.; Zainab-L, I.; Pyary, S.; Sudesh, K. A novel biological recovery approach for PHA employing selective digestion of bacterial biomass in animals. Appl. Microbiol. Biotechnol. 2018, 102, 2117–2127.
- Hodgman, C.E.; Jewett, M.C. Cell-free synthetic biology: Thinking outside the cell. Metab. Eng. 2012, 14, 261–269.
- Lu, X.; Liu, Y.; Yang, Y.; Wang, S.; Wang, Q.; Wang, X.; Yan, Z.; Cheng, J.; Liu, C.; Yang, X.; et al. Constructing a synthetic pathway for acetyl-coenzyme A from one-carbon through enzyme design. Nat. Commun. 2019, 10, 1378.
- Liu, H.; Arbing, M.A.; Bowie, J.U. Expanding the use of ethanol as a feedstock for cell-free synthetic biochemistry by implementing acetyl-CoA and ATP generating pathways. Sci. Rep. 2022, 12, 7700.
- Tajima, K.; Han, X.; Hashimoto, Y.; Satoh, Y.; Satoh, T.; Taguchi, S. In vitro synthesis of polyhydroxyalkanoates using thermostable acetyl-CoA synthetase, CoA transferase, and PHA synthase from thermotorelant bacteria. J. Biosci. Bioeng. 2016, 122, 660–665.
- Phan, H.T.; Hosoe, Y.; Guex, M.; Tomoi, M.; Tomita, H.; Zinn, M.; Matsumoto, K. Directed evolution of sequence-regulating polyhydroxyalkanoate synthase to synthesize a Medium-Chain-Length-Short-Chain-Length (MCL-SCL) block copolymer. Biomacromolecules 2022, 23, 1221–1231.
- Hori, C.; Oishi, K.; Matsumoto, K.; Taguchi, S.; Ooi, T. Site-directed saturation mutagenesis of polyhydroxylalkanoate synthase for efficient microbial production of poly. J. Biosci. Bioeng. 2018, 125, 632–636.
- Dubey, N.C.; Tripathi, B.P. Nature inspired multienzyme immobilization: Strategies and concepts. ACS Appl. Bio Mater. 2021, 4, 1077–1114.
- Zhao, H.; van der Donk, W.A. Regeneration of cofactors for use in biocatalysis. Curr. Opin. Biotechnol. 2003, 14, 583–589.
- Britton, J.; Majumdar, S.; Weiss, G.A. Continuous flow biocatalysis. Chem. Soc. Rev. 2018, 47, 5891–5918.
- Cai, T.; Sun, H.; Qiao, J.; Zhu, L.; Zhang, F.; Zhang, J.; Tang, Z.; Wei, X.; Yang, J.; Yuan, Q.; et al. Cell-free chemoenzymatic starch synthesis from carbon dioxide. Science 2021, 373, 1523–1527.
- Wu, R.; Li, F.; Cui, X.; Li, Z.; Ma, C.; Jiang, H.; Zhang, L.; Zhang, Y.P.J.; Zhao, T.; Zhang, Y.; et al. Enzymatic electrosynthesis of glycine from CO2 and NH3. Angew. Chem. Int. Ed. Engl. 2023, 62, e202218387.
- Welch, P.; Scopes, R.K. Studies on cell-free metabolism: Ethanol production by a yeast glycolytic system reconstituted from purified enzymes. J. Biotechnol. 1985, 2, 257–273.
- Opgenorth, P.H.; Korman, T.P.; Bowie, J.U. A synthetic biochemistry molecular purge valve module that maintains redox balance. Nat. Commun. 2014, 5, 4113.
- Yang, X.; Yuan, Q.; Luo, H.; Li, F.; Mao, Y.; Zhao, X.; Du, J.; Li, P.; Ju, X.; Zheng, Y.; et al. Systematic design and in vitro validation of novel one-carbon assimilation pathways. Metab. Eng. 2019, 56, 142–153.
- Wang, W.; Liu, M.; You, C.; Li, Z.; Zhang, Y.-H.P. ATP-free biosynthesis of a high-energy phosphate metabolite fructose 1,6-diphosphate by in vitro metabolic engineering. Metab. Eng. 2017, 42, 168–174.
- Paul, C.E.; Hollmann, F. A survey of synthetic nicotinamide cofactors in enzymatic processes. Appl. Microbiol. Biotechnol. 2016, 100, 4773–4778.
- Zhu, Z.; Tam, T.K.; Zhang, Y.H.P. Cell-free biosystems in the production of electricity and bioenergy. In Fundamentals and Application of New Bioproduction Systems; Zeng, A.-P., Ed.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 125–152.
- Andexer, J.N.; Richter, M. Emerging enzymes for ATP regeneration in biocatalytic processes. ChemBioChem 2015, 16, 380–386.
- Bachosz, K.; Zdarta, J.; Bilal, M.; Meyer, A.S.; Jesionowski, T. Enzymatic cofactor regeneration systems: A new perspective on efficiency assessment. Sci. Total Environ. 2023, 868, 161630.
- Han, J.; Hou, J.; Zhang, F.; Ai, G.; Li, M.; Cai, S.; Liu, H.; Wang, L.; Wang, Z.; Zhang, S.; et al. Multiple propionyl coenzyme a-supplying pathways for production of the bioplastic poly(3-hydroxybutyrate-co-3-hydroxyvalerate) in Haloferax mediterranei. Appl. Environ. Microbiol. 2013, 79, 2922–2931.
- Schwander, T.; Schada von Borzyskowski, L.; Burgener, S.; Cortina, N.S.; Erb, T.J. A synthetic pathway for the fixation of carbon dioxide in vitro. Science 2016, 354, 900–904.
- Elçin, A.E.; Elçin, Y.M. Polycation-coated polyanion microspheres of urease for urea hydrolysis. Artif. Cells Blood Substit. Biotechnol. 2000, 28, 95–111.
- Ullah, M.W.; Khattak, W.A.; Ul-Islam, M.; Khan, S.; Park, J.K. Metabolic engineering of synthetic cell-free systems: Strategies and applications. Biochem. Eng. J. 2016, 105, 391–405.
- Fu, G.; Yue, J.; Li, D.; Li, Y.; Lee, S.Y.; Zhang, D. An operator-based expression toolkit for Bacillus subtilis enables fine-tuning of gene expression and biosynthetic pathway regulation. Proc. Natl. Acad. Sci. USA 2022, 119, e2119980119.
- Luo, B.; Jin, M.M.; Li, X.; Makunga, N.P.; Hu, X. Yeast surface display for in vitro biosynthetic pathway reconstruction. ACS Synth. Biol. 2021, 10, 2938–2946.
- Chen, Z.; Duan, R.; Xiao, Y.; Wei, Y.; Zhang, H.; Sun, X.; Wang, S.; Cheng, Y.; Wang, X.; Tong, S.; et al. Biodegradation of highly crystallized poly(ethylene terephthalate) through cell surface codisplay of bacterial PETase and hydrophobin. Nat. Commun. 2022, 13, 7138.
- Dong, H.; Zhang, W.; Zhou, S.; Wang, P. Programmable biofilm-cellulose hybrid platform for specific clustering of microbial catalysts with optimized cellular synergy. Chem. Commun. 2022, 58, 8222–8225.
- van der Donk, W.A.; Zhao, H. Recent developments in pyridine nucleotide regeneration. Curr. Opin. Biotechnol. 2003, 14, 421–426.
- Nocek, B.; Kochinyan, S.; Proudfoot, M.; Brown, G.; Evdokimova, E.; Osipiuk, J.; Edwards, A.M.; Savchenko, A.; Joachimiak, A.; Yakunin, A.F. Polyphosphate-dependent synthesis of ATP and ADP by the family-2 polyphosphate kinases in bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 17730–17735.
- Karim, A.S.; Jewett, M.C. A cell-free framework for rapid biosynthetic pathway prototyping and enzyme discovery. Metab. Eng. 2016, 36, 116–126.
- Petroll, K.; Kopp, D.; Care, A.; Bergquist, P.L.; Sunna, A. Tools and strategies for constructing cell-free enzyme pathways. Biotechnol. Adv. 2019, 37, 91–108.
- Koller, M.; Novak, M.; Braunegg, G.; Horvat, P. Mathematical modelling as a tool for optimized pha production. Chem. Biochem. Eng. Q. 2015, 29, 183–220.
- Rollin, J.A.; Martin del Campo, J.; Myung, S.; Sun, F.; You, C.; Bakovic, A.; Castro, R.; Chandrayan, S.K.; Wu, C.-H.; Adams, M.W.W.; et al. High-yield hydrogen production from biomass by in vitro metabolic engineering: Mixed sugars coutilization and kinetic modeling. Proc. Natl. Acad. Sci. USA 2015, 112, 4964–4969.
- Horvath, N.; Vilkhovoy, M.; Wayman, J.A.; Calhoun, K.; Swartz, J.; Varner, J.D. Toward a genome scale sequence specific dynamic model of cell-free protein synthesis in Escherichia coli. Metab. Eng. Commun. 2020, 10, e00113.
- Domenzain, I.; Sánchez, B.; Anton, M.; Kerkhoven, E.J.; Millán-Oropeza, A.; Henry, C.; Siewers, V.; Morrissey, J.P.; Sonnenschein, N.; Nielsen, J. Reconstruction of a catalogue of genome-scale metabolic models with enzymatic constraints using GECKO 2.0. Nat. Commun. 2022, 13, 3766.
- Yang, X.; Mao, Z.; Zhao, X.; Wang, R.; Zhang, P.; Cai, J.; Xue, C.; Ma, H. Integrating thermodynamic and enzymatic constraints into genome-scale metabolic models. Metab. Eng. 2021, 67, 133–144.
- Salvy, P.; Fengos, G.; Ataman, M.; Pathier, T.; Soh, K.C.; Hatzimanikatis, V. pyTFA and matTFA: A python package and a matlab toolbox for thermodynamics-based flux analysis. Bioinformatics 2019, 35, 167–169.
- Nogales, J.; Mueller, J.; Gudmundsson, S.; Canalejo, F.J.; Duque, E.; Monk, J.; Feist, A.M.; Ramos, J.L.; Niu, W.; Palsson, B.O. High-quality genome-scale metabolic modelling of Pseudomonas putida highlights its broad metabolic capabilities. Environ. Microbiol. 2020, 22, 255–269.
- Borzou, P.; Ghaisari, J.; Izadi, I.; Eshraghi, Y.; Gheisari, Y. A novel strategy for dynamic modeling of genome-scale interaction networks. Bioinformatics 2023, 39, btad079.
- Goldberg, A.P.; Szigeti, B.; Chew, Y.H.; Sekar, J.A.; Roth, Y.D.; Karr, J.R. Emerging whole-cell modeling principles and methods. Curr. Opin. Biotechnol. 2018, 51, 97–102.
- Zampieri, G.; Vijayakumar, S.; Yaneske, E.; Angione, C. Machine and deep learning meet genome-scale metabolic modeling. PLoS Comput. Biol. 2019, 15, e1007084.
- Mezzina, M.P.; Manoli, M.T.; Prieto, M.A.; Nikel, P.I. Engineering native and synthetic pathways in Pseudomonas putida for the production of tailored polyhydroxyalkanoates. Biotechnol. J. 2021, 16, 2000165.
- Kumar, P.; Kim, B.S. Valorization of polyhydroxyalkanoates production process by co-synthesis of value-added products. Bioresour. Technol. 2018, 269, 544–556.
- Xu, M.; Qin, J.; Rao, Z.; You, H.; Zhang, X.; Yang, T.; Wang, X.; Xu, Z. Effect of polyhydroxybutyrate (PHB) storage on l-arginine production in recombinant Corynebacterium crenatum using coenzyme regulation. Microb. Cell Factories 2016, 15, 15.
More
Information
Subjects:
Biochemical Research Methods
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.8K
Revisions:
2 times
(View History)
Update Date:
30 May 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




