
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Yue Feng | -- | 2019 | 2023-05-26 02:26:09 | | | |
| 2 | Lindsay Dong | Meta information modification | 2019 | 2023-05-26 03:10:10 | | |
Video Upload Options
Legionella pneumophila is the causative agent of Legionnaires’ disease, causing fever and lung infection, with a death rate up to 15% in severe cases. In the process of infection, Legionella pneumophila secretes over 330 effectors into host cell via the Dot/Icm type IV secretion system to modulate multiple host cellular physiological processes, thereby changing the environment of the host cell and promoting the growth and propagation of the bacterium. Among these effector proteins, SidE family proteins from Legionella pneumophila catalyze a non-canonical ubiquitination reaction, which combines mono-ADP-ribosylation and phosphodiesterase activities together to attach ubiquitin onto substrates. Meanwhile, the activity of SidE family proteins is also under multiple modulations by other effectors.
1. Introduction
2. SidE Family Effectors Catalyze a Non-Canonical Ubiquitination Process
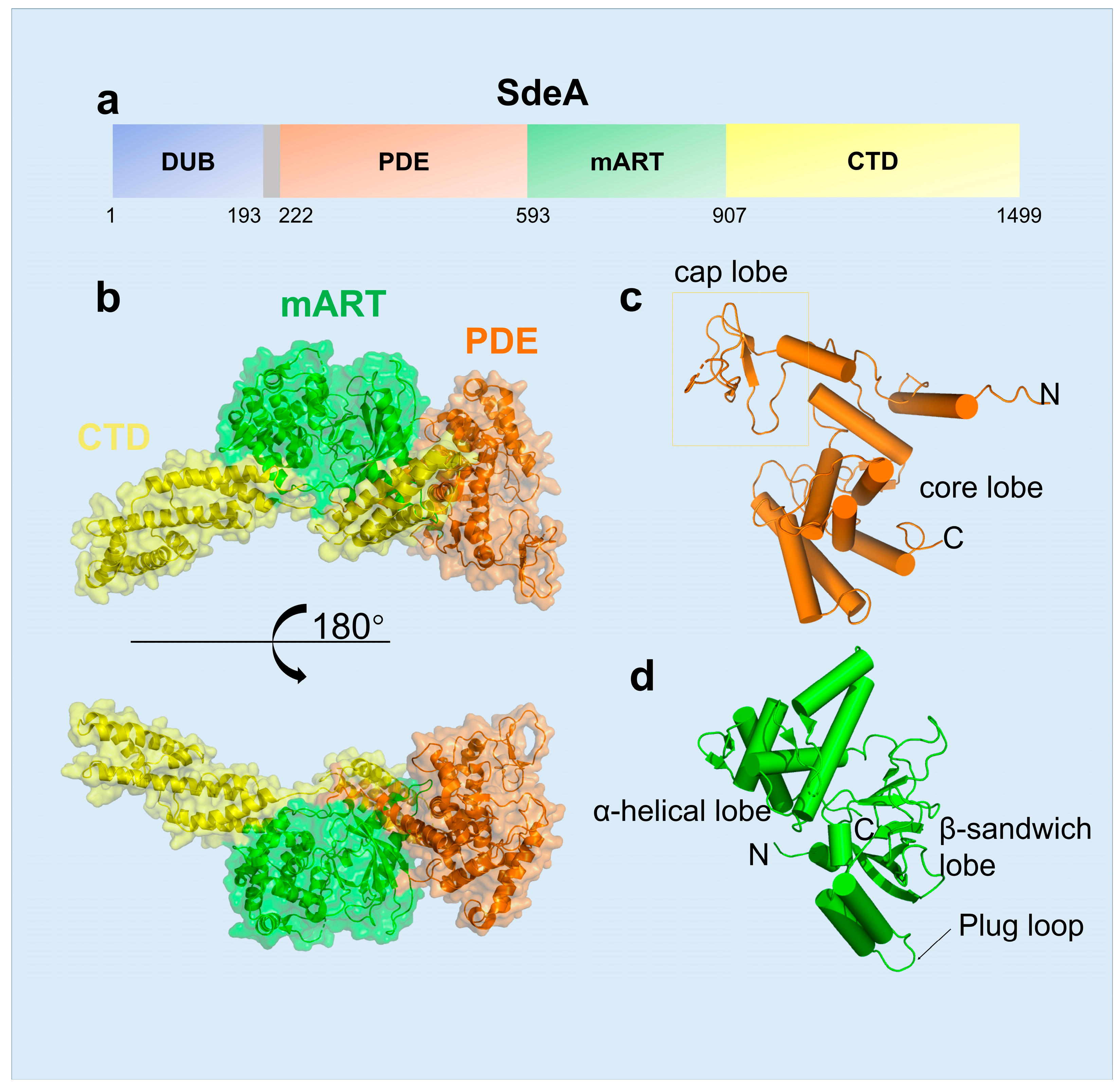
2.1. The Structural Features of SidE Family
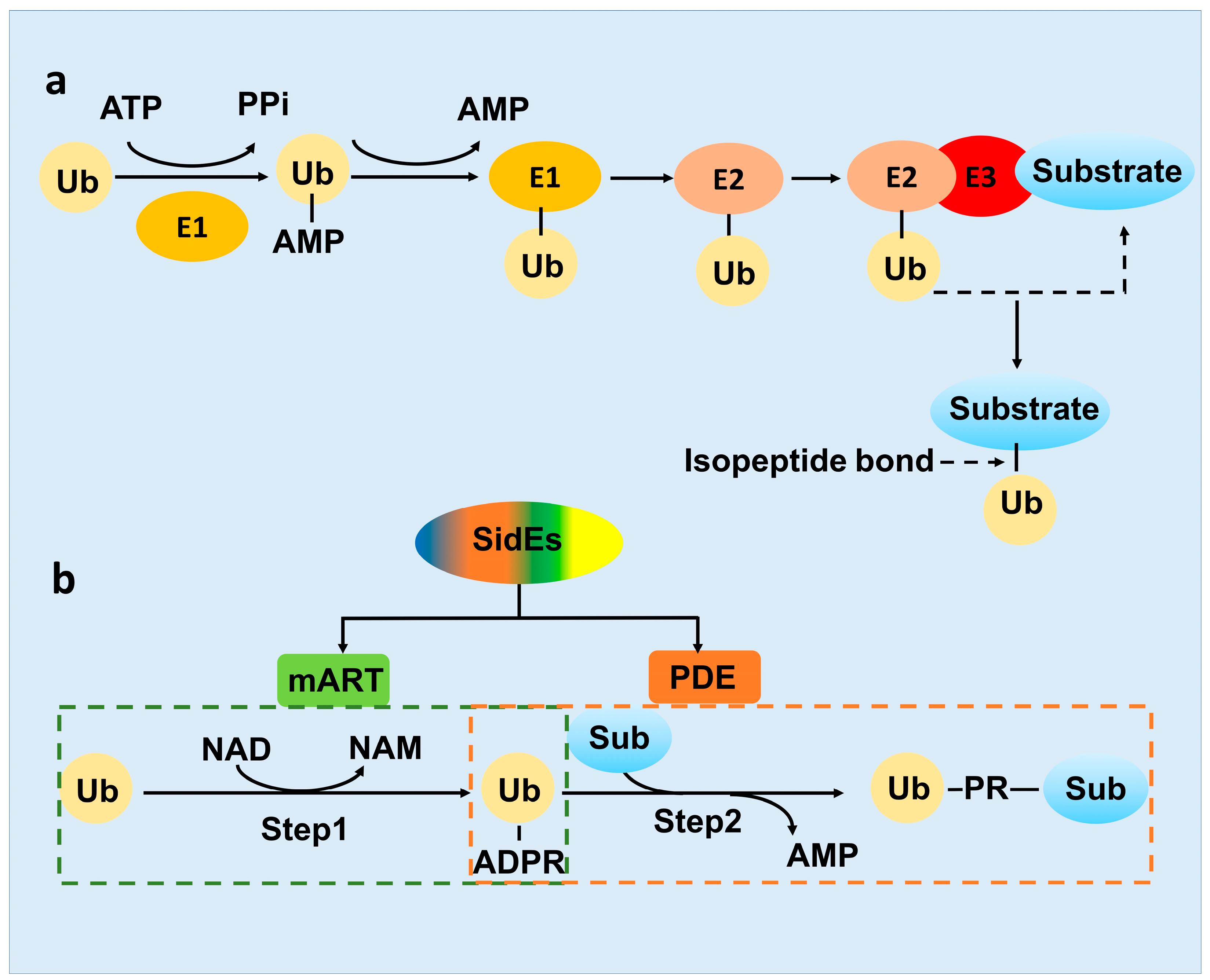
2.2. The Novel-Ubiquitination Machinery of the SidE Family
3. Activity of the SidE Family Was Strictly Modulated by Many Effectors
3.1. SidJ Interacts with Calmodulin to Modify SdeA
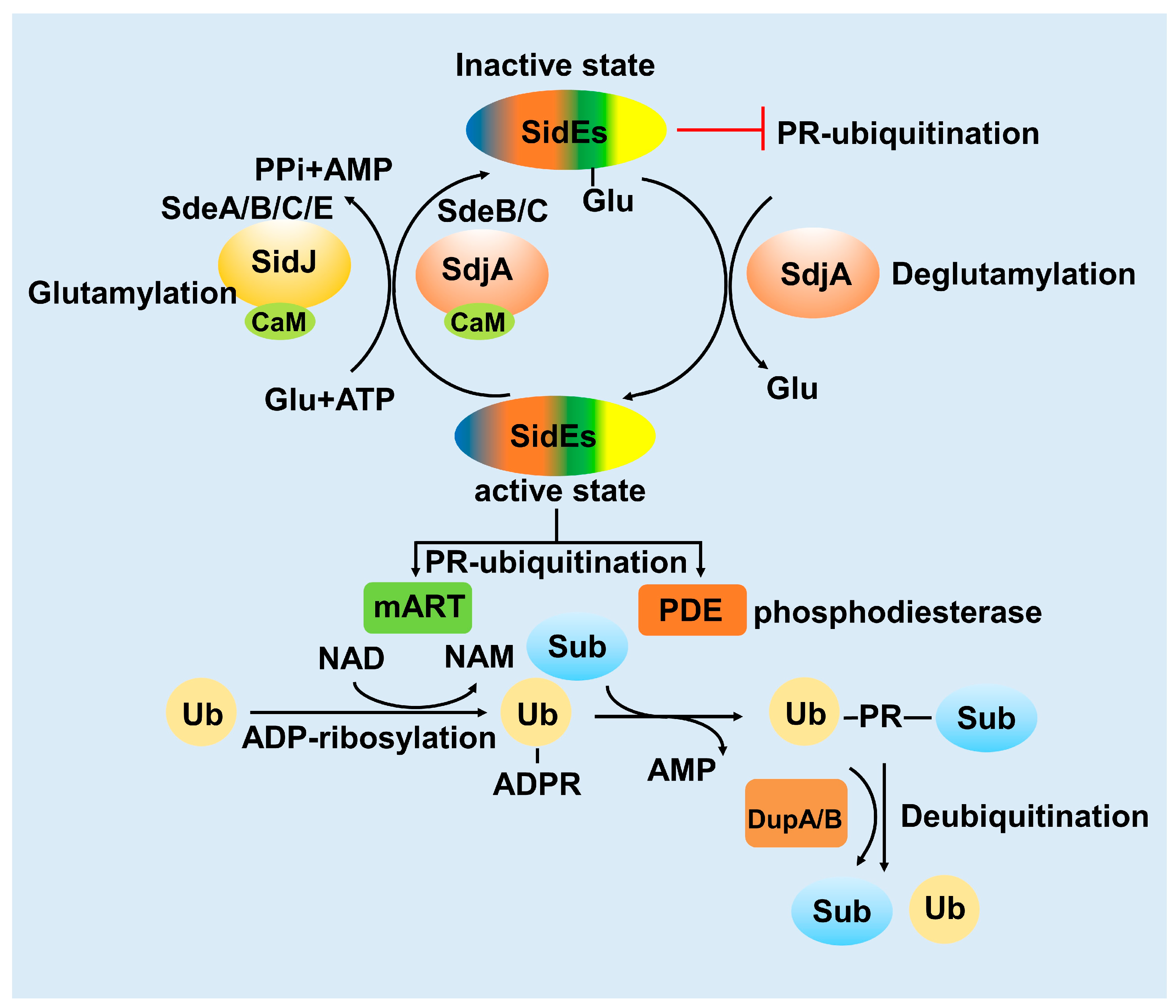
3.2. SdjA Reverses the Glutamylation Modification of SdeA
3.3. DupA and DupB Function as Deubiquitinases for PR-Ubiquitination
4. Multiple Host Proteins Targeted by SidE Family Effectors
4.1. The Effects of SidE Family Proteins on Endoplasmic Reticulum
4.2. The Effects of SidE Family Proteins on the Golgi Complex
References
- Fraser, D.W.; Tsai, T.R.; Orenstein, W.; Parkin, W.E.; Beecham, H.J.; Sharrar, R.G.; Harris, J.; Mallison, G.F.; Martin, S.M.; Mcdade, J.E.; et al. Legionnaires’ Disease. N. Engl. J. Med. 1977, 297, 1189–1197.
- Luo, Z.Q.; Isberg, R.R. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl. Acad. Sci. USA 2004, 101, 841–846.
- Ensminger, A.W. Legionella pneumophila, armed to the hilt: Justifying the largest arsenal of effectors in the bacterial world. Curr. Opin. Microbiol. 2016, 29, 74–80.
- Isberg, R.R.; O’connor, T.J.; Heidtman, M. The Legionella pneumophila replication vacuole: Making a cosy niche inside host cells. Nat. Rev. Microbiol. 2009, 7, 13–24.
- Robinson, C.G.; Roy, C.R. Attachment and fusion of endoplasmic reticulum with vacuoles containing Legionella pneumophila. Cell. Microbiol. 2006, 8, 793–805.
- Ramazi, S.; Zahiri, J. Posttranslational modifications in proteins: Resources, tools and prediction methods. Database 2021, baab012.
- Mann, M.; Jensen, O.N.J.N.B. Proteomic analysis of post-translational modifications. Nat. Biotechnol. 2003, 21, 255–261.
- Olsen, J.V.; Mann, M.J.M.; Mcp, C.P. Status of Large-scale Analysis of Post-translational Modifications by Mass Spectrometry. Mol. Cell. Proteom. 2013, 12, 3444–3452.
- Sun, S.C. Deubiquitylation and regulation of the immune response. Nat. Rev. Immunol. 2008, 8, 501–511.
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479.
- Zheng, N.; Shabek, N. Ubiquitin Ligases: Structure, Function, and Regulation. Annu. Rev. Biochem. 2017, 86, 129–157.
- Yan, F.; Huang, C.; Wang, X.; Tan, J.; Cheng, S.; Wan, M.; Wang, Z.; Wang, S.; Luo, S.; Li, A.; et al. Threonine ADP-Ribosylation of Ubiquitin by a Bacterial Effector Family Blocks Host Ubiquitination. Mol. Cell 2020, 78, 641–652.e649.
- Lin, Y.; Hu, Q.; Zhou, J.; Yin, W.; Yao, D.; Shao, Y.; Zhao, Y.; Guo, B.; Xia, Y.; Chen, Q.; et al. Phytophthora sojae effector Avr1d functions as an E2 competitor and inhibits ubiquitination activity of GmPUB13 to facilitate infection. Proc. Natl. Acad. Sci. USA 2021, 118, e2018312118.
- Gan, N.; Nakayasu, E.S.; Hollenbeck, P.J.; Luo, Z.Q. Legionella pneumophila inhibits immune signalling via MavC-mediated transglutaminase-induced ubiquitination of UBE2N. Nat. Microbiol. 2019, 4, 134–143.
- Dong, Y.; Mu, Y.; Xie, Y.; Zhang, Y.; Han, Y.; Zhou, Y.; Wang, W.; Liu, Z.; Wu, M.; Wang, H.; et al. Structural basis of ubiquitin modification by the Legionella effector SdeA. Nature 2018, 557, 674–678.
- Mu, Y.; Wang, Y.; Huang, Y.; Li, D.; Han, Y.; Chang, M.; Fu, J.; Xie, Y.; Ren, J.; Wang, H.; et al. Structural insights into the mechanism and inhibition of transglutaminase-induced ubiquitination by the Legionella effector MavC. Nat. Commun. 2020, 11, 1774.
- Bardill, J.P.; Miller, J.L.; Vogel, J.P. IcmS-dependent translocation of SdeA into macrophages by the Legionella pneumophila type IV secretion system. Mol. Microbiol. 2005, 56, 90–103.
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797.
- Han, S.; Arvai, A.S.; Clancy, S.B.; Tainer, J.A. Crystal structure and novel recognition motif of Rho ADP-ribosylating C3 exoenzyme from Clostridium botulinum: Structural insights for recognition specificity and catalysis1 1Edited by D. Rees. J. Mol. Biol. 2001, 305, 95–107.
- Qiu, J.; Sheedlo, M.J.; Yu, K.; Tan, Y.; Nakayasu, E.S.; Das, C.; Liu, X.; Luo, Z.Q. Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature 2016, 533, 120–124.
- Zhang, M.; Mcewen, J.M.; Sjoblom, N.M.; Kotewicz, K.M.; Isberg, R.R.; Scheck, R.A. Members of the Legionella pneumophila Sde family target tyrosine residues for phosphoribosyl-linked ubiquitination. RSC Chem. Biol. 2021, 2, 1509–1519.
- Sheedlo, M.J.; Qiu, J.; Tan, Y.; Paul, L.N.; Luo, Z.Q.; Das, C. Structural basis of substrate recognition by a bacterial deubiquitinase important for dynamics of phagosome ubiquitination. Proc. Natl. Acad. Sci. USA 2015, 112, 15090–15095.
- Wang, Y.; Shi, M.; Feng, H.; Zhu, Y.; Liu, S.; Gao, A.; Gao, P. Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE. Cell 2018, 173, 1231–1243.e1216.
- Ji, X.; Wu, Y.; Yan, J.; Mehrens, J.; Yang, H.; Delucia, M.; Hao, C.; Gronenborn, A.M.; Skowronski, J.; Ahn, J.; et al. Mechanism of allosteric activation of SAMHD1 by dGTP. Nat. Struct. Mol. Biol. 2013, 20, 1304–1309.
- Varshavsky, A. The ubiquitin system. Trends Biochem. Sci. 1997, 22, 383–387.
- Bhogaraju, S.; Kalayil, S.; Liu, Y.; Bonn, F.; Colby, T.; Matic, I.; Dikic, I. Phosphoribosylation of Ubiquitin Promotes Serine Ubiquitination and Impairs Conventional Ubiquitination. Cell 2016, 167, 1636–1649.e1613.
- Honjo, T.; Nishizuka, Y.; Hayaishi, O.; Kato, I. Diphtheria Toxin-dependent Adenosine Diphosphate Ribosylation of Aminoacyl Transferase II and Inhibition of Protein Synthesis. J. Biol. Chem. 1968, 243, 3553–3555.
- Corda, D.; Di Girolamo, M. Functional aspects of protein mono-ADP-ribosylation. EMBO J. 2003, 22, 1953–1958.
- Kotewicz, K.M.; Ramabhadran, V.; Sjoblom, N.; Vogel, J.P.; Haenssler, E.; Zhang, M.; Behringer, J.; Scheck, R.A.; Isberg, R.R. A Single Legionella Effector Catalyzes a Multistep Ubiquitination Pathway to Rearrange Tubular Endoplasmic Reticulum for Replication. Cell Host Microbe 2017, 21, 169–181.
- Havey, J.C.; Roy, C.R. Toxicity and SidJ-Mediated Suppression of Toxicity Require Distinct Regions in the SidE Family of Legionella pneumophila Effectors. Infect. Immun. 2015, 83, 3506–3514.
- Qiu, J.; Yu, K.; Fei, X.; Liu, Y.; Nakayasu, E.S.; Piehowski, P.D.; Shaw, J.B.; Puvar, K.; Das, C.; Liu, X.; et al. A unique deubiquitinase that deconjugates phosphoribosyl-linked protein ubiquitination. Cell Res. 2017, 27, 865–881.
- Sulpizio, A.G.; Minelli, M.E.; Mao, Y. Glutamylation of Bacterial Ubiquitin Ligases by a Legionella Pseudokinase. Trends Microbiol. 2019, 27, 967–969.
- Osinski, A.; Black, M.H.; Pawłowski, K.; Chen, Z.; Li, Y.; Tagliabracci, V.S. Structural and mechanistic basis for protein glutamylation by the kinase fold. Mol. Cell 2021, 81, 4527–4539.e4528.
- Komander, D.; Clague, M.J.; Urbé, S. Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009, 10, 550–563.
- Rowbotham, T.J. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 1980, 33, 1179–1183.
- Swanson, M.S.; Isberg, R.R. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect. Immun. 1995, 63, 3609–3620.
- Schwarz, D.S.; Blower, M.D. The endoplasmic reticulum: Structure, function and response to cellular signaling. Cell. Mol. Life Sci. CMLS 2016, 73, 79–94.
- Nixon-Abell, J.; Obara, C.J.; Weigel, A.V.; Li, D.; Legant, W.R.; Xu, C.S.; Pasolli, H.A.; Harvey, K.; Hess, H.F.; Betzig, E.; et al. Increased spatiotemporal resolution reveals highly dynamic dense tubular matrices in the peripheral ER. Science 2016, 354, aaf3928.
- Kawabata, M.; Matsuo, H.; Koito, T.; Murata, M.; Kubori, T.; Nagai, H.; Tagaya, M.; Arasaki, K. Legionella hijacks the host Golgi-to-ER retrograde pathway for the association of Legionella-containing vacuole with the ER. PLoS Pathog. 2021, 17, e1009437.
- Kulkarni-Gosavi, P.; Makhoul, C.; Gleeson, P.A. Form and function of the Golgi apparatus: Scaffolds, cytoskeleton and signalling. FEBS Lett. 2019, 593, 2289–2305.
- Li, J.; Ahat, E.; Wang, Y. Golgi Structure and Function in Health, Stress, and Diseases. Results Probl. Cell Differ. 2019, 67, 441–485.




