Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Mihai Dan Pomohaci | -- | 3111 | 2023-05-25 17:34:04 | | | |
| 2 | Alfred Zheng | Meta information modification | 3111 | 2023-05-26 04:32:38 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Pomohaci, M.D.; Grasu, M.C.; Dumitru, R.L.; Toma, M.; Lupescu, I.G. Artificial Intelligence for Liver Transplant with Hepatocarcinoma. Encyclopedia. Available online: https://encyclopedia.pub/entry/44858 (accessed on 04 March 2026).
Pomohaci MD, Grasu MC, Dumitru RL, Toma M, Lupescu IG. Artificial Intelligence for Liver Transplant with Hepatocarcinoma. Encyclopedia. Available at: https://encyclopedia.pub/entry/44858. Accessed March 04, 2026.
Pomohaci, Mihai Dan, Mugur Cristian Grasu, Radu Lucian Dumitru, Mihai Toma, Ioana Gabriela Lupescu. "Artificial Intelligence for Liver Transplant with Hepatocarcinoma" Encyclopedia, https://encyclopedia.pub/entry/44858 (accessed March 04, 2026).
Pomohaci, M.D., Grasu, M.C., Dumitru, R.L., Toma, M., & Lupescu, I.G. (2023, May 25). Artificial Intelligence for Liver Transplant with Hepatocarcinoma. In Encyclopedia. https://encyclopedia.pub/entry/44858
Pomohaci, Mihai Dan, et al. "Artificial Intelligence for Liver Transplant with Hepatocarcinoma." Encyclopedia. Web. 25 May, 2023.
Copy Citation
Hepatocellular carcinoma is the most common primary malignant hepatic tumor and occurs most often in the setting of chronic liver disease. Liver transplantation is a curative treatment option and is an ideal solution because it solves the chronic underlying liver disorder while removing the malignant lesion. Artificial intelligence is an emerging technology with multiple applications in medicine with a predilection for domains that work with medical imaging, like radiology. With the help of these technologies, laborious tasks can be automated, and new lesion imaging criteria can be developed based on pixel-level analysis.
hepatocarcinoma
liver transplantation
artificial intelligence
1. Introduction
According to the Global Cancer Observatory, primary liver cancer is ranked the third most frequent cause of death and the sixth most commonly diagnosed cancer in 2020 [1]. Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer, accounting for approximately 75–85% of cases [2], representing a significant public health burden worldwide. The incidence of HCC is most often linked with chronic liver disease, and cirrhosis is the primary risk factor, with one-third of cirrhotic patients reported to develop liver cancer during their lifetime [2]. The most common cause of chronic liver disease in Europe is the hepatitis C virus, followed by excessive alcohol intake [3]. In addition, there is a male predominance compared to women of 2:1 [2].
HCC has been the main indication for transplantation in patients with oncologic disease. Together with NASH and NAFLD, it is described as the fastest-rising indication for hepatic transplant [4]. In theory, it is the optimal treatment option because it has a double role of eliminating the underlying liver disease while removing the lesion [5]. However, the selection of transplant candidates that have developed HCC needs to be rigorous as there is a general organ shortage. The United Network for Organ Sharing (UNOS) has described a drop-out of 20% in patients awaiting transplantation [6]. Therefore, extended donor criteria have been adopted to reduce these figures, like older donors, fatty liver, and cardiac arrest donors with inevitable inferior post-transplant outcomes [6]. These factors further stress the importance of patient selection and organ allocation to reduce mortality and improve post-transplantation survival.
The demand for precision medicine and personalized treatments, together with technological advances, has led to an increasing amount of research regarding the application of artificial intelligence (AI) to medical images. The term and technology are not new, as the first artificial neuron was described in 1943 [7]. Today, AI is a large field of study incorporating algorithms capable of solving tasks that normally require human intelligence. Machine learning (ML) is a subset of AI that involves extracting patterns from data without explicit programming [8]. As the algorithm has a more complex structure with multiple components, or “layers”, the term deep learning (DL) is used as a subset of ML [8].
2. AI-Aided Evaluations in Candidates for LT with HCC
2.1. Detection
Detection involves applying a bounding box to the region of interest in the processed images (e.g., lesions, organs, etc.). It is often a preliminary step of more complex algorithms that use a combination of detection-segmentation classification.
Major international guidelines recommend ultrasound (US) as the main imaging surveillance tool for patients with cirrhosis [9][10]. US has a detection sensitivity for HCC of around 84%, but with a substantial drop for early-stage lesions, to almost 47% [11]. This is essential as these lesions have the highest likelihood of long-term cures using radical treatments [5]. However, contrast-enhanced (CE) CT and MRI are not cost-effective for the general surveillance of HCC, except for patients awaiting transplants, according to the EASL guidelines [9]. Therefore, CT is more widely used as it has lower costs, faster acquisition times and less susceptibility to motion artifacts. However, it uses ionizing radiation with lower soft tissue contrast [12]. Although MRI has increased costs and acquisition times, it offers superior tissue contrast and can also use hepatospecific contrast agents, increasing sensitivity [12].
An AI-detection tool would be essential in monitoring patients on the transplant list since the presence of HCC implies the accordance with MELD (Model for End-Stage Liver Disease) exception points [13], which changes the prioritisation on the transplant list and can permit an earlier treatment. Furthermore, the importance of early detection cannot be overstated, as it can impact survival. This is relevant as more patients will be identified with smaller lesions within Milan criteria, with favourable 5-year survival rates of around 65–80% [9]. Thus, an integrated AI tool for lesion detection could improve diagnostic accuracy and improve transplant patient stratification.
In US imaging, the number of applications dedicated to focal liver lesions is reduced mainly due to limited datasets available and because liver lesion characteristics often overlap [14]. However, such a tool can aid those performing US examinations, especially in centres with limited experience. Tiyarattanachai et al. [15] prospectively evaluated US images from 334 patients using a RetinaNet DL model, obtaining detection rates for focal liver lesions as high as 89.8%, surpassing that of clinicians. For HCC, the detection rate was 100%, but only 23 cases were included in the study. Lee et al. [16] used a CNN to detect HCC in multiphase CECT imaging from 302 CT studies using all three phases (arterial, venous, and delayed) with a sensitivity of 93.88%. Using multiphase CECT (pre-contrast, arterial, venous, and delayed), Kim et al. [17] trained and tested a DL model using data from 1320 patients with either cirrhosis or chronic B virus hepatitis to detect HCC. The sensitivity varied according to size, with 33.3% for lesions <10 mm, 74.7% for those between 10–20 mm and 95.9% for lesions >20 mm, with an overall sensitivity of 84.8%. The most frequent cause of the error was an atypical enhancement pattern. Kim et al. [18] studied data from 549 patients with HCC who underwent MR imaging with gadoxetic acid (Gd-EOB-DTPA) to train and test a DL model for HCC detection. Using the hepatobiliary phase, the application had a sensitivity of 87%. Fabijańska et al. [19] obtained a sensitivity of 90.8% for HCC detection in cirrhotic patients. The DL model used integrated T1 dynamic acquisitions with extracellular contrast (non-contrast, arterial and late phase), but the dataset was small, with only nine patients. Integrating all three post-contrast acquisition phases proved superior to using either phase alone. An example of how a detection algorithm works is pictured in Figure 1, with bounding boxes (red) being applied to detected lesions.
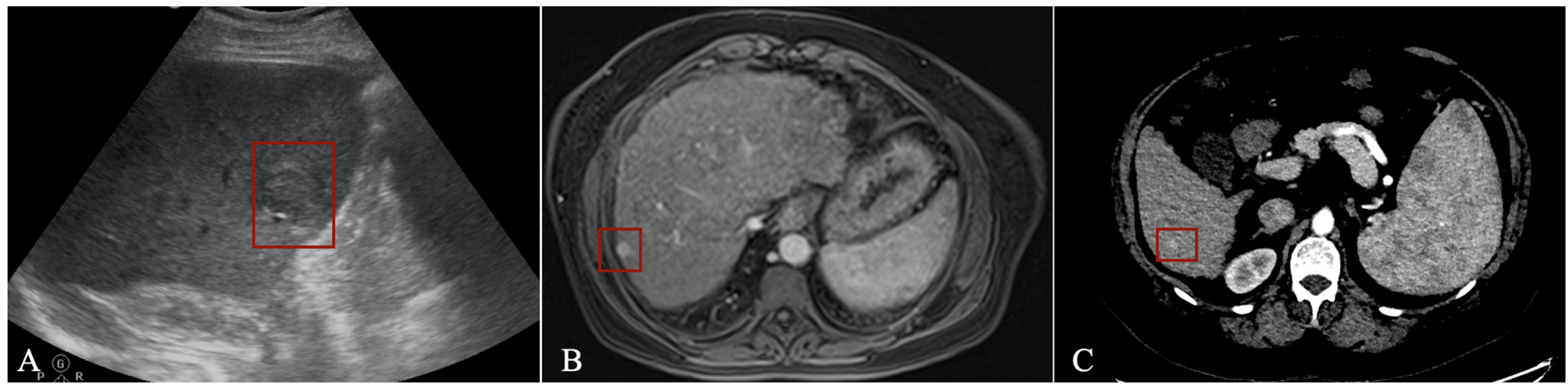
Figure 1. Detection algorithm using ultrasound (A), MRI (B) and CT (C) imaging in the arterial phase.
2.2. Segmentation
Segmentation involves the labelling of pixels in an image to delineate with great precision a region of interest (e.g., lesions, viable tissue in tumour, organs, etc.). The gold standard is represented by manual segmentation done by radiologists. However, this is time-consuming and prone to inter-reader variability [20]. Therefore, the evaluation of segmentation performance is most often done using the Dice-Sørensen coefficient, with results varying between 0–1, with 1 meaning complete overlap.
Liver/liver lesion segmentation with CT represents the main interest regarding AI applications to hepatic imaging [21] as it shows great promise to optimize this process and provide fast, standardized segmentations. Some of the first grand challenges for liver segmentation were organised during the Medical Image Computing and Computer Assisted Intervention Conference (MICCAI) in 2007 [22] and 2008 [23], where only conventional ML methods were used. A shift was seen during the Liver Tumor Segmentation Challenge (LITS) in 2017 [24], where most applications were based on DL. The difficulty lies in the variable liver and liver lesion density and shape, similar densities with surrounding organs like the spleen, gastrointestinal tract, and heart, and the presence of artefacts. Furthermore, anatomical variants are common imaging findings, like accessory fissures or lobes, elongated left liver lobe and Riedel lobe [25]. With cirrhosis, the structure is even more heterogenous, and the contours are irregular, which makes segmentations even more difficult.
Hepatic segmentation is the preferred method for liver volumetry [20]. As living-donor liver transplantation (LDLT) becomes more widespread, it is mandatory to do volumetric evaluations before surgery, as inadequate graft volume is the main contraindication to LDLT [26]. An inaccurate transplanted liver size can cause a small-for-size syndrome with functional insufficiency, leading to death [20]. The minimum remnant liver volume for the adult population is 30%, provided there is no underlying liver dysfunction [27]. Therefore, the recipient’s ratio of graft size to standard liver volume according to body surface area should be over 40–50% [20][26]. An example of whole liver segmentation (A) and right-hemiliver/left-hemiliver segmentation (B) is provided in Figure 2.
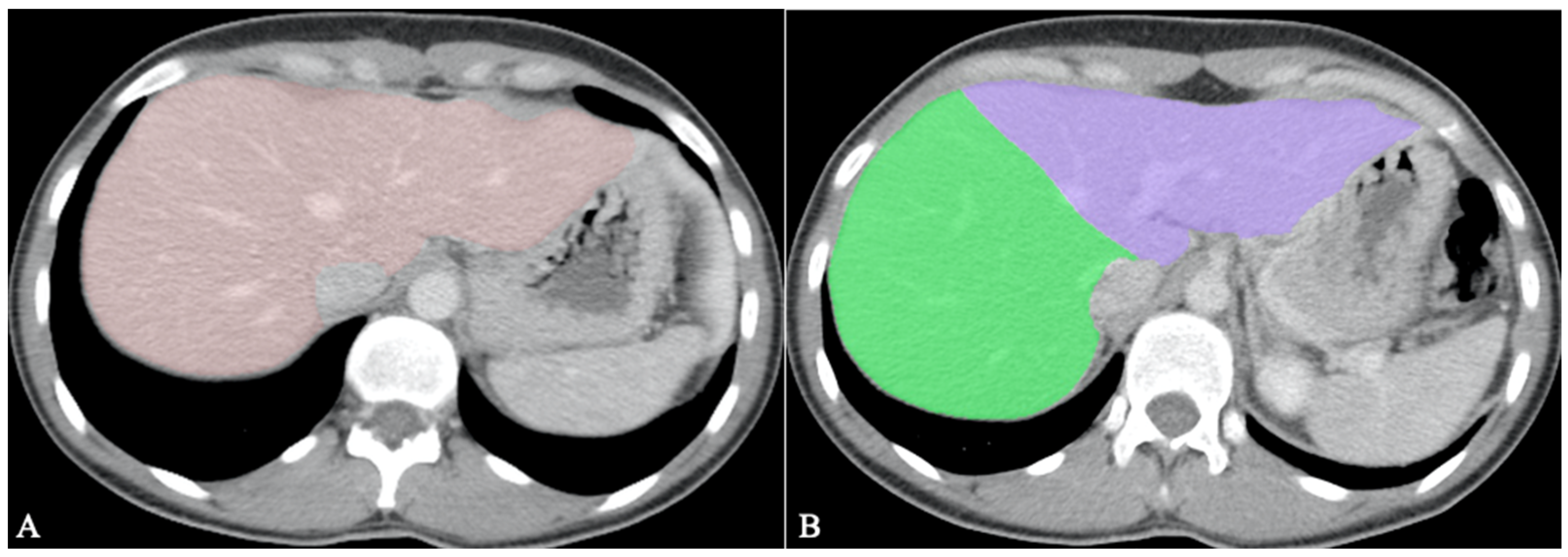
Figure 2. Volumetric measurements before LDLT; (A). Whole liver segmentation; (B). Right hemiliver/left hemiliver segmentation.
Although there is great interest in developing DL models for hepatic segmentation, respecting the Couinaud [28] functional liver segmentation according to vascular supply is mandatory for clinical applications. Tian et al. [29] developed a DL method (GLC-UNet) to segment the liver according to Couinaud using 193 CT scans manually annotated by radiologists. The model obtained a DICE score of 92.46%. Wang et al. [30] used a cascaded neural network (ARH-CNet) to segment the liver according to Couinaud from 193 CT scans manually annotated by radiologists. The model obtained a DICE score of 84%. Using MR imaging, Han et al. [31] developed a 3D convolutional neural network on portal phase acquisitions from 744 scans. The average DICE score was 90.2%, and the dataset included cirrhotic patients. The authors also experimented with the localization of lesions according to segments with a 93.4% accuracy.
Another factor that influences transplant outcome is the presence of steatosis in the donor liver, which may lead to graft dysfunction and biliary and vascular complications [32]. The cut-off varies between 10 to 30% [26]. The gold standard for steatosis diagnosis is biopsy which only evaluates a tiny portion of parenchyma and is subject to inter-pathologist subjectivity [33]. MRI proton density fat fraction (PDFF) has been shown to have a very good diagnostic performance for liver fat assessment and grading [34], evaluating the whole liver structure. Thus, there is a need to develop AI models with more complex roles of both whole liver segmentation and fat quantification. Jimenez-Pastor et al. [35] developed a DL method on 183 MRI multi-echo chemical shift encoded (MECSE) liver studies with the ability to segment the liver and provide fat and iron quantifications. The DICE score for segmentation was 93%, and the model showed a high correlation and low relative error compared to manual fat and iron quantifications. An example of a model for PDFF segmentation and quantification is presented in Figure 3, with an analysis of the whole liver structure on multiple slices.
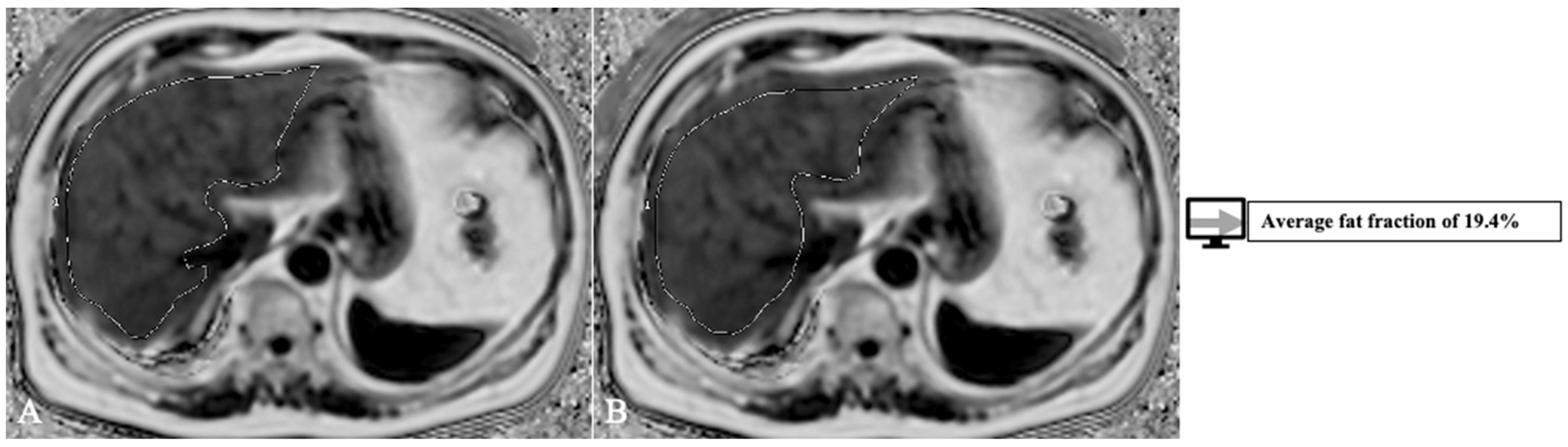
Figure 3. Exemplification of fat-fraction automatic quantification on MRI PDFF acquisitions with whole liver segmentation (1) at two different levels (A,B).
HCC lesion segmentation with volumetric data extraction can also provide an aid to better select patients eligible for transplant with total tumour volume (TTV) as an inclusion criterion with a threshold of 115 cm3 [36]. Bousabarah et al. [37] analysed 174 patients with HCC scanned with MR imaging using a DL method (U-Net) to segment the liver and the lesions. The model used T1 postcontrast acquisitions (arterial, venous, and delayed) using extracellular agents and obtained a DICE score of 91% for liver segmentation and 68% for HCC segmentation. As volumetric assessment becomes automatic with an AI model, more precise and quantifiable inclusion criteria can be developed. For example, the LITS challenge [24] included a tumor burden metric as part of the AI algorithm segmentation accuracy evaluation (calculated as voxels of tumour/voxels of the liver). An example of HCC segmentation and volumetric measurements using CECT is provided in Figure 4.
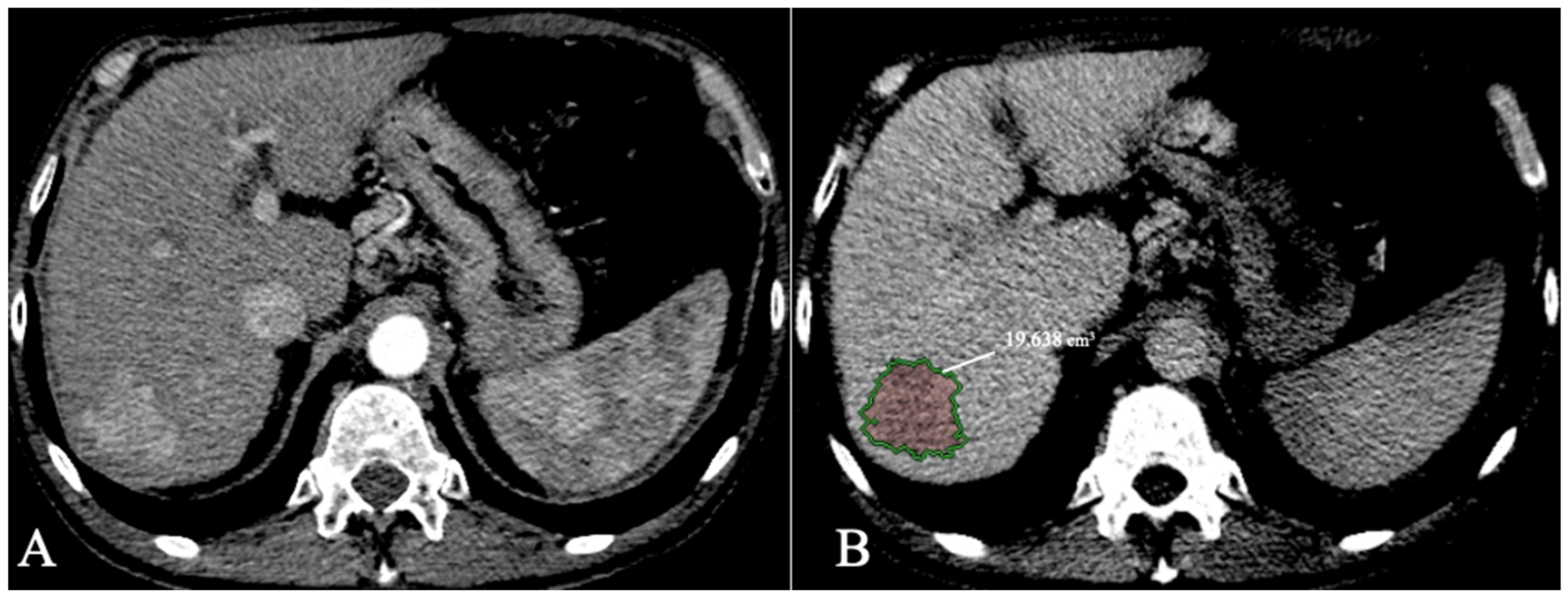
Figure 4. HCC with volumetric measurements; (A). Arterial phase; (B). Delayed phase segmentation.
Automatic segmentation algorithms that help with transplant recipients can also be applied to assess sarcopenia, characterised by the loss of skeletal muscle mass and function. Sarcopenia impacts survival in the liver transplant setting as it is an independent predictor of orthotopic liver transplantation outcome [38], associated with higher mortality [39]. The quantitative assessment of body composition (defined as the percentage of muscle, fat, bone, and water) is usually done at the level of the third lumbar vertebrae [39] by segmentation of muscle, adipose tissue, and bone. AI can reduce segmentation times by providing automatic measurements and more standardised assessment techniques. Most of the research regarding sarcopenia evaluation using AI uses DL methods [40]. For example, blanc-Durand et al. [41] used a convolutional neural network and obtained a Dice score of 97% in a study done on 1025 CT scans.
2.3. Classification
2.3.1. Microvascular Invasion
Microvascular invasion (MVI) is recognised as an essential factor for survival in patients with HCC after LT, and its presence doubles the risk of recurrence [42]. It is defined as tumour present in a vessel lined by endothelium seen by microscopy [43]. The ability to detect this feature before a transplant would allow for better transplant list stratification and risk assessment.
Chen et al. [44] studied 415 patients with small HCC (<3 cm) from three independent institutions. They developed a radiomics signature from DCE MRI with an intracellular agent (Gd-EOB-DTPA) and DWI that predicted MVI with an AUC of 0.971. The HBP and DWI images were most relevant for MVI prediction. Jiang et al. [45] analysed 405 patients with HCC and triple-phase CT acquisitions (arterial, porto-venous, and delayed phase) using a 3D CNN with an AUC of 0.906. They compared these results with a radiomics model that included clinicopathological data, which obtained a lower AUC of 0.887. Sun et al. [46] used DL and AFP information to study 321 patients with HCC and DCE MR imaging with intracellular contrast (Gd-EOB-DTPA). They obtained an AUC of 0.824 in predicting MVI. An ablation study was done to demonstrate which is the most relevant acquisition for determining MVI. A combination of non-contrast T1, delayed, and porto-venous phases showed the best results, while DWI had less impact. Zhou et al. [47] analysed 114 patients undergoing MR imaging using extracellular contrast (Gd-DTPA) with T1, arterial and venous phases. The data from all three contrast acquisitions was processed using a 3D CNN that obtained an AUC for predicting an MVI of 0.926. They also tested the acquisition phases separately and observed that the arterial phase had the best performance (AUC of 0.855).
2.3.2. HCC Grading Prediction
Hepatocarcinoma grade is a biological marker for the aggressiveness of tumors, and, like MVI, it is an important prognostic indicator of recurrence for transplanted patients [48]. The most common classification is the Edmondson and Steiner (ES) according to the degree of differentiation (from well to undifferentiated) [49]. In transplant patients, a biopsy is indicated for excluding undifferentiated and poorly differentiated HCC in the Hangzhou criteria for lesions >8 cm, AFP < 400 ng/mL, and the Toronto Criteria for lesions beyond Milan. This marker can impact prognosis and allow for better patient stratification, as size, and histopathological differentiation are significant independent factors for survival [48].
Mao et al. [50] analysed 297 patients with HCC to develop a radiomics model that classifies lesions according to ES into low-grade or high-grade. CECT imaging data from dual-phase acquisitions (arterial and venous) and clinicopathological data were processed, and the application reached an AUC of 0.801. Even though arterial phase features showed more relevance for the prediction task, using both arterial and venous phases proved superior. Using non-contrast MR imaging and clinical data, Wu et al. [51] studied 170 patients with HCC to train a radiomics model that classifies lesions according to ES grade into low or high. The imaging protocol consisted of non-contrast T1 and T2 weighted images combined with clinical data and obtained an AUC of 0.8. Compared to a model that relied on imaging alone, the combined clinico-radiological model proved superior (0.742 versus 0.800). Han et al. [52] developed a combined clinical and imaging radiomics model to assess HCC grade using hepatospecific DCE MRI (Gd-EOB-DTPA) with T1-weighted, T2-weighted, hepatobilliary and portovenous imaging. The model analysed 137 patients with the hepatobiliary phase having the most significant impact on prediction, obtaining the highest AUC of 0.8. Zhou et al. [53] developed a model to predict ES grading on DWI MR images using a 3D CNN on a cohort of 98 patients obtaining an AUC of 0.83. MR acquisitions consisted of DWI using 0, 100 and 600 s/mm2 b-values and generated ADC maps. The highest b-values proved to be more valuable for classification. Zhou et al. [54] used a deep neural network (SE-DenseNet) to grade HCC lesions using the ES system from DCE MR images from a dataset of 75 patients. They used arterial, venous, and delayed phase images and obtained an AUC of 0.83. They focused more on comparing their model to other neural networks like DenseNet, ResNet and AlexNet, which performed worse.
2.3.3. Molecular Evaluation
Several immunohistochemical markers can offer further information regarding prognosis or improve the positive diagnosis of HCC. Unfortunately, these can only be obtained from biopsy specimens or resected lesions.
Glypican 3 (GPC3) is present on the cell surface and has been included in the panel of markers for HCC diagnosis in highly differentiated small lesions [9]. It can also act as a marker of poor prognosis [55]. The presence of GPC3 in HCC lesions impacts survival as it has been associated with a higher incidence of MVI [56], a reduced 5-year survival rate and disease-free survival in patients with LT [56][57]. Gu et al. [58] analysed a cohort of 293 patients with HCC that underwent MR imaging and developed a radiomics model based on clinical and imaging data to predict the presence of GPC3. MR protocol consisted of T1-weighted postcontrast acquisitions using extracellular contrast (Gd-DTPA) with arterial, venous, and delayed phases. The model that used only imaging data obtained an AUC of 0.871, and when combined with AFP, the AUC increased to 0.914. In a recent study, Chong et al. [59] studied 259 patients with HCC that underwent MR imaging with intracellular contrast (Gd-EOB-DTPA) to develop a radiomics model that predicts the presence of GPC3. The study showed that the most relevant imaging features were from T2 weighted images and T1 hepatobiliary phase, and together with clinical data, a nomogram was created that obtained an AUC of 0.943.
Cytokeratin 19 (CK19) is normally expressed in hepatic progenitor cells but not in healthy hepatocytes, and its presence in HCC lesions is a marker of aggressiveness and poor prognosis [9][60]. In patients with HCC that have undergone transplants beyond Milan criteria, CK19 has been associated with recurrence. In contrast, patients without expression of CK19 showed similar survival rates as those within the Milan criteria [61]. Zhang et al. [62] studied 214 patients and developed a radiomics model using ultrasound imaging and clinical data to predict the presence of CK19. The combined model obtained an AUC of 0.867, while the one that used only imaging data had a lower AUC of 0.789, showing the relevance of combining multiple types of input data. Yang et al. [63] analysed 257 patients with HCC from multiple centres that underwent MR imaging and developed a radiomics model to determine the presence of CK19+ lesions. The model with the best predictive performance used features from T2 and DWI with an AUC of 0.790. Chen et al. [64] developed a radiomics model using data from 80 patients with HCC to determine the presence of CK19. The imaging protocol consisted of MR imaging with intracellular contrast (Gd-EOB-DTPA), and the data was obtained from two institutions. They obtained an AUC of 0.833 by adding clinical data like AFP to improve performance while relying on imaging alone resulted in an AUC of 0.82. Analysing the data, they found that targetoid features on imaging were correlated with the presence of CK19.
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021, 71, 209–249.
- Singal, A.G.; Lampertico, P.; Nahon, P. Epidemiology and surveillance for hepatocellular carcinoma: New trends. J. Hepatol. 2020, 72, 250–261.
- Akinyemiju, T.; Abera, S.; Ahmed, M.; Alam, N.; Alemayohu, M.A.; Allen, C.; Al-Raddadi, R.; Alvis-Guzman, N.; Amoako, Y.; Artaman, A.; et al. Global Burden of Disease Liver Cancer Collaboration. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level: Results from the global burden of disease study 2015. JAMA Oncol. 2017, 3, 1683–1691.
- Kim, W.R.; Lake, J.R.; Smith, J.M.; Schladt, D.P.; Skeans, M.A.; Harper, A.M.; Wainright, J.L.; Snyder, J.J.; Israni, A.K.; Kasiske, B.L. OPTN/SRTR 2016 Annual Data Report: Liver. Am. J. Transpl. 2018, 18 (Suppl. S1), 172–253.
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular carcinoma. Lancet 2018, 391, 1301–1314.
- Mehta, N.; Dodge, J.L.; Hirose, R.; Roberts, J.P.; Yao, F.Y. Predictors of low risk for dropout from the liver transplant waiting list for hepatocellular carcinoma in long wait time regions: Implications for organ allocation. Am. J. Transpl. 2019, 19, 2210–2218.
- McCulloch, W.S.; Pitts, W. A logical calculus of the ideas immanent in nervous activity. Bull. Math. Biophys. 1943, 5, 115–133.
- Do, S.; Song, K.D.; Chung, J.W. Basics of deep learning: A radiologist’s guide to understanding published radiology articles on deep learning. Korean J. Radiol. 2020, 21, 33–41.
- Galle, P.R.; Forner, A.; Llovet, J.M.; Mazzaferro, V.; Piscaglia, F.; Raoul, J.L.; Schirmacher, P.; Vilgrain, V. EASL clinical practice guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2018, 69, 182–236.
- Heimbach, J.K.; Kulik, L.M.; Finn, R.S.; Sirlin, C.B.; Abecassis, M.M.; Roberts, L.R.; Zhu, A.X.; Murad, M.H.; Marrero, J.A. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology 2018, 67, 358–380.
- Tzartzeva, K.; Obi, J.; Rich, N.E.; Parikh, N.D.; Marrero, J.A.; Yopp, A.; Waljee, A.K.; Singal, A.G. Surveillance imaging and alpha fetoprotein for early detection of hepatocellular carcinoma in patients with cirrhosis: A meta-analysis. Gastroenterology 2018, 154, 1706–1718.
- Roberts, L.R.; Sirlin, C.B.; Zaiem, F.; Almasri, J.; Prokop, L.J.; Heimbach, J.K.; Murad, M.H.; Mohammed, K. Imaging for the diagnosis of hepatocellular carcinoma: A systematic review and meta-analysis. Hepatology 2018, 67, 401–421.
- Goldberg, D.S.; Olthoff, K.M. Standardizing MELD exceptions: Current challenges and future directions. Curr. Transplant. Rep. 2014, 1, 232–237.
- Song, K.D. Current status of deep learning applications in abdominal ultrasonography. Ultrasonography 2021, 40, 177.
- Tiyarattanachai, T.; Apiparakoon, T.; Marukatat, S.; Sukcharoen, S.; Yimsawad, S.; Chaichuen, O.; Bhumiwat, S.; Tanpowpong, N.; Pinjaroen, N.; Rerknimitr, R.; et al. The feasibility to use artificial intelligence to aid detecting focal liver lesions in real-time ultrasound: A preliminary study based on videos. Sci. Rep. 2022, 12, 7749.
- Lee, G.; Kim, J.; Lee, J.G.; Ahn, G.; Park, S.H.; Kim, S.Y.; Kim, K.W.; Lee, S.S.; Kim, N. Automatic hepatocellular carcinoma lesion detection with dynamic enhancement characteristic from multi-phase CT images. In International Forum on Medical Imaging in Asia 2019; SPIE: Bellingham, DC, USA, 2019; Volume 11050, pp. 203–208.
- Kim, D.W.; Lee, G.; Kim, S.Y.; Ahn, G.; Lee, J.G.; Lee, S.S.; Kim, K.W.; Park, S.H.; Lee, Y.J.; Kim, N. Deep learning–based algorithm to detect primary hepatic malignancy in multiphase CT of patients at high risk for HCC. Eur. Radiol. 2021, 31, 7047–7057.
- Kim, J.; Min, J.H.; Kim, S.K.; Shin, S.Y.; Lee, M.W. Detection of hepatocellular carcinoma in contrast-enhanced magnetic resonance imaging using deep learning classifier: A multi-center retrospective study. Sci. Rep. 2020, 10, 9458.
- Fabijańska, A.; Vacavant, A.; Lebre, M.A.; Pavan, A.L.; de Pina, D.R.; Abergel, A.; Chabrot, P.; Magnin, B. U-CatcHCC: An accurate HCC detector in hepatic DCE-MRI sequences based on an U-Net framework. In Proceedings of the International Conference on Computer Vision, ICCVG 2018, Warsaw, Poland, 17–19 September 2018; Springer International Publishing: Berlin/Heidelberg, Germany, 2018; pp. 319–328.
- Gotra, A.; Sivakumaran, L.; Chartrand, G.; Vu, K.N.; Vandenbroucke-Menu, F.; Kauffmann, C.; Kadoury, S.; Gallix, B.; de Guise, J.A.; Tang, A. Liver segmentation: Indications, techniques and future directions. Insights Into Imaging 2017, 8, 377–392.
- Survarachakan, S.; Prasad, P.J.R.; Naseem, R.; de Frutos, J.P.; Kumar, R.P.; Langø, T.; Cheikh, F.A.; Elle, O.J.; Lindseth, F. Deep learning for image-based liver analysis—A comprehensive review focusing on malignant lesions. Artif. Intell. Med. 2022, 130, 102331.
- Van Ginneken, B.; Heimann, T.; Styner, M. 3D Segmentation in the Clinic: A Grand Challenge. In MICCAI Workshop on 3D Segmentation in the Clinic: A Grand Challenge; 2007; Volume 1, pp. 7–15. Available online: http://rumc-gcorg-p-public.s3.amazonaws.com/f/challenge/8/5db8e512-75f2-4878-a6a1-b426f43104a5/p7.pdf (accessed on 7 March 2023).
- Deng, X.; Du, G. 3D Segmentation in the Clinic: A Grand Challenge II-Liver Tumor Segmentation. In MICCAI Workshop; 2008. Available online: https://www.researchgate.net/profile/Guangwei-Du-2/publication/267699971_Editorial_3D_Segmentation_in_the_Clinic_A_Grand_Challenge_II_-Liver_Tumor_Segmentation/links/549a28f40cf2b8037135913b/Editorial-3D-Segmentation-in-the-Clinic-A-Grand-Challenge-II-Liver-Tumor-Segmentation.pdf (accessed on 7 March 2023).
- Bilic, P.; Christ, P.F.; Vorontsov, E.; Chlebus, G.; Chen, H.; Dou, Q.; Fu, C.; Han, X.; Heng, P.; Hesser, J.W.; et al. The Liver Tumor Segmentation Benchmark (LiTS). Med. Image Anal. 2019, 84, 102680.
- Singh, H.R.; Rabi, S. Study of morphological variations of liver in human. Transl. Res. Anat. 2019, 14, 1–5.
- Vernuccio, F.; Whitney, S.A.; Ravindra, K.; Marin, D. CT and MR imaging evaluation of living liver donors. Abdom. Radiol. 2021, 46, 17–28.
- Lim, M.C.; Tan, C.H.; Cai, J.; Zheng, J.; Kow, A.W.C. CT volumetry of the liver: Where does it stand in clinical practice? Clin. Radiol. 2014, 69, 887–895.
- Couinaud, C. Le Foie: Études Anatomiques et Chirurgicales; Masson: Paris, France, 1957.
- Tian, J.; Liu, L.; Shi, Z.; Xu, F. Automatic couinaud segmentation from CT volumes on liver using GLC-UNet. In Proceedings of the 10th International Workshop, MLMI 2019, Shenzhen, China, 13 October 2019; pp. 274–282.
- Wang, M.; Jin, R.; Lu, J.; Song, E.; Ma, G. Automatic CT liver Couinaud segmentation based on key bifurcation detection with attentive residual hourglass-based cascaded network. Comput. Biol. Med. 2022, 144, 105363.
- Han, X.; Wu, X.; Wang, S.; Xu, L.; Xu, H.; Zheng, D.; Yu, N.; Hong, Y.; Yu, Z.; Yang, D.; et al. Automated segmentation of liver segment on portal venous phase MR images using a 3D convolutional neural network. Insights Into Imaging 2022, 13, 26.
- Hamar, M.; Selzner, M. Steatotic donor livers: Where is the risk-benefit maximized? Liver Transplant. 2017, 23, S34–S39.
- Yersiz, H.; Lee, C.; Kaldas, F.M.; Hong, J.C.; Rana, A.; Schnickel, G.T.; Wertheim, J.A.; Zarrinpar, A.; Agopian, V.G.; Gornbein, J.; et al. Assessment of hepatic steatosis by transplant surgeon and expert pathologist: A prospective, double-blind evaluation of 201 donor livers. Liver Transplant. 2013, 19, 437–449.
- Gu, J.; Liu, S.; Du, S.; Zhang, Q.; Xiao, J.; Dong, Q.; Xin, Y. Diagnostic value of MRI-PDFF for hepatic steatosis in patients with non-alcoholic fatty liver disease: A meta-analysis. Eur. Radiol. 2019, 29, 3564–3573.
- Jimenez-Pastor, A.; Alberich-Bayarri, A.; Lopez-Gonzalez, R.; Marti-Aguado, D.; França, M.; Bachmann, R.S.M.; Mazzucco, J.; Marti-Bonmati, L. Precise whole liver automatic segmentation and quantification of PDFF and R2* on MR images. Eur. Radiol. 2021, 31, 7876–7887.
- Toso, C.; Trotter, J.; Wei, A.; Bigam, D.L.; Shah, S.; Lancaster, J.; Grant, D.R.; Greig, P.D.; Shapiro, A.J.; Kneteman, N.M. Total tumor volume predicts risk of recurrence following liver transplantation in patients with hepatocellular carcinoma. Liver Transplant. 2008, 14, 1107–1115.
- Bousabarah, K.; Letzen, B.; Tefera, J.; Savic, L.; Schobert, I.; Schlachter, T.; Staib, L.H.; Kocher, M.; Chapiro, J.; Lin, M. Automated detection and delineation of hepatocellular carcinoma on multiphasic contrast-enhanced MRI using deep learning. Abdom. Radiol. 2021, 46, 216–225.
- Van Vugt, J.L.A.; Levolger, S.; De Bruin, R.W.F.; van Rosmalen, J.; Metselaar, H.J.; IJzermans, J.N.M. Systematic review and meta-analysis of the impact of computed tomography–assessed skeletal muscle mass on outcome in patients awaiting or undergoing liver transplantation. Am. J. Transplant. 2016, 16, 2277–2292.
- Shafaat, O.; Liu, Y.; Jackson, K.R.; Motter, J.D.; Boyarsky, B.J.; Latif, M.A.; Yuan, F.; Khalil, A.; King, E.A.; Zaheer, A.; et al. Association between abdominal CT measurements of body composition before deceased donor liver transplant with posttransplant outcomes. Radiology 2022, 306, 212403.
- Rozynek, M.; Kucybała, I.; Urbanik, A.; Wojciechowski, W. Use of artificial intelligence in the imaging of sarcopenia: A narrative review of current status and perspectives. Nutrition 2021, 89, 111227.
- Blanc-Durand, P.; Schiratti, J.B.; Schutte, K.; Jehanno, P.; Herent, P.; Pigneur, F.; Lucidarme, O.; Benaceur, Y.; Sadate, A.; Luciani, A.; et al. Abdominal musculature segmentation and surface prediction from CT using deep learning for sarcopenia assessment. Diagn. Interv. Imaging 2020, 101, 789–794.
- Mazzaferro, V.; Llovet, J.M.; Miceli, R.; Bhoori, S.; Schiavo, M.; Mariani, L.; Camerini, T.; Roayaie, S.; Schwartz, M.E.; Grazi, G.L.; et al. Predicting survival after liver transplantation in patients with hepatocellular carcinoma beyond the Milan criteria: A retrospective, exploratory analysis. Lancet Oncol. 2009, 10, 35–43.
- Roayaie, S.; Blume, I.N.; Thung, S.N.; Guido, M.; Fiel, M.I.; Hiotis, S.; Labow, D.M.; Llovet, J.M.; Schwartz, M.E. A system of classifying microvascular invasion to predict outcome after resection in patients with hepatocellular carcinoma. Gastroenterology 2009, 137, 850–855.
- Chen, Y.D.; Zhang, L.; Zhou, Z.P.; Lin, B.; Jiang, Z.J.; Tang, C.; Dang, Y.W.; Xia, Y.W.; Song, B.; Long, L.L. Radiomics and nomogram of magnetic resonance imaging for preoperative prediction of microvascular invasion in small hepatocellular carcinoma. World J. Gastroenterol. 2022, 28, 4399.
- Jiang, Y.Q.; Cao, S.E.; Cao, S.; Chen, J.N.; Wang, G.Y.; Shi, W.Q.; Deng, Y.N.; Cheng, N.; Ma, K.; Zeng, K.N.; et al. Preoperative identification of microvascular invasion in hepatocellular carcinoma by XGBoost and deep learning. J. Cancer Res. Clin. Oncol. 2021, 147, 821–833.
- Sun, B.Y.; Gu, P.Y.; Guan, R.Y.; Zhou, C.; Lu, J.W.; Yang, Z.F.; Pan, C.; Zhou, P.Y.; Zhu, Y.P.; Li, J.R.; et al. Deep-learning-based analysis of preoperative MRI predicts microvascular invasion and outcome in hepatocellular carcinoma. World J. Surg. Oncol. 2022, 20, 189.
- Zhou, W.; Jian, W.; Cen, X.; Zhang, L.; Guo, H.; Liu, Z.; Liang, C.; Wang, G. Prediction of microvascular invasion of hepatocellular carcinoma based on contrast-enhanced MR and 3D convolutional neural networks. Front. Oncol. 2021, 11, 189.
- Tamura, S.; Kato, T.; Berho, M.; Misiakos, E.P.; O’Brien, C.; Reddy, K.R.; Nery, J.R.; Burke, G.W.; Schiff, E.R.; Miller, J.; et al. Impact of histological grade of hepatocellular carcinoma on the outcome of liver transplantation. Arch. Surg. 2001, 136, 25–30.
- Martins-Filho, S.N.; Paiva, C.; Azevedo, R.S.; Alves, V.A.F. Histological grading of hepatocellular carcinoma—A systematic review of literature. Front. Med. 2017, 4, 193.
- Mao, B.; Zhang, L.; Ning, P.; Ding, F.; Wu, F.; Lu, G.; Geng, Y.; Ma, J. Preoperative prediction for pathological grade of hepatocellular carcinoma via machine learning–based radiomics. Eur. Radiol. 2020, 30, 6924–6932.
- Wu, M.; Tan, H.; Gao, F.; Hai, J.; Ning, P.; Chen, J.; Zhu, S.; Wang, M.; Dou, S.; Shi, D. Predicting the grade of hepatocellular carcinoma based on non-contrast-enhanced MRI radiomics signature. Eur. Radiol. 2019, 29, 2802–2811.
- Han, Y.E.; Cho, Y.; Kim, M.J.; Park, B.J.; Sung, D.J.; Han, N.Y.; Sim, K.C.; Park, Y.S.; Park, B.N. Hepatocellular carcinoma pathologic grade prediction using radiomics and machine learning models of gadoxetic acid-enhanced MRI: A two-center study. Abdom. Radiol. 2023, 48, 244–256.
- Zhou, W.; Wang, G.; Xie, G.; Zhang, L. Grading of hepatocellular carcinoma based on diffusion weighted images with multiple b-values using convolutional neural networks. Med. Phys. 2019, 46, 3951–3960.
- Zhou, Q.; Zhou, Z.; Chen, C.; Fan, G.; Chen, G.; Heng, H.; Ji, J.; Dai, Y. Grading of hepatocellular carcinoma using 3D SE-DenseNet in dynamic enhanced MR images. Comput. Biol. Med. 2019, 107, 47–57.
- Xiao, W.K.; Qi, C.Y.; Chen, D.; Li, S.Q.; Fu, S.J.; Peng, B.G.; Liang, L.J. Prognostic significance of glypican-3 in hepatocellular carcinoma: A meta-analysis. BMC Cancer 2014, 14, e9702.
- Wang, Y.L.; Zhu, Z.J.; Teng, D.H.; Yao, Z.; Gao, W.; Shen, Z.Y. Glypican-3 expression and its relationship with recurrence of HCC after liver transplantation. World J. Gastroenterol. WJG 2012, 18, 2408.
- Cui, X.; Li, Z.; Gao, P.J.; Gao, J.; Zhu, J.Y. Prognostic value of glypican-3 in patients with HBV-associated hepatocellular carcinoma after liver transplantation. Hepatobiliary Pancreat. Dis. Int. 2015, 14, 157–163.
- Gu, D.; Xie, Y.; Wei, J.; Li, W.; Ye, Z.; Zhu, Z.; Tian, J.; Li, X. MRI-based radiomics signature: A potential biomarker for identifying glypican 3-positive hepatocellular carcinoma. J. Magn. Reson. Imaging 2020, 52, 1679–1687.
- Chong, H.; Gong, Y.; Zhang, Y.; Dai, Y.; Sheng, R.; Zeng, M. Radiomics on Gadoxetate Disodium-enhanced MRI: Non-invasively Identifying Glypican 3-Positive Hepatocellular Carcinoma and Postoperative Recurrence. Acad. Radiol. 2023, 30, 49–63.
- Vogel, A.; Cervantes, A.; Chau, I.; Daniele, B.; Llovet, J.M.; Meyer, T.; Nault, J.C.; Neumann, U.; Ricke, J.; Sangro, B.; et al. Hepatocellular carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2018, 29, iv238–iv255.
- Miltiadous, O.; Sia, D.; Hoshida, Y.; Fiel, M.I.; Harrington, A.N.; Thung, S.N.; Tan, P.S.; Dong, H.; Revill, K.; Chang, C.Y.; et al. Progenitor cell markers predict outcome of patients with hepatocellular carcinoma beyond Milan criteria undergoing liver transplantation. J. Hepatol. 2015, 63, 1368–1377.
- Zhang, L.; Qi, Q.; Li, Q.; Ren, S.; Liu, S.; Mao, B.; Li, X.; Wu, Y.; Yang, L.; Liu, L.; et al. Ultrasomics prediction for cytokeratin 19 expression in hepatocellular carcinoma: A multicenter study. Front. Oncol. 2022, 12, 994456.
- Yang, F.; Wan, Y.; Xu, L.; Wu, Y.; Shen, X.; Wang, J.; Lu, D.; Shao, C.; Zheng, S.; Niu, T.; et al. MRI-Radiomics prediction for cytokeratin 19-Positive hepatocellular carcinoma: A multicenter study. Front. Oncol. 2021, 11, 672126.
- Chen, Y.; Chen, J.; Zhang, Y.; Lin, Z.; Wang, M.; Huang, L.; Huang, M.; Tang, M.; Zhou, X.; Peng, Z.; et al. Preoperative Prediction of Cytokeratin 19 Expression for Hepatocellular Carcinoma with Deep Learning Radiomics Based on Gadoxetic Acid-Enhanced Magnetic Resonance Imaging. J. Hepatocell. Carcinoma 2021, 8, 795–808.
More
Information
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
770
Revisions:
2 times
(View History)
Update Date:
26 May 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





