Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | François Bachand | -- | 3309 | 2023-05-23 15:59:20 | | | |
| 2 | Rita Xu | Meta information modification | 3309 | 2023-05-24 03:57:21 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Landry-Voyer, A.; Mir Hassani, Z.; Avino, M.; Bachand, F. Ribosomal Protein uS5 and Friends. Encyclopedia. Available online: https://encyclopedia.pub/entry/44726 (accessed on 07 February 2026).
Landry-Voyer A, Mir Hassani Z, Avino M, Bachand F. Ribosomal Protein uS5 and Friends. Encyclopedia. Available at: https://encyclopedia.pub/entry/44726. Accessed February 07, 2026.
Landry-Voyer, Anne-Marie, Zabih Mir Hassani, Mariano Avino, François Bachand. "Ribosomal Protein uS5 and Friends" Encyclopedia, https://encyclopedia.pub/entry/44726 (accessed February 07, 2026).
Landry-Voyer, A., Mir Hassani, Z., Avino, M., & Bachand, F. (2023, May 23). Ribosomal Protein uS5 and Friends. In Encyclopedia. https://encyclopedia.pub/entry/44726
Landry-Voyer, Anne-Marie, et al. "Ribosomal Protein uS5 and Friends." Encyclopedia. Web. 23 May, 2023.
Copy Citation
Ribosomal proteins are fundamental components of the ribosomes in all living cells. The ribosomal protein uS5 (Rps2) is a stable component of the small ribosomal subunit within all three domains of life. In addition to its interactions with proximal ribosomal proteins and rRNA inside the ribosome, uS5 has a surprisingly complex network of evolutionarily conserved non-ribosome-associated proteins.
uS5
dedicated chaperone
PDCD2
PDCD2L
1. Introduction
Despite the expanding number of roles played by noncoding RNAs, proteins remain key actors involved in nearly every operation required for cellular life, from proliferation to differentiation, internal organization, intercellular communication, and cell death. In order to set in motion the synthesis of new proteins, the information encoded by genes as messenger RNAs (mRNAs) is decoded into polymers of amino acids by a highly complex cellular machine, the ribosome, in a process known as translation. The ribosome is one of the most important ribonucleoprotein complexes in the cell, as demonstrated by its essential role in protein synthesis, its highly conserved nature, and its dominating abundance in most cell types. In fact, the fundamental structure and function of the ribosome were highly conserved throughout the evolution from bacteria to humans [1][2].
The 80S ribosome is a large RNA–protein complex with a molecular mass of 4.3 megadalton in humans [3] and is composed of two independent subunits: the 40S (or small) and 60S (or large) ribosomal subunits. The 40S ribosomal subunit consists of 33 different ribosomal proteins (RPs) and a single ribosomal RNA (rRNA), the 1869-nt-long 18S rRNA, whereas the 60S ribosomal subunit contains 47 RPs and three different RNA molecules: the 5S (121-nt), 5.8S (157-nt), and 28S (5070-nt) rRNAs [3]. While the 40S subunit contains the decoding center that monitors the complementarity of mRNA and tRNA during translation, the peptidyl-transferase center and the exit tunnel, in which the nascent polypeptide emerges out the ribosome, are at the heart of the 60S ribosomal subunit [4].
The synthesis of new ribosomes is one of the most energy demanding and complex processes occurring in eukaryotic cells. In addition to the four rRNAs and 80 RPs, ribosome biogenesis involves the coordinated action of the three cellular RNA polymerases, several hundred ribosome biogenesis factors (RBFs), as well as about 200 small nucleolar RNAs (snoRNAs) [5]. Ribosome synthesis begins in the nucleolus, where nascent rRNA is transcribed by RNA polymerase I (RNAPI) and assembled co-transcriptionally into a 90S pre-ribosomal particle via the spatio-temporal recruitment of several RPs, RBFs, and snoRNPs. Following endonucleolytic cleavage of the primary transcript between 18S and 5.8S rRNAs sequences, pre-40S and pre-60S particles will subsequently follow distinct maturation pathways. Whereas this endonucleolytic cleavage step mainly occurs co-transcriptionally in budding yeast [6], the extent to which this internal cleavage step is co-transcriptional in mammalian cells remains unclear. RNAPIII transcribes the fourth rRNA, the 5S rRNA, which joins the pre-60S particle in the nucleolus as part of the 5S RNP complex [7]. After transiting through the nucleoplasm, pre-40S and pre-60S particles are independently exported to the cytoplasm where they will be further processed to ultimately become competent for translation [5][7][8].
While the main role of the 80 RPs is to assist in the folding of the four rRNA molecules into a three-dimensional structure required for the precise interaction of mRNA codons with tRNA anticodons, the coordinated incorporation of RPs into their corresponding pre-ribosomal particle is critical for the stepwise assembly of mature ribosomal subunits. Specifically, functional studies in yeast and human cells show that deficiency of most RPs affects specific steps of pre-rRNA processing associated with pre-90S, pre-40S, and/or pre-60S maturation, which usually coincides with the timing of RP incorporation into pre-ribosomes [9][10][11][12]. Accordingly, most genes that code for RPs are essential for cellular proliferation and viability as well as for embryonic development in multi-cellular organisms [4].
2. Structural Features of Eukaryotic uS5 and Role in Translation
Human uS5 is a 293-amino-acids-long protein with a molecular mass of approximately 31 kDa that shows cytoplasmic expression in most tissues [13]. Analyses of actively translating ribosomes by cryo-electron microscopy (cryo-EM) [14] reveal that uS5 is located on the solvent-exposed side of the 40S ribosomal subunit (Figure 1A). Specifically, in the context of the mature 40S ribosomal subunit, uS5 is physically connected with ribosomal proteins eS21, uS2, uS8, uS4, and uS3. Residues of uS5 also interact with the 18S rRNA via a double-stranded RNA-binding-like domain (Figure 1B, magenta; PROSITE entry PS50881) and the conserved S5 C-terminal domain (Figure 1B, orange; PROSITE entry PS00585). Like many other ribosomal proteins, uS5 adopts a globular structure that is associated with disordered N- and C-terminal extensions [4][14] (Figure 1B). More precisely, the first 56 and last 14 amino acids of uS5 were not modeled from the cryo-EM analysis of active ribosomes [14] and show very low structural confidence scores as predicted by AlphaFold [15], consistent with disordered regions (Figure 1B). Notably, both N- and C-terminal extensions are unique to eukaryotic uS5 and absent in the E. coli homolog (Figure 1C). As shown in Figure 1D, the eukaryotic-specific N-terminal extension of uS5 is rich in arginine and glycine residues. Arginine-glycine (RG)-rich motifs have been associated with mediating interactions with RNA and protein as well as contributing to nuclear localization [16]. Interestingly, several arginine residues in the N-terminal RG-rich extension of uS5 are targeted by asymmetric dimethylation (see section on PRMT3), a uS5 post-translational modification that appears to be evolutionarily conserved [17][18][19]. Finally, human uS5 would include an unconventional nuclear localization signal (NLS), between lysine-159 and threonine-232, which would allow uS5 to be transported to the nucleus by various import receptors [20].
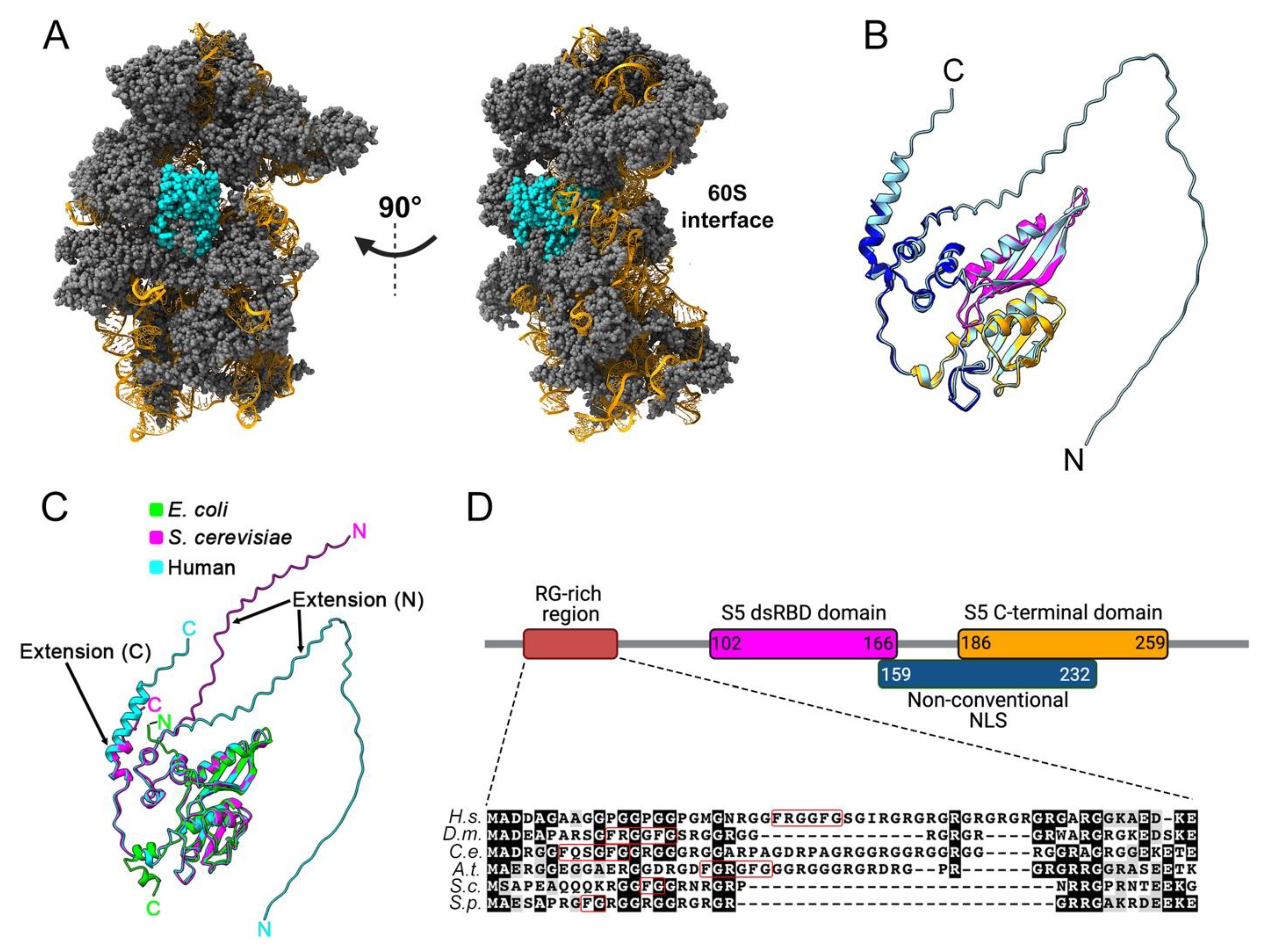
Figure 1. Structural characteristics of the 40S ribosomal protein uS5. (A) Cryo-EM structure of the actively translating 40 ribosomal subunit (PDB entry 5AJ0), left, and rotated 90 degrees, right. uS5 is shown in cyan, while the other 40S ribosomal proteins are colored in grey. The 18S rRNA is shown in orange. (B) Superposition of the tertiary structures of uS5 extracted from the active 40S ribosomal subunit (PDB entry 5AJ0; dark blue, magenta, and orange colors) and predicted by AlphaFold (pale blue). Note that the uS5 structure from the 40S subunit (PDB 5AJ0) represents only amino acids D57 to T278. The double-stranded RNA-binding-like domain and the conserved S5 C-terminal domain are shown in pink and orange, respectively, while the N- and C-terminal extensions are only seen in the AlphaFold model (pale blue). (C) Superposition of the AlphaFold tertiary structures of uS5 from E. coli (lime, P0A7W1), S. cerevisiae (magenta, P25443), and human (cyan, P15880) showing eukaryotic-specific N- and C-terminal extensions. (D) Motifs and functional domains of uS5 are shown. Numbers indicate the amino acid positions of each domain. Alignment and shading were generated using ClustalW and Boxshade software. Sequences are from Homo sapiens (H.s.), Drosophila melanogaster (D.m.), Caenorhabditis elegans (C.e.). Arabidopsis thaliana (A.t.), Saccharomyces cerevisiae (S.c.), and Schizosaccharomyces pombe (S.p.). The FXXXFG and FG motifs are boxed in red.
uS5 has been shown to be important for translation fidelity in E. coli, especially for a conserved glycine at position 28 [21][22][23][24]. E. coli uS5, together with uS3 and uS4, form part of the tunnel through which mRNA enters the small subunit of the ribosome to reach the interface between the two subunits [25]. While uS3 and uS4 act as RNA helicases, uS5 orients the incoming mRNA for proper codon reading in the ribosome A site [26]. In eukaryotes, based on cryo-EM structures of the yeast pre-initiation complex following AUG recognition [27], recent findings using Saccharomyces cerevisiae also support a role for eukaryotic uS5 in translation fidelity, especially at the level of translational initiation [28]. Accordingly, substitutions of uS5 residues identified as proximal to mRNA nucleotides 8 to 13 downstream of the AUG start codon [27] were shown to enhance translation initiation at suboptimal start codons [28], suggesting that uS5–mRNA contacts may contribute to the stability and thermodynamics of the eukaryotic preinitiation complex. Recent studies in mammals also support the role of uS5 in translation fidelity, as a substitution of alanine-226 for a tyrosine in uS5 leads to increased mistranslation in human cells [29] and muscle atrophy in mice [30].
3. uS5 Is an Essential Protein Required for 40s Ribosomal Subunit Production
Whereas most yeast RPs are encoded by duplicated paralogous genes, uS5 is one of the few RPs encoded by a single gene in both budding and fission yeast. uS5 is an essential gene in budding yeast as its deletion in S. cerevisiae yields inviable spores [31]. Accordingly, uS5 expression is required for ribosome biogenesis. A conditional mutant strain of uS5 in S. cerevisiae results in the accumulation of 20S rRNA precursors; yet, it also shows a reduction in newly made 20S pre-rRNA molecules in the cytoplasm, suggesting a role for uS5 in the export of pre-40S particles [10]. As for budding yeast, uS5 is also essential for cell viability in fission yeast, and knockdown of uS5 results in the complete inhibition of 40S ribosomal subunit production [32]. Notably, Schizosaccharomyces pombe cells depleted of uS5 showed only a small fraction of pre-rRNA matured into 20S precursors, suggesting that a large fraction of pre-40S is actively turned-over in the absence of uS5 [32]. In Drosophila, uS5 was identified as the allele associated with the “string of pearl (sop)” recessive female sterile mutants [33]. The sop allele is associated with reduced uS5 mRNA levels, oogenesis and early development defects, larval lethality, and a Minute-like phenotype [33]. The Minute syndrome in Drosophila—which includes delayed development, low fertility and viability, and decreased body size—is thought to arise as a consequence of suboptimal protein synthesis that results from reduced levels of cellular ribosomes [34]. In mammals, most of the knowledge about uS5 comes from studies performed on immortalized cell lines. Consistent with findings in yeast and Drosophila, uS5 is an essential gene in most tested human cancer cell lines [35], thereby making uS5 a potentially interesting target for cancer vulnerabilities [36]. Biochemical and structural data obtained from human cells indicate that uS5 is incorporated at late stages of pre-40S particle assembly prior to nuclear export [11][37]. Accordingly, knockdown of uS5 in human cell lines results in the accumulation of 21S pre-rRNA, suggesting the uncoupling of cleavage at sites A0–1 in the 5′ external transcribed spacer sequence [11][38], as well as increase detection of 18S-E precursors in the nucleus, consistent with delayed nuclear export of pre-40S particles [11].
Collectively, the current data support a conserved role for uS5 in the late stages of 40S ribosomal subunits assembly. Consistent with this conclusion, recent cryo-EM analyses of pre-40S intermediates isolated prior to nuclear export suggest that uS5 is incorporated into nucleoplasmic pre-40S particles [37]. Interestingly, although data generally support that the ribosome assembly process is largely conserved between yeast and human cells [7], recent results suggest that uS5 may incorporate pre-40S particles at different time points between yeast and humans [37]. Specifically, S. cerevisiae uS5 was detected in pre-40S particles before the incorporation of uS2 and eS21, whereas, in human cells, the timing of uS5 incorporation coincided with the insertion of uS2 and eS21, suggesting that uS2–uS5–eS21 are incorporated as a cluster in humans [37].
A Multifaceted Network of uS5-Associated Proteins
The identification of evolutionarily conserved uS5-associated proteins has been the focus of several studies in the past two decades. Such studies have provided new insights into the processes and mechanisms that promote uS5 expression and incorporation into ribosomes, as well as possible yet-to-be-defined extra-ribosomal functions.
4. PDCD2 and PDCD2L: uS5-Associated Paralogs That Take Part in Human Ribosome Assembly
During the process of establishing that the uS5–PRMT3 complex, which was originally identified in fission yeast [17], is conserved in humans, a set of novel and highly specific PRMT3 interactors were identified in addition to uS5, including strong enrichments of the PDCD2 and PDCD2-like (PDCD2L) proteins [39]. Biochemical assays further demonstrated that uS5 bridges the association between PRMT3 and PDCD2/PDCD2L, as depletion of uS5 totally prevented the copurification of PRMT3 and PDCD2/PDCD2L [39]. PDCD2 and PDCD2L are paralogous genes conserved through evolution, with homologs from bacteria to animals but not in archaebacteria. Based on sequence analysis, PDCD2 is thought to have arisen from the duplication of the PDCD2L gene prior to the divergence of animals, fungi, and plants from a common ancestor [40]. Homologs of human PDCD2 and PDCD2L paralogs are also found in mice (Pdcd2 and Pdcd2l), in Drosophila (Zfrp8 and Trus), and in fission yeast (Trs401 and Trs402); however, a single homolog is found in budding yeast (Tsr4). As shown in Figure 2A, PDCD2 and PDCD2L (34% identical; 52% similar) belong to a family of proteins containing N- and C-terminal TYPP (Tsr4, YwqG, PDCD2L, PDCD2) domains [40], each consisting of GGxP and Cx1-2C-like motifs as well as a highly conserved glutamine (Q) residue (see Figure 2A). In PDCD2, the N- and C-terminal TYPP motifs are interrupted by the insertion of a MYND-type zinc finger, which was shown to be involved in transcriptional repression via protein–protein interactions [41][42]. On the other hand, PDCD2L lacks the MYND zinc finger but contains a leucine-rich nuclear export sequence (NES) that enables PDCD2L to exit the nucleus in a CRM1-dependent manner [39] (Figure 2A). While the MYND domain is conserved in Drosophila Zfrp8 (Figure 2B), it is not found in the S. cerevisiae homolog of PDCD2 (Tsr4). The C-terminal TYPP domain also appears to be degenerated in yeast Tsr4 (Figure 2B, note lack of Cx1-2C motif), resulting in a predicted structure that is markedly different from other PDCD2 homologs (Figure 2C). The functional role of the TYPP domain has not been well studied, though it is thought to facilitate chaperoning activity and protein–protein interactions [40]. Indeed, substitutions that modify key residues conserved in the TYPP domain of PDCD2 completely abolish the association between PDCD2 and uS5 in human cells [38].
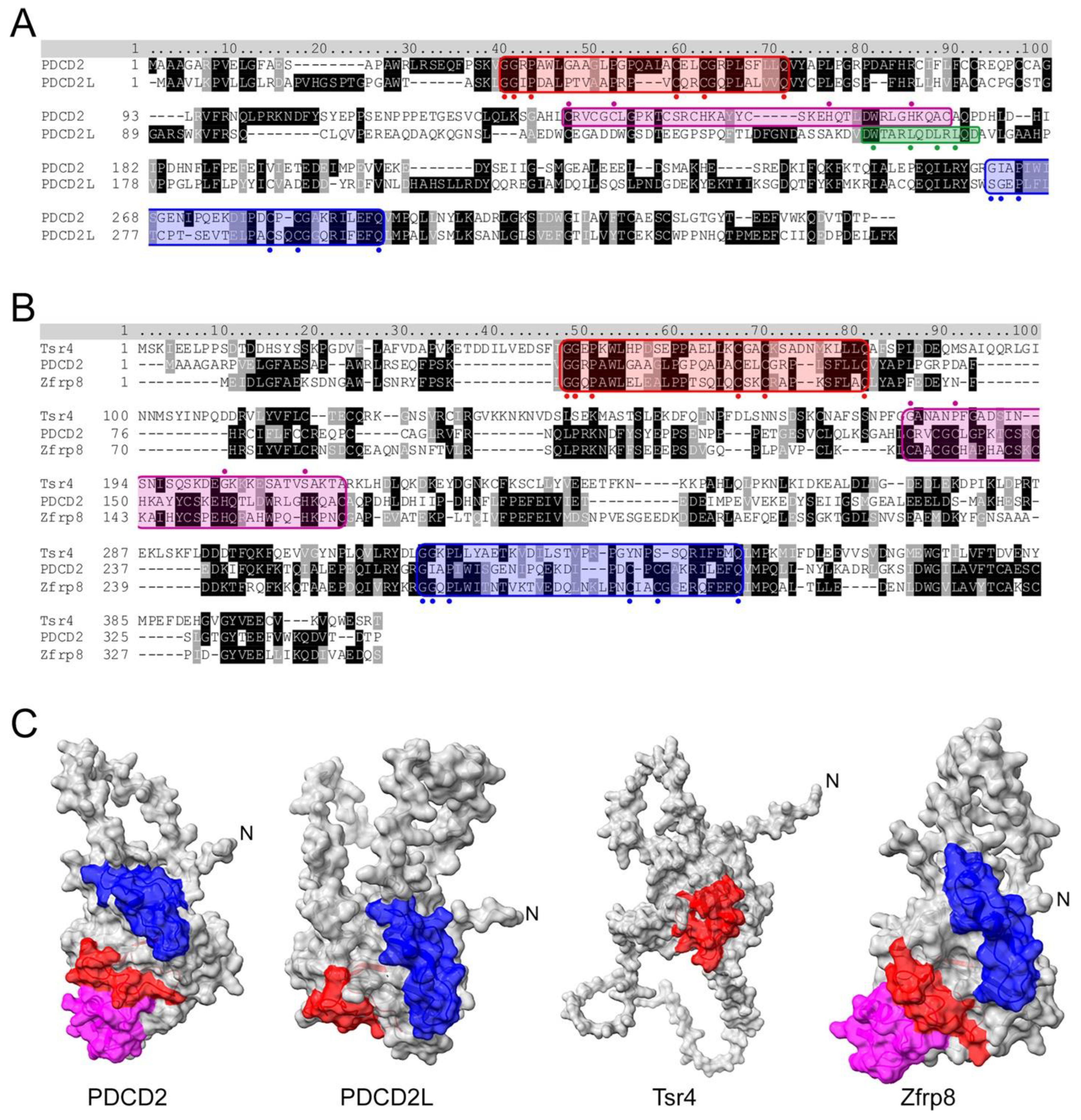
Figure 2. Sequence and structural analysis of PDCD2 and PDCD2L paralogs. (A) Amino acid sequence alignment of human PDCD2 and PDCD2L. Both proteins harbor N- and C-terminal TYPP domains (highlighted in red and blue, respectively) with conserved GxxP, Cx1-2C, and Q residues highlighted with circles. Whereas PDCD2 contains a MYND zinc finger domain (in magenta with critical cysteine and histidine residues indicated by circles marked above), PDCD2L harbors a leucine-rich NES consensus sequence (green), Φx2-3Φx2-3ΦxΦ, where Φ represents large hydrophobic residues (indicated by green circles marked underneath). (B) S. cerevisiae Tsr4 lacks the MYND zinc finger domain and its C-terminal TYPP domain is degenerated. (C) AlphaFold structures for human PDCD2 (Q16342), human PDCD2L (Q9BRP1), yeast Tsr4 (P25040), and Drosophila Zfrp8 (Q9W1A3). Red: N-terminal TYPP domain; Blue: C-terminal TYPP domain; Magenta: MYND domain.
5. PDCD2 Is a Conserved Dedicated Chaperone for uS5
The PDCD2 gene was originally identified in a screen for mRNAs upregulated upon apoptosis in rat cells [43]. However, subsequent experiments failed to support a correlation between PDCD2 mRNA expression and apoptosis [44][45]. Since then, PDCD2 has been associated with the pathogenesis of several disorders, including cancer [44][46][47][48][49][50][51][52], Parkinson’s disease [53], and fragile X syndrome [54]. Along with its potential role in diseases, PDCD2 is also implicated in development. In mice, PDCD2 is essential for stem cell viability and proliferation, and its absence leads to early embryonic lethality [55]. Although the aforementioned studies establish a clear role for PDCD2 in the development of human disorders as well as during embryonic development, the molecular function of PDCD2 had remained largely elusive until recently. Indeed, a set of elegant studies in budding yeast and human cell lines both support the conclusion that Tsr4/PDCD2 functions as an evolutionarily conserved chaperone dedicated for uS5 [38][56][57].
Previous work had already suggested the involvement of the yeast homolog of PDCD2/PDCD2L in ribosome biogenesis. Specifically, a screen for candidate genes involved in ribosome biogenesis identified TSR4 (Twenty S rRNA accumulation 4) as a gene required for 40S ribosomal subunit production [58]. A few years later, it was reported that Zfrp8 (Tsr4/PDCD2/PDCD2L homolog in Drosophila) depletion results in reduced cytoplasmic level of three RPs, including uS5 [59]. Consistent with these observations, PDCD2 copurifies with uS5 in both yeast and human cells and show binding via two-hybrid assays [38][56][57], suggesting a direct interaction between PDCD2 and uS5 that is evolutionarily conserved. Although the structure of the uS5–PDCD2 complex remains to be determined experimentally, researchers used AlphaFold-Multimer [60] to generate models of the human uS5–PDCD2 complex. Figure 3A shows the best confident relaxed structure with the highest predicted Local Distance Difference Test (pLDDT). Alternative predicted models showed highly similar pLDDT values, indicating uniformity among the predicted structures. Whereas the overall globular structure of uS5 remained largely unchanged in the context of the uS5–PDCD2 heterodimer relative to the uS5 monomer, residues 20–50 in the N-terminal disordered region of uS5 exhibited an increased confidence score and a considerably reduced predicted position error in the uS5–PDCD2 complex compared to the same region in the uS5 monomer (Figure 3B). In contrast, the C-terminal region of uS5 (aa 273–293) appears to be more disordered in the context of the uS5–PDCD2 complex relative to the uS5 monomer (Figure 3B). Interestingly, the disordered N-terminal extension of uS5 (see Figure 1B) is predicted to fold into a hydrophobic pocket located in the N-terminal half of human PDCD2 (Figure 3C). Notably, two phenylalanine residues of human uS5 (Phe25 and Phe29) are buried inside hydrophobic core regions of PDCD2 (Figure 3D). Consistent with this model, an FXXXFG motif can be found in the N-terminal extension of uS5 from humans, fruit flies, nematodes, and plants, whereas a single phenylalanine-glycine (FG motif) is found in uS5 from budding and fission yeasts (see Figure 1D). Although this remains a predicted model, the rearrangement of the uS5 unstructured N-terminal extension into a relatively stable structure in the context of the uS5–PDCD2 heterodimer is consistent with data in yeast showing that the first 42 amino acids of uS5 appear sufficient for interaction with S. cerevisiae Tsr4 [56][57]. The minimal PDCD2 interaction domain of uS5 in metazoans remains to be determined. The AlphaFold-Multimer prediction of the human uS5–PDCD2 complex also suggests the insertion of the uS5 dsRBD into a C-shaped opening formed by amino acids 204 to 239 of PDCD2 (Figure 3A,E), which is likely to stabilize the heterodimer.
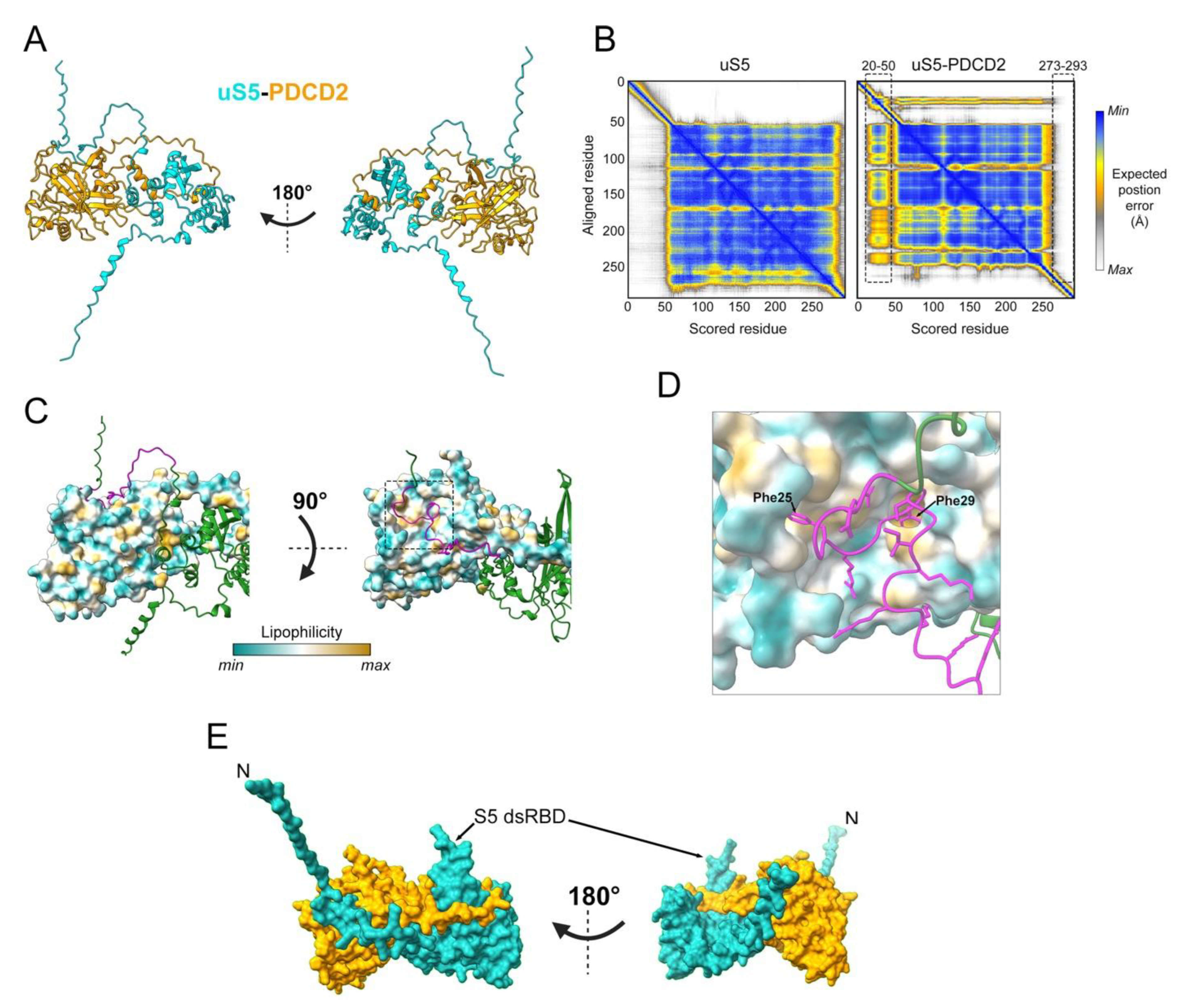
Figure 3. Predicted model of the human uS5–PDCD2 complex. (A) AlphaFold-Multimer prediction of the human uS5 (cyan)–PDCD2 (orange) complex, left, and rotated 180 degrees, right. (B) AlphaFold-predicted aligned error plot for the uS5 monomer (left) and uS5–PDCD2 complex (right), highlighting residues 20–50 of uS5 confidently predicted to interact with PDCD2 and residues 273–293 that show reduced predicted position error. (C) Surface representation of PDCD2 lipophilicity with ribbon-like structure of uS5 (green), left, and rotated 90 degrees, right. Residues 20–50 of uS5 are colored in magenta. (D) Phe25 and Phe29 residues of human uS5 are predicted to be embedded in hydrophobic core regions of PDCD2. (E) Surface representation of the uS5 (cyan)–PDCD2 (orange) complex, left, and rotated 180 degrees, right. A C-shaped region of PDCD2 (aa 204–239) wraps around the S5 dsRBD of uS5.
Studies in both yeast and human cells indicate that Tsr4/PDCD2 recognizes uS5 co-translationally and that Tsr4/PDCD2 is required for the accumulation of newly synthesized uS5 [38][56][57]. Consistent with the view that Tsr4/PDCD2 recognizes nascent uS5 is the fact that the N-terminal disordered region of uS5 is required for the formation of a stable Tsr4–uS5 complex in yeast [56][57], which is also suggested by the prediction of the human PDCD2–uS5 complex shown in Figure 3. The underlying mechanism of the specific co-translational recruitment remains unclear, however. It is possible that PDCD2/Tsr4 might have some degree of affinity for the uS5 mRNA, and thus, that the recruitment is initiated prior to uS5 translation initiation. The loss of function of PDCD2/Tsr4 phenocopies that of uS5 deficiency: reduced 40S production; 20S and 21S pre-rRNA accumulation in yeast and humans, respectively; and reduced incorporation of uS5 into pre-40S particles [38][56][57]. These findings, the co-translational binding of PDCD2 to nascent uS5, and the lack of identification of ribosome assembly factors in the interaction network PDCD2 support a conserved role of PDCD2 as a dedicated chaperone to uS5 [38][56][57].
References
- Anger, A.M.; Armache, J.P.; Berninghausen, O.; Habeck, M.; Subklewe, M.; Wilson, D.N.; Beckmann, R. Structures of the human and Drosophila 80S ribosome. Nature 2013, 497, 80–85.
- Petrov, A.S.; Bernier, C.R.; Hsiao, C.; Norris, A.M.; Kovacs, N.A.; Waterbury, C.C.; Stepanov, V.G.; Harvey, S.C.; Fox, G.E.; Wartell, R.M.; et al. Evolution of the ribosome at atomic resolution. Proc. Natl. Acad. Sci. USA 2014, 111, 10251–10256.
- Khatter, H.; Myasnikov, A.G.; Natchiar, S.K.; Klaholz, B.P. Structure of the human 80S ribosome. Nature 2015, 520, 640–645.
- Melnikov, S.; Manakongtreecheep, K.; Soll, D. Revising the Structural Diversity of Ribosomal Proteins Across the Three Domains of Life. Mol. Biol. Evol. 2018, 35, 1588–1598.
- Pelletier, J.; Thomas, G.; Volarevic, S. Ribosome biogenesis in cancer: New players and therapeutic avenues. Nat. Rev. Cancer 2018, 18, 51–63.
- Kos, M.; Tollervey, D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell 2010, 37, 809–820.
- Dorner, K.; Ruggeri, C.; Zemp, I.; Kutay, U. Ribosome biogenesis factors-from names to functions. EMBO J. 2023, 42, e112699.
- Henras, A.K.; Plisson-Chastang, C.; O’Donohue, M.F.; Chakraborty, A.; Gleizes, P.E. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA 2015, 6, 225–242.
- Ferreira-Cerca, S.; Poll, G.; Gleizes, P.E.; Tschochner, H.; Milkereit, P. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol. Cell 2005, 20, 263–275.
- Ferreira-Cerca, S.; Poll, G.; Kuhn, H.; Neueder, A.; Jakob, S.; Tschochner, H.; Milkereit, P. Analysis of the in vivo assembly pathway of eukaryotic 40S ribosomal proteins. Mol. Cell 2007, 28, 446–457.
- O’Donohue, M.F.; Choesmel, V.; Faubladier, M.; Fichant, G.; Gleizes, P.E. Functional dichotomy of ribosomal proteins during the synthesis of mammalian 40S ribosomal subunits. J. Cell Biol. 2010, 190, 853–866.
- Poll, G.; Braun, T.; Jakovljevic, J.; Neueder, A.; Jakob, S.; Woolford, J.L., Jr.; Tschochner, H.; Milkereit, P. rRNA maturation in yeast cells depleted of large ribosomal subunit proteins. PLoS ONE 2009, 4, e8249.
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419.
- Behrmann, E.; Loerke, J.; Budkevich, T.V.; Yamamoto, K.; Schmidt, A.; Penczek, P.A.; Vos, M.R.; Burger, J.; Mielke, T.; Scheerer, P.; et al. Structural snapshots of actively translating human ribosomes. Cell 2015, 161, 845–857.
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589.
- Chowdhury, M.N.; Jin, H. The RGG motif proteins: Interactions, functions, and regulations. Wiley Interdiscip. Rev. RNA 2023, 14, e1748.
- Bachand, F.; Silver, P.A. PRMT3 is a ribosomal protein methyltransferase that affects the cellular levels of ribosomal subunits. EMBO J. 2004, 23, 2641–2650.
- Lipson, R.S.; Webb, K.J.; Clarke, S.G. Rmt1 catalyzes zinc-finger independent arginine methylation of ribosomal protein Rps2 in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2010, 391, 1658–1662.
- Swiercz, R.; Person, M.D.; Bedford, M.T. Ribosomal protein S2 is a substrate for mammalian PRMT3 (protein arginine methyltransferase 3). Biochem. J. 2005, 386, 85–91.
- Antoine, M.; Reimers, K.; Wirz, W.; Gressner, A.M.; Muller, R.; Kiefer, P. Identification of an unconventional nuclear localization signal in human ribosomal protein S2. Biochem. Biophys. Res. Commun. 2005, 335, 146–153.
- Agarwal, D.; Kamath, D.; Gregory, S.T.; O’Connor, M. Modulation of decoding fidelity by ribosomal proteins S4 and S5. J. Bacteriol. 2015, 197, 1017–1025.
- Piepersberg, W.; Bock, A.; Wittmann, H.G. Effect of different mutations in ribosomal protein S5 of Escherichia coli on translational fidelity. Mol. Gen. Genet. 1975, 140, 91–100.
- Piepersberg, W.; Bock, A.; Yaguchi, M.; Wittmann, H.G. Genetic position and amino acid replacements of several mutations in ribosomal protein S5 from Escherichia coli. Mol. Gen. Genet. 1975, 143, 43–52.
- Rosset, R.; Gorini, L. A ribosomal ambiguity mutation. J. Mol. Biol. 1969, 39, 95–112.
- Culver, G.M. Meanderings of the mRNA through the ribosome. Structure 2001, 9, 751–758.
- Kurkcuoglu, O.; Doruker, P.; Sen, T.Z.; Kloczkowski, A.; Jernigan, R.L. The ribosome structure controls and directs mRNA entry, translocation and exit dynamics. Phys. Biol. 2008, 5, 046005.
- Llacer, J.L.; Hussain, T.; Saini, A.K.; Nanda, J.S.; Kaur, S.; Gordiyenko, Y.; Kumar, R.; Hinnebusch, A.G.; Lorsch, J.R.; Ramakrishnan, V. Translational initiation factor eIF5 replaces eIF1 on the 40S ribosomal subunit to promote start-codon recognition. eLife 2018, 7, e39273.
- Dong, J.; Hinnebusch, A.G. uS5/Rps2 residues at the 40S ribosome entry channel enhance initiation at suboptimal start codons in vivo. Genetics 2022, 220, iyab176.
- Shcherbakov, D.; Teo, Y.; Boukari, H.; Cortes-Sanchon, A.; Mantovani, M.; Osinnii, I.; Moore, J.; Juskeviciene, R.; Brilkova, M.; Duscha, S.; et al. Ribosomal mistranslation leads to silencing of the unfolded protein response and increased mitochondrial biogenesis. Commun. Biol. 2019, 2, 381.
- Moore, J.; Akbergenov, R.; Nigri, M.; Isnard-Petit, P.; Grimm, A.; Seebeck, P.; Restelli, L.; Frank, S.; Eckert, A.; Thiam, K.; et al. Random errors in protein synthesis activate an age-dependent program of muscle atrophy in mice. Commun. Biol. 2021, 4, 703.
- Steffen, K.K.; McCormick, M.A.; Pham, K.M.; MacKay, V.L.; Delaney, J.R.; Murakami, C.J.; Kaeberlein, M.; Kennedy, B.K. Ribosome deficiency protects against ER stress in Saccharomyces cerevisiae. Genetics 2012, 191, 107–118.
- Perreault, A.; Bellemer, C.; Bachand, F. Nuclear export competence of pre-40S subunits in fission yeast requires the ribosomal protein Rps2. Nucleic Acids Res. 2008, 36, 6132–6142.
- Cramton, S.E.; Laski, F.A. String of pearls encodes Drosophila ribosomal protein S2, has Minute-like characteristics, and is required during oogenesis. Genetics 1994, 137, 1039–1048.
- Marygold, S.J.; Roote, J.; Reuter, G.; Lambertsson, A.; Ashburner, M.; Millburn, G.H.; Harrison, P.M.; Yu, Z.; Kenmochi, N.; Kaufman, T.C.; et al. The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol. 2007, 8, R216.
- Tsherniak, A.; Vazquez, F.; Montgomery, P.G.; Weir, B.A.; Kryukov, G.; Cowley, G.S.; Gill, S.; Harrington, W.F.; Pantel, S.; Krill-Burger, J.M.; et al. Defining a Cancer Dependency Map. Cell 2017, 170, 564–576.e516.
- Wang, M.; Hu, Y.; Stearns, M.E. RPS2: A novel therapeutic target in prostate cancer. J. Exp. Clin. Cancer Res. CR 2009, 28, 6.
- Cheng, J.; Lau, B.; Thoms, M.; Ameismeier, M.; Berninghausen, O.; Hurt, E.; Beckmann, R. The nucleoplasmic phase of pre-40S formation prior to nuclear export. Nucleic Acids Res. 2022, 50, 11924–11937.
- Landry-Voyer, A.M.; Bergeron, D.; Yague-Sanz, C.; Baker, B.; Bachand, F. PDCD2 functions as an evolutionarily conserved chaperone dedicated for the 40S ribosomal protein uS5 (RPS2). Nucleic Acids Res. 2020, 48, 12900–12916.
- Landry-Voyer, A.M.; Bilodeau, S.; Bergeron, D.; Dionne, K.L.; Port, S.A.; Rouleau, C.; Boisvert, F.M.; Kehlenbach, R.H.; Bachand, F. Human PDCD2L Is an Export Substrate of CRM1 That Associates with 40S Ribosomal Subunit Precursors. Mol. Cell. Biol. 2016, 36, 3019–3032.
- Burroughs, A.M.; Aravind, L. Analysis of two domains with novel RNA-processing activities throws light on the complex evolution of ribosomal RNA biogenesis. Front. Genet. 2014, 5, 424.
- Lutterbach, B.; Sun, D.; Schuetz, J.; Hiebert, S.W. The MYND motif is required for repression of basal transcription from the multidrug resistance 1 promoter by the t(8;21) fusion protein. Mol. Cell. Biol. 1998, 18, 3604–3611.
- Melnick, A.M.; Westendorf, J.J.; Polinger, A.; Carlile, G.W.; Arai, S.; Ball, H.J.; Lutterbach, B.; Hiebert, S.W.; Licht, J.D. The ETO protein disrupted in t(8;21)-associated acute myeloid leukemia is a corepressor for the promyelocytic leukemia zinc finger protein. Mol. Cell. Biol. 2000, 20, 2075–2086.
- Owens, G.P.; Hahn, W.E.; Cohen, J.J. Identification of mRNAs associated with programmed cell death in immature thymocytes. Mol. Cell. Biol. 1991, 11, 4177–4188.
- Fan, C.W.; Chan, C.C.; Chao, C.C.; Fan, H.A.; Sheu, D.L.; Chan, E.C. Expression patterns of cell cycle and apoptosis-related genes in a multidrug-resistant human colon carcinoma cell line. Scand. J. Gastroenterol. 2004, 39, 464–469.
- Kawakami, T.; Furukawa, Y.; Sudo, K.; Saito, H.; Takami, S.; Takahashi, E.; Nakamura, Y. Isolation and mapping of a human gene (PDCD2) that is highly homologous to Rp8, a rat gene associated with programmed cell death. Cytogenet. Cell Genet. 1995, 71, 41–43.
- Baron, B.W.; Zeleznik-Le, N.; Baron, M.J.; Theisler, C.; Huo, D.; Krasowski, M.D.; Thirman, M.J.; Baron, R.M.; Baron, J.M. Repression of the PDCD2 gene by BCL6 and the implications for the pathogenesis of human B and T cell lymphomas. Proc. Natl. Acad. Sci. USA 2007, 104, 7449–7454.
- Kusam, S.; Munugalavadla, V.; Sawant, D.; Dent, A. BCL6 cooperates with CD40 stimulation and loss of p53 function to rapidly transform primary B cells. Int. J. Cancer J. Int. Cancer 2009, 125, 977–981.
- Liu, H.; Wang, M.; Liang, N.; Guan, L. PDCD2 sensitizes HepG2 cells to sorafenib by suppressing epithelial-mesenchymal transition. Mol. Med. Rep. 2019, 19, 2173–2179.
- Steinemann, D.; Gesk, S.; Zhang, Y.; Harder, L.; Pilarsky, C.; Hinzmann, B.; Martin-Subero, J.I.; Calasanz, M.J.; Mungall, A.; Rosenthal, A.; et al. Identification of candidate tumor-suppressor genes in 6q27 by combined deletion mapping and electronic expression profiling in lymphoid neoplasms. Genes Chromosomes Cancer 2003, 37, 421–426.
- Wang, W.; Song, X.W.; Bu, X.M.; Zhang, N.; Zhao, C.H. PDCD2 and NCoR1 as putative tumor suppressors in gastric gastrointestinal stromal tumors. Cell. Oncol. 2016, 39, 129–137.
- Yang, X.; Lee, Y.; Fan, H.; Sun, X.; Lussier, Y.A. Identification of common microRNA-mRNA regulatory biomodules in human epithelial cancers. Chin. Sci. Bull. 2010, 55, 3576–3589.
- Yang, Y.; Jin, Y.; Du, W. Programmed cell death 2 functions as a tumor suppressor in osteosarcoma. Int. J. Clin. Exp. Pathol. 2015, 8, 10894–10900.
- Fukae, J.; Sato, S.; Shiba, K.; Sato, K.; Mori, H.; Sharp, P.A.; Mizuno, Y.; Hattori, N. Programmed cell death-2 isoform1 is ubiquitinated by parkin and increased in the substantia nigra of patients with autosomal recessive Parkinson’s disease. FEBS Lett. 2009, 583, 521–525.
- Tan, W.; Schauder, C.; Naryshkina, T.; Minakhina, S.; Steward, R. Zfrp8 forms a complex with fragile-X mental retardation protein and regulates its localization and function. Dev. Biol. 2016, 410, 202–212.
- Mu, W.; Munroe, R.J.; Barker, A.K.; Schimenti, J.C. PDCD2 is essential for inner cell mass development and embryonic stem cell maintenance. Dev. Biol. 2010, 347, 279–288.
- Black, J.J.; Musalgaonkar, S.; Johnson, A.W. Tsr4 Is a Cytoplasmic Chaperone for the Ribosomal Protein Rps2 in Saccharomyces cerevisiae. Mol. Cell. Biol. 2019, 39, e00019–e00094.
- Rossler, I.; Embacher, J.; Pillet, B.; Murat, G.; Liesinger, L.; Hafner, J.; Unterluggauer, J.J.; Birner-Gruenberger, R.; Kressler, D.; Pertschy, B. Tsr4 and Nap1, two novel members of the ribosomal protein chaperOME. Nucleic Acids Res. 2019, 47, 6984–7002.
- Li, Z.; Lee, I.; Moradi, E.; Hung, N.J.; Johnson, A.W.; Marcotte, E.M. Rational extension of the ribosome biogenesis pathway using network-guided genetics. PLoS Biol. 2009, 7, e1000213.
- Minakhina, S.; Naryshkina, T.; Changela, N.; Tan, W.; Steward, R. Zfrp8/PDCD2 Interacts with RpS2 Connecting Ribosome Maturation and Gene-Specific Translation. PLoS ONE 2016, 11, e0147631.
- Evans, R.; O’Neill, M.; Pritzel, A.; Antropova, N.; Senior, A.; Green, T.; Žídek, A.; Bates, R.; Blackwell, S.; Yim, J.; et al. Protein complex prediction with AlphaFold-Multimer. BioRxiv 2022.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
575
Revisions:
2 times
(View History)
Update Date:
24 May 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




