
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Bindu Subhadra | -- | 3872 | 2023-05-20 04:32:18 | | | |
| 2 | Jason Zhu | Meta information modification | 3872 | 2023-05-22 03:47:14 | | |
Video Upload Options
Biofilm formation in healthcare is an issue of considerable concern, as it results in increased morbidity and mortality, imposing a significant financial burden on the healthcare system. Biofilms are highly resistant to conventional antimicrobial therapies and lead to persistent infections. Hence, there is a high demand for novel strategies other than conventional antibiotic therapies to control biofilm-based infections. There are two approaches which have been employed so far to control biofilm formation in healthcare settings: one is the development of biofilm inhibitors based on the understanding of the molecular mechanism of biofilm formation, and the other is to modify the biomaterials which are used in medical devices to prevent biofilm formation.
1. Introduction
2. Inhibition of Initial Attachment
2.1. By Altering the Chemical Properties of Biomaterials
2.2. By Changing the Physical Properties of Biomaterials
3. Biofilm Removal
3.1. Matrix-Degrading Enzymes
3.2. Surfactants
3.3. Free Fatty Acids, Amino Acids and Nitric Oxide Donors
4. Biofilm Inhibition by Quorum Quenching
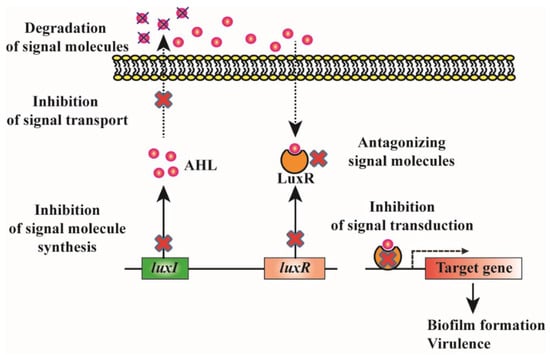
4.1. Degradation of QS Signals
4.2. Inhibition of Signal Synthesis
4.3. Antagonizing the Signal Molecules
4.4. Inhibition of Signal Transduction by Interfering with Response Regulator Activity
4.5. Inhibition of Signal Transport
References
- Kalia, V.C.; Purohit, H.J. Quenching the quorum sensing system: Potential antibacterial drug targets. Crit. Rev. Microbiol. 2011, 37, 121–140.
- Wright, J.S., III; Lyon, G.J.; George, E.A.; Muir, T.W.; Novick, R.P. Hydrophobic interactions drive ligand-receptor recognition for activation and inhibition of staphylococcal quorum sensing. Proc. Natl. Acad. Sci. USA 2004, 101, 16168–16173.
- Conrady, D.G.; Brescia, C.C.; Horii, K.; Weiss, A.A.; Hassett, D.J.; Herr, A.B. A zinc-dependent adhesion module is responsible for intercellular adhesion in staphylococcal biofilms. Proc. Natl. Acad. Sci. USA 2008, 105, 19456–19461.
- Maira-Litrán, T.; Kropec, A.; Abeygunawardana, C.; Joyce, J.; Mark, G., 3rd; Goldmann, D.A.; Pier, G.B. Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect. Immun. 2002, 70, 4433–4440.
- Mittelman, M.W. Adhesion to biomaterials. In Bacterial Adhesion: Molecular and Ecological Diversity; Fletcher, M., Ed.; Wiley-Liss, Inc.: New York, NY, USA, 1996.
- Characklis, W.G.; McFeters, G.A.; Marshall, K.C. Physiological ecology in biofilm systems. In Biofilms; Characklis, W.G., Marshall, K.C., Eds.; John Wiley & Sons: New York, NY, USA, 1990.
- Fletcher, M.; Loeb, G.I. Influence of substratum characteristics on the attachment of a marine pseudomonad to solid surfaces. Appl. Environ. Microbiol. 1979, 37, 67–72.
- Dror, N.; Mandel, M.; Hazan, Z.; Lavie, G. Advances in microbial biofilm prevention on indwelling medical devices with emphasis on usage of acoustic energy. Sensors 2009, 9, 2538–2554.
- Ramos, E.R.; Reitzel, R.; Jiang, Y.; Hachem, R.Y.; Chaftari, A.M.; Chemaly, R.F.; Hackett, B.; Pravinkumar, S.E.; Nates, J.; Tarrand, J.J.; et al. Clinical effectiveness and risk of emerging resistance associated with prolonged use of antibiotic-impregnated catheters: More than 0.5 million catheter days and 7 years of clinical experience. Crit. Care Med. 2011, 39, 245–251.
- Schumm, K.; Lam, T.B. Types of urethral catheters for management of short-term voiding problems in hospitalised adults. Cochrane Database Syst. Rev. 2008, CD004013.
- Sun, H.; Xu, Y.; Sitkiewicz, I.; Ma, Y.; Wang, X.; Yestrepsky, B.D.; Huang, Y.; Lapadatescu, M.C.; Larsen, M.J.; Larsen, S.D.; et al. Inhibitor of streptokinase gene expression improves survival after group A Streptococcus infection in mice. Proc. Natl. Acad. Sci. USA 2012, 109, 3469–3474.
- Ma, Y.; Xu, Y.; Yestrepsky, B.D.; Sorenson, R.J.; Chen, M.; Larsen, S.D.; Sun, H. Novel inhibitors of Staphylococcus aureus virulence gene expression and biofilm formation. PLoS ONE 2012, 7, e47255.
- Opperman, T.J.; Kwasny, S.M.; Williams, J.D.; Khan, A.R.; Peet, N.P.; Moir, D.T.; Bowlin, T.L. Aryl rhodanines specifically inhibit staphylococcal and enterococcal biofilm formation. Antimicrob. Agents Chemother. 2009, 53, 4357–4367.
- Jenal, U.; Dorman, C.J. Small molecule signaling. Curr. Opin. Microbiol. 2009, 12, 125–128.
- Römling, U.; Balsalobre, C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J. Intern. Med. 2012, 272, 541–561.
- Pérez-Giraldo, C.; Rodríguez-Benito, A.; Morán, F.J.; Hurtado, C.; Blanco, M.T.; Gómez-García, A.C. Influence of N-acetylcysteine on the formation of biofilm by Staphylococcus epidermidis. J. Antimicrob. Chemother. 1997, 39, 643–646.
- De la Fuente-Núñez, C.; Reffuveille, F.; Haney, E.F.; Straus, S.K.; Hancock, R.E. Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathog. 2014, 10, e1004152.
- Parisot, J.; Carey, S.; Breukink, E.; Chan, W.C.; Narbad, A.; Bonev, B. Molecular mechanism of target recognition by subtilin, a class I lanthionine antibiotic. Antimicrob. Agents Chemother. 2008, 52, 612–618.
- Saising, J.; Dube, L.; Ziebandt, A.K.; Voravuthikunchai, S.P.; Nega, M.; Götz, F. Activity of gallidermin on Staphylococcus aureus and Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 2012, 56, 5804–5810.
- Abraham, N.M.; Lamlertthon, S.; Fowler, V.G.; Jefferson, K.K. Chelating agents exert distinct effects on biofilm formation in Staphylococcus aureus depending on strain background: Role for clumping factor B. J. Med. Microbiol. 2012, 61, 1062–1070.
- Chernousova, S.; Epple, M. Silver as antibacterial agent: Ion, nanoparticle, and metal. Angew. Chem. Int. Ed. 2013, 52, 1636–1653.
- Besinis, A.; Hadi, S.D.; Le, H.R.; Tredwin, C.; Handy, R.D. Antibacterial activity and biofilm inhibition by surface modified titanium alloy medical implants following application of silver, titanium dioxide and hydroxyapatite nanocoatings. Nanotoxicology 2017, 11, 327–338.
- Feng, Q.L.; Wu, J.; Chen, G.Q.; Cui, F.Z.; Kim, T.N.; Kim, J.O. A mechanistic study of the antibacterial effect of silver ions on Escherichia coli and Staphylococcus aureus. J. Biomed. Mater. Res. 2000, 52, 662–668.
- Yamanaka, M.; Hara, K.; Kudo, J. Bactericidal actions of a silver ion solution on Escherichia coli, studied by energy-filtering transmission electron microscopy and proteomic analysis. Appl. Environ. Microbiol. 2005, 71, 7589–7593.
- Klasen, H.J. A historical review of the use of silver in the treatment of burns. II. Renewed interest for silver. Burns 2000, 26, 131–138.
- Secinti, K.D.; Özalp, H.; Attar, A.; Sargon, M.F. Nanoparticle silver ion coatings inhibit biofilm formation on titanium implants. J. Clin. Neurosci. 2011, 18, 391–395.
- Shahverdi, A.R.; Fakhimi, A.; Shahverdi, H.R.; Minaian, S. Synthesis and effect of silver nanoparticles on the antibacterial activity of different antibiotics against Staphylococcus aureus and Escherichia coli. Nanomedicine 2007, 3, 168–171.
- Jansen, B.; Kohnen, W. Prevention of biofilm formation by polymer modification. J. Ind. Microbiol. 1995, 15, 391–396.
- Marlow, V.L.; Porter, M.; Hobley, L.; Kiley, T.B.; Swedlow, J.R.; Davidson, F.A.; Stanley-Wall, N.R. Phosphorylated DegU manipulates cell fate differentiation in the Bacillus subtilis biofilm. J. Bacteriol. 2014, 196, 16–27.
- Cassinelli, C.; Morra, M.; Pavesio, A.; Renier, D. Evaluation of interfacial properties of hyaluronan coated poly(methylmethacrylate) intraocular lenses. J. Biomater. Sci. Polym. Ed. 2000, 11, 961–977.
- Boelens, J.J.; Tan, W.F.; Dankert, J.; Zaat, S.A. Antibacterial activity of antibiotic-soaked polyvinylpyrrolidone-grafted silicon elastomer hydrocephalus shunts. J. Antimicrob. Chemother. 2000, 45, 221–224.
- John, T.; Rajpurkar, A.; Smith, G.; Fairfax, M.; Triest, J. Antibiotic pretreatment of hydrogel ureteral stent. J. Endourol. 2007, 21, 1211–1216.
- Falde, E.J.; Yohe, S.T.; Colson, Y.L.; Grinstaff, M.W. Superhydrophobic materials for biomedical applications. Biomaterials 2016, 104, 87–103.
- Liu, T.; Yin, B.; He, T.; Guo, N.; Dong, L.; Yin, Y. Complementary effects of nanosilver and superhydrophobic coatings on the prevention of marine bacterial adhesion. ACS Appl. Mater. Interfaces 2012, 4, 4683–4690.
- Crick, C.R.; Ismail, S.; Pratten, J.; Parkin, I.P. An investigation into bacterial attachment to an elastomeric superhydrophobic surface prepared via aerosol assisted deposition. Thin Solid Films 2011, 519, 3722–3727.
- Tang, P.; Zhang, W.; Wang, Y.; Zhang, B.; Wang, H.; Lin, C.; Zhang, L. Effect of superhydrophobic surface of titanium on Staphylococcus aureus adhesion. J. Nanomater. 2011, 2011, 2.
- Privett, B.J.; Youn, J.; Hong, S.A.; Lee, J.; Han, J.; Shin, J.H.; Schoenfisch, M.H. Antibacterial fluorinated silica colloid superhydrophobic surfaces. Langmuir 2011, 27, 9597–9601.
- Appelgren, P.; Ransjö, U.; Bindslev, L.; Espersen, F.; Larm, O. Surface heparinization of central venous catheters reduces microbial colonization in vitro and in vivo: Results from a prospective, randomized trial. Crit. Care Med. 1996, 24, 1482–1489.
- Russell, P.B.; Kline, J.; Yoder, M.C.; Polin, R.A. Staphylococcal adherence to polyvinyl chloride and heparin-bonded polyurethane catheters is species dependent and enhanced by fibronectin. J. Clin. Microbiol. 1987, 25, 1083–1087.
- Abdelkefi, A.; Achour, W.; Ben Othman, T.; Ladeb, S.; Torjman, L.; Lakhal, A.; Ben Hassen, A.; Hsairi, M.; Ben Abdeladhim, A. Use of heparin-coated central venous lines to prevent catheter-related bloodstream infection. J. Support. Oncol. 2007, 5, 273–278.
- Meiron, T.S.; Saguy, I.S. Adhesion modeling on rough low linear density polyethylene. J. Food Sci. 2007, 72, E485–E491.
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193.
- Ito, A.; Taniuchi, A.; May, T.; Kawata, K.; Okabe, S. Increased antibiotic resistance of Escherichia coli in mature biofilms. Appl. Environ. Microbiol. 2009, 75, 4093–4100.
- Savage, V.J.; Chopra, I.; O’Neill, A.J. Staphylococcus aureus biofilms promote horizontal transfer of antibiotic resistance. Antimicrob. Agents Chemother. 2013, 57, 1968–1970.
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633.
- Mann, E.E.; Wozniak, D.J. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol. Rev. 2012, 36, 893–916.
- Whitchurch, C.B.; Tolker-Nielsen, T.; Ragas, P.C.; Mattick, J.S. Extracellular DNA required for bacterial biofilm formation. Science 2002, 295, 1487.
- Mulcahy, H.; Charron-Mazenod, L.; Lewenza, S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2008, 4, e1000213.
- Mah, T.F.C.; O’Toole, G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001, 9, 34–39.
- Kaplan, J.B. Therapeutic potential of biofilm-dispersing enzymes. Int. J. Artif. Organs 2009, 32, 545–554.
- Li, X.H.; Lee, J.H. Antibiofilm agents: A new perspective for antimicrobial strategy. J. Microbiol. 2017, 55, 753–766.
- Ramasubbu, N.; Thomas, L.M.; Ragunath, C.; Kaplan, J.B. Structural analysis of dispersin B, a biofilm-releasing glycoside hydrolase from the periodontopathogen Actinobacillus actinomycetemcomitans. J. Mol. Biol. 2005, 349, 475–486.
- Darouiche, R.O.; Mansouri, M.D.; Gawande, P.V.; Madhyastha, S. Antimicrobial and antibiofilm efficacy of triclosan and dispersinb combination. J. Antimicrob. Chemother. 2009, 64, 88–93.
- Shen, Y.; Köller, T.; Kreikemeyer, B.; Nelson, D.C. Rapid degradation of Streptococcus pyogenes biofilms by PlyC, a bacteriophage-encoded endolysin. J. Antimicrob. Chemother. 2013, 68, 1818–1824.
- Kaplan, J.B.; LoVetri, K.; Cardona, S.T.; Madhyastha, S.; Sadovskaya, I.; Jabbouri, S.; Izano, E.A. Recombinant human DNase I decreases biofilm and increases antimicrobial susceptibility in Staphylococci. J. Antibiot. 2012, 65, 73–77.
- Tetz, G.V.; Artemenko, N.K.; Tetz, V.V. Effect of DNase and antibiotics on biofilm characteristics. Antimicrob. Agents Chemother. 2009, 53, 1204–1209.
- Chaignon, P.; Sadovskaya, I.; Ragunah, C.; Ramasubbu, N.; Kaplan, J.B.; Jabbouri, S. Susceptibility of staphylococcal biofilms to enzymatic treatments depends on their chemical composition. Appl. Microbiol. Biotechnol. 2007, 75, 125–132.
- Chen, M.; Yu, Q.; Sun, H. Novel strategies for the prevention and treatment of biofilm related infections. Int. J. Mol. Sci. 2013, 14, 18488–18501.
- Van Hamme, J.D.; Singh, A.; Ward, O.P. Physiological aspects. Part 1 in a series of papers devoted to surfactants in microbiology and biotechnology. Biotechnol. Adv. 2006, 24, 604–620.
- Simões, M.; Pereira, M.O.; Vieira, M.J. Action of a cationic surfactant on the activity and removal of bacterial biofilms formed under different flow regimes. Water Res. 2005, 39, 478–486.
- Boles, B.R.; Thoendel, M.; Singh, P.K. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol. Microbiol. 2005, 57, 1210–1223.
- Chen, X.; Stewart, P.S. Biofilm removal caused by chemical treatments. Water Res. 2000, 34, 4229–4233.
- Mireles, J.R., 2nd; Toguchi, A.; Harshey, R.M. Salmonella enterica serovar typhimurium swarming mutants with altered biofilm-forming abilities: Surfactin inhibits biofilm formation. J. Bacteriol. 2001, 183, 5848–5854.
- Abdel-Mawgoud, A.M.; Lépine, F.; Déziel, E. Rhamnolipids: Diversity of structures, microbial origins and roles. Appl. Microbiol. Biotechnol. 2010, 86, 1323–1336.
- Davies, D.G.; Marques, C.N. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 2009, 191, 1393–1403.
- Deng, Y.; Lim, A.; Lee, J.; Chen, S.; An, S.; Dong, Y.H.; Zhang, L.H. Diffusible signal factor (DSF) quorum sensing signal and structurally related molecules enhance the antimicrobial efficacy of antibiotics against some bacterial pathogens. BMC Microbiol. 2014, 14, 51.
- Pedrido, M.E.; de Oña, P.; Ramirez, W.; Leñini, C.; Goñi, A.; Grau, R. Spo0a links de novo fatty acid synthesis to sporulation and biofilm development in Bacillus subtilis. Mol. Microbiol. 2013, 87, 348–367.
- Kolodkin-Gal, I.; Romero, D.; Cao, S.; Clardy, J.; Kolter, R.; Losick, R. d-amino acids trigger biofilm disassembly. Science 2010, 328, 627–629.
- Jermy, A. Biofilms: Disassembly instructions included. Nat. Rev. Microbiol. 2012, 10, 376.
- Bernier, S.P.; Ha, D.G.; Khan, W.; Merritt, J.H.; O’Toole, G.A. Modulation of Pseudomonas aeruginosa surface-associated group behaviors by individual amino acids through c-di-GMP signaling. Res. Microbiol. 2011, 162, 680–688.
- Brandenburg, K.S.; Rodriguez, K.J.; McAnulty, J.F.; Murphy, C.J.; Abbott, N.L.; Schurr, M.J.; Czuprynski, C.J. Tryptophan inhibits biofilm formation by Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2013, 57, 1921–1925.
- Barraud, N.; Schleheck, D.; Klebensberger, J.; Webb, J.S.; Hassett, D.J.; Rice, S.A.; Kjelleberg, S. Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J. Bacteriol. 2009, 191, 7333–7342.
- Howlin, R.P.; Cathie, K.; Hall-Stoodley, L.; Cornelius, V.; Duignan, C.; Allan, R.N.; Fernandez, B.O.; Barraud, N.; Bruce, K.D.; Jefferies, J.; et al. Low-dose nitric oxide as targeted anti-biofilm adjunctive therapy to treat chronic Pseudomonas aeruginosa infection in cystic fibrosis. Mol. Ther. 2017, 25, 2104–2116.
- Socransky, S.S.; Haffajee, A.D. Dental biofilms: Difficult therapeutic targets. Periodontol 2000, 28, 12–55.
- Frias, J.; Olle, E.; Alsina, M. Periodontal pathogens produce quorum sensing signal molecules. Infect. Immun. 2001, 69, 3431–3434.
- Eberhard, A.; Burlingame, A.L.; Eberhard, C.; Kenyon, G.L.; Nealson, K.H.; Oppenheimer, N.J. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry 1981, 20, 2444–2449.
- Waters, C.M.; Bassler, B.L. Quorum sensing: Cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346.
- Brözel, V.S.; Strydom, G.M.; Cloete, T.E. A method for the study of de novo protein synthesis in Pseudomonas aeruginosa after attachment. Biofouling 1995, 8, 195–210.
- Davies, D.G.; Chakrabarty, A.M.; Geesey, G.G. Exopolysaccharide production in biofilms: Substratum activation of alginate gene expression by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 1993, 59, 1181–1186.
- Sauer, K.; Camper, A.K. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J. Bacteriol. 2001, 183, 6579–6589.
- Parsek, M.R.; Greenberg, E.P. Sociomicrobiology: The connections between quorum sensing and biofilms. Trends Microbiol. 2005, 13, 27–33.
- Brackman, G.; Coenye, T. Quorum sensing inhibitors as anti-biofilm agents. Curr. Pharm. Des. 2015, 21, 5–11.
- Hentzer, M.; Riedel, K.; Rasmussen, T.B.; Heydorn, A.; Andersen, J.B.; Parsek, M.R.; Rice, S.A.; Eberl, L.; Molin, S.; Høiby, N.; et al. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 2002, 148, 87–102.
- Hentzer, M.; Wu, H.; Andersen, J.B.; Riedel, K.; Rasmussen, T.B.; Bagge, N.; Kumar, N.; Schembri, M.A.; Song, Z.; Kristoffersen, P.; et al. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 2003, 22, 3803–3815.
- Uroz, S.; Chhabra, S.R.; Cámara, M.; Williams, P.; Oger, P.; Dessaux, Y. N-acylhomoserine lactone quorum-sensing molecules are modified and degraded by Rhodococcus erythropolis W2 by both amidolytic and novel oxidoreductase activities. Microbiology 2005, 151, 3313–3322.
- Tang, K.; Zhang, X.H. Quorum quenching agents: Resources for antivirulence therapy. Mar. Drugs 2014, 12, 3245–3282.
- Yang, F.; Wang, L.H.; Wang, J.; Dong, Y.H.; Hu, J.Y.; Zhang, L.H. Quorum quenching enzyme activity is widely conserved in the sera of mammalian species. FEBS Lett. 2005, 579, 3713–3717.
- Uroz, S.; Heinonsalo, J. Degradation of N-acyl homoserine lactone quorum sensing signal molecules by forest root-associated fungi. FEMS Microbiol. Ecol. 2008, 65, 271–278.
- dos Reis Ponce, A.; Martins, M.L.; de Araujo, E.F.; Mantovani, H.C.; Vanetti, M.C. Aiia quorum-sensing quenching controls proteolytic activity and biofilm formation by Enterobacter cloacae. Curr. Microbiol. 2012, 65, 758–763.
- Ivanova, K.; Fernandes, M.M.; Francesko, A.; Mendoza, E.; Guezguez, J.; Burnet, M.; Tzanov, T. Quorum-quenching and matrix-degrading enzymes in multilayer coatings synergistically prevent bacterial biofilm formation on urinary catheters. ACS Appl. Mater. Interfaces 2015, 7, 27066–27077.
- Jo, S.J.; Kwon, H.; Jeong, S.Y.; Lee, S.H.; Oh, H.S.; Yi, T.; Lee, C.H.; Kim, T.G. Effects of quorum quenching on the microbial community of biofilm in an anoxic/oxic mbr for wastewater treatment. J. Microbiol. Biotechnol. 2016, 26, 1593–1604.
- Lee, J.; Lee, I.; Nam, J.; Hwang, D.S.; Yeon, K.M.; Kim, J. Immobilization and stabilization of acylase on carboxylated polyaniline nanofibers for highly effective antifouling application via quorum quenching. ACS Appl. Mater. Interfaces 2017, 9, 15424–15432.
- Vinoj, G.; Vaseeharan, B.; Thomas, S.; Spiers, A.J.; Shanthi, S. Quorum-quenching activity of the AHL-lactonase from Bacillus licheniformis DAHB1 inhibits vibrio biofilm formation in vitro and reduces shrimp intestinal colonisation and mortality. Mar. Biotechnol. 2014, 16, 707–715.
- Weiland-Bräuer, N.; Kisch, M.J.; Pinnow, N.; Liese, A.; Schmitz, R.A. Highly effective inhibition of biofilm formation by the first metagenome-derived AI-2 quenching enzyme. Front. Microbiol. 2016, 7, 1098.
- Kiran, S.; Sharma, P.; Harjai, K.; Capalash, N. Enzymatic quorum quenching increases antibiotic susceptibility of multidrug resistant Pseudomonas aeruginosa. Iran. J. Microbiol. 2011, 3, 1–12.
- Bjarnsholt, T.; Tolker-Nielsen, T.; Høiby, N.; Givskov, M. Interference of Pseudomonas aeruginosa signalling and biofilm formation for infection control. Expert Rev. Mol. Med. 2010, 12, e11.
- Tomlin, K.L.; Malott, R.J.; Ramage, G.; Storey, D.G.; Sokol, P.A.; Ceri, H. Quorum-sensing mutations affect attachment and stability of Burkholderia cenocepacia biofilms. Appl. Environ. Microbiol. 2005, 71, 5208–5218.
- Huber, B.; Riedel, K.; Hentzer, M.; Heydorn, A.; Gotschlich, A.; Givskov, M.; Molin, S.; Eberl, L. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 2001, 147, 2517–2528.
- Lynch, M.J.; Swift, S.; Kirke, D.F.; Keevil, C.W.; Dodd, C.E.; Williams, P. The regulation of biofilm development by quorum sensing in Aeromonas hydrophila. Environ. Microbiol. 2002, 4, 18–28.
- Labbate, M.; Queck, S.Y.; Koh, K.S.; Rice, S.A.; Givskov, M.; Kjelleberg, S. Quorum sensing-controlled biofilm development in Serratia liquefaciens MG1. J. Bacteriol. 2004, 186, 692–698.
- Parsek, M.R.; Val, D.L.; Hanzelka, B.L.; Cronan, J.E., Jr.; Greenberg, E.P. Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl. Acad. Sci. USA 1999, 96, 4360–4365.
- Hanzelka, B.L.; Greenberg, E.P. Quorum sensing in Vibrio fischeri: Evidence that S-adenosylmethionine is the amino acid substrate for autoinducer synthesis. J. Bacteriol. 1996, 178, 5291–5294.
- Favre-Bonté, S.; Köhler, T.; Van Delden, C. Biofilm formation by Pseudomonas aeruginosa: Role of the C4-HSL cell-to-cell signal and inhibition by azithromycin. J. Antimicrob. Chemother. 2003, 52, 598–604.
- Hoffmann, N.; Lee, B.; Hentzer, M.; Rasmussen, T.B.; Song, Z.; Johansen, H.K.; Givskov, M.; Høiby, N. Azithromycin blocks quorum sensing and alginate polymer formation and increases the sensitivity to serum and stationary-growth-phase killing of Pseudomonas aeruginosa and attenuates chronic P. aeruginosa lung infection in Cftr−/− mice. Antimicrob. Agents Chemother. 2007, 51, 3677–3687.
- Shen, G.; Rajan, R.; Zhu, J.; Bell, C.E.; Pei, D. Design and synthesis of substrate and intermediate analogue inhibitors of S-ribosylhomocysteinase. J. Med. Chem. 2006, 49, 3003–3011.
- Gutierrez, J.A.; Crowder, T.; Rinaldo-Matthis, A.; Ho, M.C.; Almo, S.C.; Schramm, V.L. Transition state analogs of 5′-methylthioadenosine nucleosidase disrupt quorum sensing. Nat. Chem. Biol. 2009, 5, 251–257.
- Vega, L.M.; Mathieu, J.; Yang, Y.; Pyle, B.H.; McLean, R.J.C.; Alvarez, P.J.J. Nickel and cadmium ions inhibit quorum sensing and biofilm formation without affecting viability in Burkholderia multivorans. Int. Biodeterior. Biodegrad. 2014, 91, 82–87.
- Thornhill, S.G.; Kumar, M.; Vega, L.M.; McLean, R.J.C. Cadmium ion inhibition of quorum signalling in Chromobacterium violaceum. Microbiology 2017, 163, 1429–1435.
- Smith, K.M.; Bu, Y.; Suga, H. Induction and inhibition of Pseudomonas aeruginosa quorum sensing by synthetic autoinducer analogs. Chem. Biol. 2003, 10, 81–89.
- Smith, K.M.; Bu, Y.; Suga, H. Library screening for synthetic agonists and antagonists of a Pseudomonas aeruginosa autoinducer. Chem. Biol. 2003, 10, 563–571.
- Kim, C.; Kim, J.; Park, H.Y.; Park, H.J.; Kim, C.K.; Yoon, J.; Lee, J.H. Development of inhibitors against TraR quorum-sensing system in Agrobacterium tumefaciens by molecular modeling of the ligand-receptor interaction. Mol. Cells 2009, 28, 447–453.
- Ishida, T.; Ikeda, T.; Takiguchi, N.; Kuroda, A.; Ohtake, H.; Kato, J. Inhibition of quorum sensing in Pseudomonas aeruginosa by N-acyl cyclopentylamides. Appl. Environ. Microbiol. 2007, 73, 3183–3188.
- Morohoshi, T.; Shiono, T.; Takidouchi, K.; Kato, M.; Kato, N.; Kato, J.; Ikeda, T. Inhibition of quorum sensing in Serratia marcescens AS-1 by synthetic analogs of N-acylhomoserine lactone. Appl. Environ. Microbiol. 2007, 73, 6339–6344.
- Kim, C.; Kim, J.; Park, H.Y.; Park, H.J.; Lee, J.H.; Kim, C.K.; Yoon, J. Furanone derivatives as quorum-sensing antagonists of Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 2008, 80, 37–47.
- Rasmussen, T.B.; Skindersoe, M.E.; Bjarnsholt, T.; Phipps, R.K.; Christensen, K.B.; Jensen, P.O.; Andersen, J.B.; Koch, B.; Larsen, T.O.; Hentzer, M.; et al. Identity and effects of quorum-sensing inhibitors produced by Penicillium species. Microbiology 2005, 151, 1325–1340.
- Galloway, W.R.; Hodgkinson, J.T.; Bowden, S.D.; Welch, M.; Spring, D.R. Quorum sensing in Gram-negative bacteria: Small-molecule modulation of AHL and AI-2 quorum sensing pathways. Chem. Rev. 2011, 111, 28–67.
- Bjarnsholt, T.; Jensen, P.Ø.; Rasmussen, T.B.; Christophersen, L.; Calum, H.; Hentzer, M.; Hougen, H.P.; Rygaard, J.; Moser, C.; Eberl, L.; et al. Garlic blocks quorum sensing and promotes rapid clearing of pulmonary Pseudomonas aeruginosa infections. Microbiology 2005, 151, 3873–3880.
- Rasmussen, T.B.; Bjarnsholt, T.; Skindersoe, M.E.; Hentzer, M.; Kristoffersen, P.; Köte, M.; Nielsen, J.; Eberl, L.; Givskov, M. Screening for quorum-sensing inhibitors (QSI) by use of a novel genetic system, the QSI selector. J. Bacteriol. 2005, 187, 1799–1814.
- Jakobsen, T.H.; van Gennip, M.; Phipps, R.K.; Shanmugham, M.S.; Christensen, L.D.; Alhede, M.; Skindersoe, M.E.; Rasmussen, T.B.; Friedrich, K.; Uthe, F.; et al. Ajoene, a sulfur rich molecule from garlic, inhibits genes controlled by quorum sensing. Antimicrob. Agents Chemother. 2012, 56, 2314–2325.
- Brackman, G.; Cos, P.; Maes, L.; Nelis, H.J.; Coenye, T. Quorum sensing inhibitors increase the susceptibility of bacterial biofilms to antibiotics in vitro and in vivo. Antimicrob. Agents Chemother. 2011, 55, 2655–2661.
- Brackman, G.; Hillaert, U.; Van Calenbergh, S.; Nelis, H.J.; Coenye, T. Use of quorum sensing inhibitors to interfere with biofilm formation and development in Burkholderia multivorans and Burkholderia cenocepacia. Res. Microbiol. 2009, 160, 144–151.
- Huber, B.; Eberl, L.; Feucht, W.; Polster, J. Influence of polyphenols on bacterial biofilm formation and quorum-sensing. Z. Naturforsch. C 2003, 58, 879–884.
- Fong, J.; Zhang, C.; Yang, R.; Boo, Z.Z.; Tan, S.K.; Nielsen, T.E.; Givskov, M.; Wu, B.; Su, H.; Yang, L. Synergy of quorum quenching enzyme and quorum sensing inhibitor in inhibiting P. aeruginosa quorum sensing. bioRxiv 2017, 182543.
- Ren, D.; Zuo, R.; González Barrios, A.F.; Bedzyk, L.A.; Eldridge, G.R.; Pasmore, M.E.; Wood, T.K. Differential gene expression for investigation of Escherichia coli biofilm inhibition by plant extract ursolic acid. Appl. Environ. Microbiol. 2005, 71, 4022–4034.
- Roy, V.; Meyer, M.T.; Smith, J.A.; Gamby, S.; Sintim, H.O.; Ghodssi, R.; Bentley, W.E. AI-2 analogs and antibiotics: A synergistic approach to reduce bacterial biofilms. Appl. Microbiol. Biotechnol. 2013, 97, 2627–2638.
- Ni, N.T.; Choudhary, G.; Li, M.Y.; Wang, B.H. Pyrogallol and its analogs can antagonize bacterial quorum sensing in Vibrio harveyi. Bioorg. Med. Chem. Lett. 2008, 18, 1567–1572.
- Brackman, G.; Celen, S.; Baruah, K.; Bossier, P.; Van Calenbergh, S.; Nelis, H.J.; Coenye, T. AI-2 quorum-sensing inhibitors affect the starvation response and reduce virulence in several Vibrio species, most likely by interfering with LuxPQ. Microbiology 2009, 155, 4114–4122.
- George, E.A.; Novick, R.P.; Muir, T.W. Cyclic peptide inhibitors of staphylococcal virulence prepared by Fmoc-based thiolactone peptide synthesis. J. Am. Chem. Soc. 2008, 130, 4914–4924.
- Li, J.; Wang, W.; Xu, S.X.; Magarvey, N.A.; McCormick, J.K. Lactobacillus reuteri-produced cyclic dipeptides quench agr-mediated expression of toxic shock syndrome toxin-1 in Staphylococci. Proc. Natl. Acad. Sci. USA 2011, 108, 3360–3365.
- Scott, R.J.; Lian, L.Y.; Muharram, S.H.; Cockayne, A.; Wood, S.J.; Bycroft, B.W.; Williams, P.; Chan, W.C. Side-chain-to-tail thiolactone peptide inhibitors of the staphylococcal quorum-sensing system. Bioorg. Med. Chem. Lett. 2003, 13, 2449–2453.
- Gov, Y.; Bitler, A.; Dell’Acqua, G.; Torres, J.V.; Balaban, N. RNAIII inhibiting peptide (RIP), a global inhibitor of Staphylococcus aureus pathogenesis: Structure and function analysis. Peptides 2001, 22, 1609–1620.
- Cirioni, O.; Mocchegiani, F.; Cacciatore, I.; Vecchiet, J.; Silvestri, C.; Baldassarre, L.; Ucciferri, C.; Orsetti, E.; Castelli, P.; Provinciali, M.; et al. Quorum sensing inhibitor FS3-coated vascular graft enhances daptomycin efficacy in a rat model of staphylococcal infection. Peptides 2013, 40, 77–81.
- Kiran, M.D.; Adikesavan, N.V.; Cirioni, O.; Giacometti, A.; Silvestri, C.; Scalise, G.; Ghiselli, R.; Saba, V.; Orlando, F.; Shoham, M.; et al. Discovery of a quorum-sensing inhibitor of drug-resistant staphylococcal infections by structure-based virtual screening. Mol. Pharmacol. 2008, 73, 1578–1586.
- El-Naggar, M.H.; Elgaml, A.; Abdel Bar, F.M.; Badria, F.A. Antimicrobial and antiquorum-sensing activity of Ricinus communis extracts and ricinine derivatives. Nat. Prod. Res. 2018, 32, 1–7.
- Pejin, B.; Ciric, A.; Dimitric Markovic, J.; Glamoclija, J.; Nikolic, M.; Sokovic, M. An insight into anti-biofilm and anti-quorum sensing activities of the selected anthocyanidins: The case study of Pseudomonas aeruginosa PAO1. Nat. Prod. Res. 2017, 31, 1177–1180.
- Pejin, B.; Ciric, A.; Glamoclija, J.; Nikolic, M.; Sokovic, M. In vitro anti-quorum sensing activity of phytol. Nat. Prod. Res. 2015, 29, 374–377.
- Pejin, B.; Talevska, A.; Ciric, A.; Glamoclija, J.; Nikolic, M.; Talevski, T.; Sokovic, M. Anti-quorum sensing activity of selected sponge extracts: A case study of Pseudomonas aeruginosa. Nat. Prod. Res. 2014, 28, 2330–2333.
- Pejin, B.; Ciric, A.; Karaman, I.; Horvatovic, M.; Glamoclija, J.; Nikolic, M.; Sokovic, M. In vitro antibiofilm activity of the freshwater bryozoan Hyalinella punctata: A case study of Pseudomonas aeruginosa PAO1. Nat. Prod. Res. 2016, 30, 1847–1850.
- Ren, D.; Sims, J.J.; Wood, T.K. Inhibition of biofilm formation and swarming of Bacillus subtilis by (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone. Lett. Appl. Microbiol. 2002, 34, 293–299.
- Hume, E.B.H.; Baveja, J.; Muir, B.W.; Schubert, T.L.; Kumar, N.; Kjelleberg, S.; Griesser, H.J.; Thissen, H.; Read, R.; Poole-Warren, L.A.; et al. The control of Staphylococcus epidermidis biofilm formation and in vivo infection rates by covalently bound furanones. Biomaterials 2004, 25, 5023–5030.
- He, Z.Y.; Wang, Q.; Hu, Y.J.; Liang, J.P.; Jiang, Y.T.; Ma, R.; Tang, Z.S.; Huang, Z.W. Use of the quorum sensing inhibitor furanone C-30 to interfere with biofilm formation by Streptococcus mutans and its luxS mutant strain. Int. J. Antimicrob. Agents 2012, 40, 30–35.
- Janssens, J.C.; Steenackers, H.; Robijns, S.; Gellens, E.; Levin, J.; Zhao, H.; Hermans, K.; De Coster, D.; Verhoeven, T.L.; Marchal, K.; et al. Brominated furanones inhibit biofilm formation by Salmonella enterica serovar typhimurium. Appl. Environ. Microbiol. 2008, 74, 6639–6648.
- Niu, C.; Afre, S.; Gilbert, E.S. Subinhibitory concentrations of cinnamaldehyde interfere with quorum sensing. Lett. Appl. Microbiol. 2006, 43, 489–494.
- Defoirdt, T.; Miyamoto, C.M.; Wood, T.K.; Meighen, E.A.; Sorgeloos, P.; Verstraete, W.; Bossier, P. The natural furanone (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone disrupts quorum sensing-regulated gene expression in Vibrio harveyi by decreasing the DNA-binding activity of the transcriptional regulator protein luxR. Environ. Microbiol. 2007, 9, 2486–2495.
- Brackman, G.; Defoirdt, T.; Miyamoto, C.; Bossier, P.; Van Calenbergh, S.; Nelis, H.; Coenye, T. Cinnamaldehyde and cinnamaldehyde derivatives reduce virulence in Vibrio spp. by decreasing the DNA-binding activity of the quorum sensing response regulator LuxR. BMC Microbiol. 2008, 8, 149.
- Manefield, M.; de Nys, R.; Kumar, N.; Read, R.; Givskov, M.; Steinberg, P.; Kjelleberg, S. Evidence that halogenated furanones from Delisea pulchra inhibit acylated homoserine lactone (AHL)-mediated gene expression by displacing the AHL signal from its receptor protein. Microbiology 1999, 145, 283–291.
- Kuehl, R.; Al-Bataineh, S.; Gordon, O.; Luginbuehl, R.; Otto, M.; Textor, M.; Landmann, R. Furanone at subinhibitory concentrations enhances staphylococcal biofilm formation by luxS repression. Antimicrob. Agents Chemother. 2009, 53, 4159–4166.
- Manefield, M.; Rasmussen, T.B.; Henzter, M.; Andersen, J.B.; Steinberg, P.; Kjelleberg, S.; Givskov, M. Halogenated furanones inhibit quorum sensing through accelerated LuxR turnover. Microbiology 2002, 148, 1119–1127.
- Han, Y.; Hou, S.; Simon, K.A.; Ren, D.; Luk, Y.Y. Identifying the important structural elements of brominated furanones for inhibiting biofilm formation by Escherichia coli. Bioorg. Med. Chem. Lett. 2008, 18, 1006–1010.
- Oh, M.H.; Choi, C.H. Role of LuxIR homologue AnoIR in Acinetobacter nosocomialis and the effect of virstatin on the expression of anoR gene. J. Microbiol. Biotechnol. 2015, 25, 1390–1400.
- Zhang, L.; Murphy, P.J.; Kerr, A.; Tate, M.E. Agrobacterium conjugation and gene regulation by N-acyl-l-homoserine lactones. Nature 1993, 362, 446–448.
- Park, S.Y.; Lee, S.J.; Oh, T.K.; Oh, J.W.; Koo, B.T.; Yum, D.Y.; Lee, J.K. AhlD, an N-acylhomoserine lactonase in Arthrobacter sp., and predicted homologues in other bacteria. Microbiology 2003, 149, 1541–1550.
- Pearson, J.P.; Van Delden, C.; Iglewski, B.H. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol. 1999, 181, 1203–1210.
- Evans, K.; Passador, L.; Srikumar, R.; Tsang, E.; Nezezon, J.; Poole, K. Influence of the MexAB-OprM multidrug efflux system on quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 1998, 180, 5443–5447.
- Rahmati, S.; Yang, S.; Davidson, A.L.; Zechiedrich, E.L. Control of the AcrAB multidrug efflux pump by quorum-sensing regulator SdiA. Mol. Microbiol. 2002, 43, 677–685.
- Pumbwe, L.; Skilbeck, C.A.; Wexler, H.M. Presence of quorum-sensing systems associated with multidrug resistance and biofilm formation in Bacteroides fragilis. Microb. Ecol. 2008, 56, 412–419.
- Chan, Y.Y.; Chua, K.L. The Burkholderia pseudomallei BpeAB-OprB efflux pump: Expression and impact on quorum sensing and virulence. J. Bacteriol. 2005, 187, 4707–4719.
- Chan, Y.Y.; Bian, H.S.; Tan, T.M.; Mattmann, M.E.; Geske, G.D.; Igarashi, J.; Hatano, T.; Suga, H.; Blackwell, H.E.; Chua, K.L. Control of quorum sensing by a Burkholderia pseudomallei multidrug efflux pump. J. Bacteriol. 2007, 189, 4320–4324.
- Kvist, M.; Hancock, V.; Klemm, P. Inactivation of efflux pumps abolishes bacterial biofilm formation. Appl. Environ. Microbiol. 2008, 74, 7376–7382.
- Baugh, S.; Ekanayaka, A.S.; Piddock, L.J.; Webber, M.A. Loss of or inhibition of all multidrug resistance efflux pumps of Salmonella enterica serovar typhimurium results in impaired ability to form a biofilm. J. Antimicrob. Chemother. 2012, 67, 2409–2417.
- Baugh, S.; Phillips, C.R.; Ekanayaka, A.S.; Piddock, L.J.; Webber, M.A. Inhibition of multidrug efflux as a strategy to prevent biofilm formation. J. Antimicrob. Chemother. 2014, 69, 673–681.
- Christena, L.R.; Mangalagowri, V.; Pradheeba, P.; Ahmed, K.B.; Shalini, B.I.; Vidyalakshmi, M.; Anbazhagan, V.; Sai subramanian, N. Copper nanoparticles as an efflux pump inhibitor to tackle drug resistant bacteria. RSC Adv. 2015, 5, 12899–12909.
- Nallathamby, P.D.; Lee, K.J.; Desai, T.; Xu, X.H. Study of the multidrug membrane transporter of single living Pseudomonas aeruginosa cells using size-dependent plasmonic nanoparticle optical probes. Biochemistry 2010, 49, 5942–5953.
- Rampioni, G.; Pillai, C.R.; Longo, F.; Bondì, R.; Baldelli, V.; Messina, M.; Imperi, F.; Visca, P.; Leoni, L. Effect of efflux pump inhibition on Pseudomonas aeruginosa transcriptome and virulence. Sci. Rep. 2017, 7, 11392.
- Gupta, D.; Singh, A.; Khan, A.U. Nanoparticles as efflux pump and biofilm inhibitor to rejuvenate bactericidal effect of conventional antibiotics. Nanoscale Res. Lett. 2017, 12, 454.
- Maseda, H.; Sawada, I.; Saito, K.; Uchiyama, H.; Nakae, T.; Nomura, N. Enhancement of the mexAB-oprM efflux pump expression by a quorum-sensing autoinducer and its cancellation by a regulator, MexT, of the mexEF-oprN efflux pump operon in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2004, 48, 1320–1328.




