You're using an outdated browser. Please upgrade to a modern browser for the best experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Piao Yang | -- | 2070 | 2023-05-18 15:08:04 | | | |
| 2 | Conner Chen | Meta information modification | 2070 | 2023-05-22 04:39:57 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Yang, P.; Zhao, L.; Gao, Y.G.; Xia, Y. Pseudomonas syringae as a Bacterial Plant Pathogen. Encyclopedia. Available online: https://encyclopedia.pub/entry/44511 (accessed on 26 December 2025).
Yang P, Zhao L, Gao YG, Xia Y. Pseudomonas syringae as a Bacterial Plant Pathogen. Encyclopedia. Available at: https://encyclopedia.pub/entry/44511. Accessed December 26, 2025.
Yang, Piao, Lijing Zhao, Yu Gary Gao, Ye Xia. "Pseudomonas syringae as a Bacterial Plant Pathogen" Encyclopedia, https://encyclopedia.pub/entry/44511 (accessed December 26, 2025).
Yang, P., Zhao, L., Gao, Y.G., & Xia, Y. (2023, May 18). Pseudomonas syringae as a Bacterial Plant Pathogen. In Encyclopedia. https://encyclopedia.pub/entry/44511
Yang, Piao, et al. "Pseudomonas syringae as a Bacterial Plant Pathogen." Encyclopedia. Web. 18 May, 2023.
Copy Citation
Plant diseases caused by the pathogen Pseudomonas syringae are serious problems for various plant species worldwide. Accurate detection and diagnosis of P. syringae infections are critical for the effective management of these plant diseases.
Pseudomonas syringae
pathogen detection
pathogen diagnosis
plant disease triangle
plant fitness tetrahedron
plant disease management hexagon
1. Introduction
Plant pathogen detection recognizes the presence of plant pathogens in a specific location or area. The process involves observing visible disease symptoms in plants, collecting plant samples for further laboratory analysis, or using remote sensing techniques to detect the presence of pathogens [1][2]. Plant pathogen detection aims to identify the presence of plant pathogens as early as possible so control measures can be implemented to alleviate their impacts on crop production [3]. On the other hand, plant pathogen diagnosis refers to identifying the specific cause of a plant disease. It involves the identification of specific disease-causing pathogens by biochemical, molecular, and other techniques [4]. Plant pathogen diagnosis aims to find the specific pathogen responsible for the specific disease so proper control measures can be implemented to limit the further spread of the pathogen and disease. Plant pathogen detection and diagnosis are critical to understanding and managing plant diseases. They are associated with applying multiple techniques and approaches to identify and understand the presence and cause of plant diseases [5][6].
Plant diseases are a significant constraint to crop production worldwide and exert particularly severe impacts in developing countries, where agricultural systems may be less resilient than in developed ones [7][8][9][10]. Although bacteria evolved billions of years ago [11], they had not been demonstrated to cause plant diseases until the late 19th century [12]. Bacterial plant diseases can reduce crop yields and debase the quality of harvested crops, thus leading to significant quality and economic losses for farmers and agricultural industries [13][14][15]. Studying bacterial plant pathogens helps identify the ways to detect, diagnose, prevent, and control those destructive plant diseases, such as using resistant crop varieties, applying chemicals or biological control agents, and implementing good agricultural practices [9][16]. By further understanding the biology and epidemiology of bacterial plant diseases, researchers can upgrade their strategies to reduce the impacts of these diseases on crop production and improve global food security. Alongside developing control measures, it is also essential to study bacterial plant diseases to understand the factors contributing to their emergence and spread [17]. These efforts can involve the identification of the genetic and environmental factors that influence plant disease development and the roles that different plant hosts, vectors, and reservoirs play in the transmission of bacterial plant pathogens.
Rapid detection and correct diagnosis of bacterial plant pathogens and diseases are increasingly essential for protecting global food security. By detecting and diagnosing these pathogens and diseases early, it is possible to implement control measures such as the application of chemicals or biological control agents or the implementation of other agricultural practices to reduce the impacts of these diseases on crop production [18]. Furthermore, bacterial plant pathogens can sometimes result in the contamination of human food with harmful pathogens [19]. By detecting and diagnosing these pathogens early, it is possible to implement the control measures to prevent food contamination and improve food safety [20]. Bacterial plant diseases can also sometimes lead to the extinction of plant species, particularly rare or endangered species [21]. By noticing these pathogens earlier, it is helpful to develop strategies to protect cultures or production directly.
There are challenges in detecting and diagnosing bacterial plant pathogens, such as P. syringae [22]. One challenge is the need for rapid and correct diagnosis of bacterial plant pathogens. However, traditional methods, such as biochemical or molecular techniques, can be time-consuming and may not provide rapid results [23]. Another challenge is adapting to the fluctuating environmental conditions [24]. Bacterial plant pathogens can be influenced by various factors, including temperature, humidity, soil conditions, etc., which can vary over time and space [20]. Therefore, it is difficult to accurately diagnose and control bacterial plant diseases as the effective control measures may vary, depending on the specific environmental conditions. Diagnosis and control require the development of flexible and adaptable diagnostic and control strategies tailored to the specific environmental conditions in which the diseases are occurring. There is also a need to account for the diversity of bacterial plant pathogens. A wide range of pathogens can cause bacterial plant diseases [25]. This diversity can make it difficult to accurately diagnose and control bacterial plant pathogens and diseases. The effective diagnostic and control strategies may vary depending on the pathogen involved.
2. P. syringae as a Bacterial Plant Pathogen
P. syringae, a Gram-negative, rod-shaped bacterium that can cause severe damage to many plant species, is a significant concern for plant health and crop production [26]. It is classified as a hemibiotrophic pathogen that initially feeds on living plant tissues and later causes the death of plant cells [27]. The P. syringae phylogenetic group includes more than 60 pathovars and 15 recognized bacterial species [28]. Each pathovar of P. syringae infects a distinct group of host plants and is known for its diverse host-specific interactions with the plants [29][30]. As early as 1939, the P. syringae pv. primulae was reported to cause necrotic leaf spots on primrose plants in the USA (Figure 1A) [31]. In 1961, the P. syringae pv. tomato was reported to cause necrotic leaf spots on tomato plants in the UK (Figure 1A) [32]. The P. syringae pv. tomato DC3000 is also pathogenic to Arabidopsis plants and has become a model pathogen for probing disease susceptibility and hormone signaling in plants [27]. Up to 2009, Japan witnessed the highest level of occurrence of plant diseases caused by P. syringae, followed by the USA (Figure 1B). Japan reported/deposited 18 different pathovars of P. syringae to the National Collection of Plant Pathogenic Bacteria (NCPPB), and the USA reported/deposited 9 different pathovars of P. syringae to NCPPB (Figure 1B), which increased our understanding of the occurrence/distribution of P. syringae on a world-scale view.
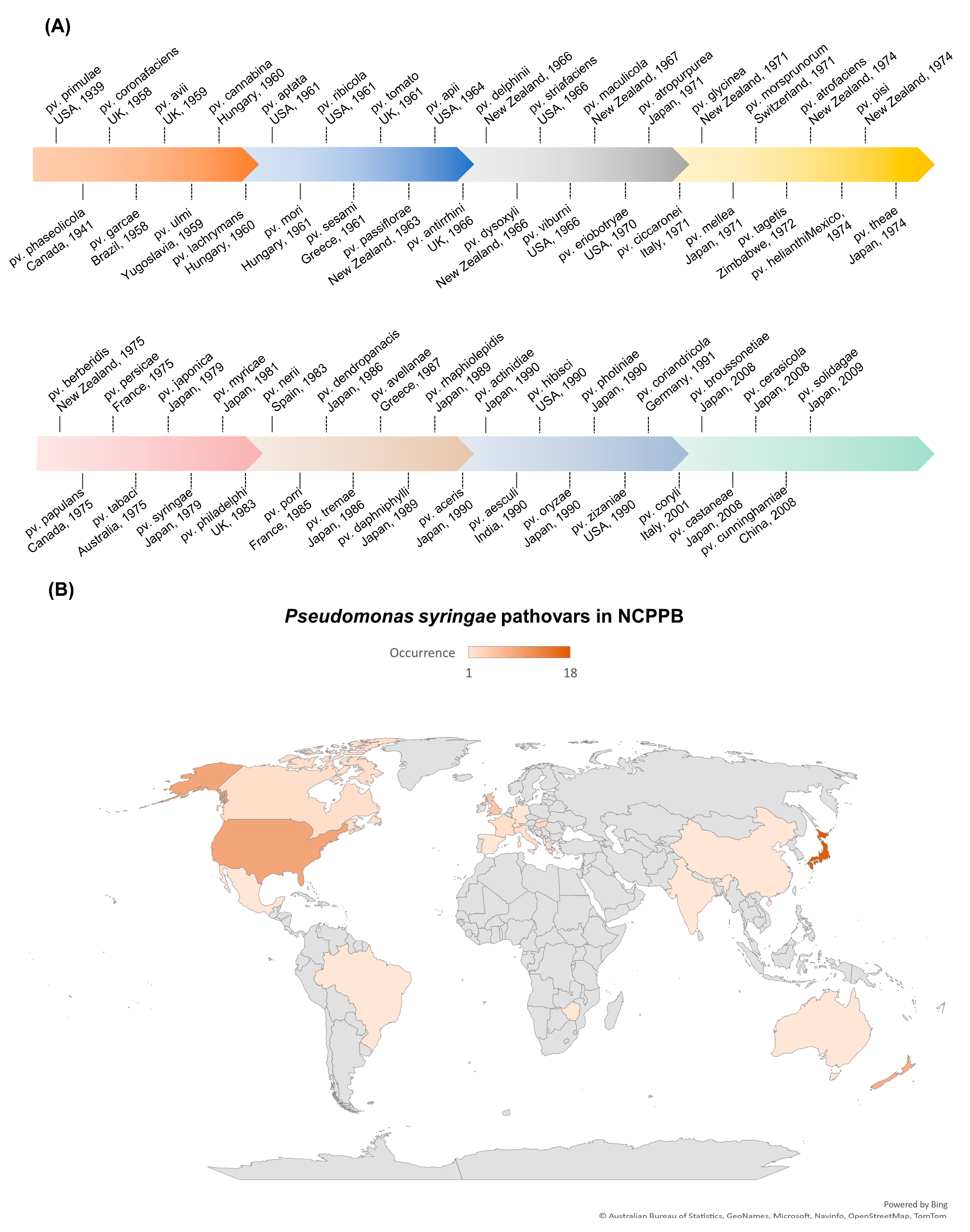
Figure 1. Occurrence of plant diseases caused by P. syringae. Data were retrieved from NCPPB (National Collection of Plant Pathogenic Bacteria, https://www.fera.co.uk/ncppb, accessed on 15 February 2023). (A) Landmark discoveries of pathovars of P. syringae.
The life cycle of P. syringae involves a range of different stages and modes of transmission [33]. P. syringae can be transmitted through seeds, water, vector insects, and infected plant debris. Once inside the plants, P. syringae can multiply and produce toxins that harm plant tissues. The infected plants can develop characteristic symptoms, such as lesions or discoloration on diseased leaves and necrosis spots on diseased fruits. P. syringae can survive in plant debris in the environment for extended periods and easily infect susceptible host plants through wounds or natural openings (Figure 2). It is worth noting that the life cycle of P. syringae can vary depending on the pathovar (strain) of the bacterium and the plant species it infects (Table 1). P. syringae is typically characterized by its ability to infect only specific areas of plants, such as foliar tissues and fruits. Some pathovars of P. syringae are more virulent or have a broader host range than others, affecting how the bacterium spreads and causes diseases [34].
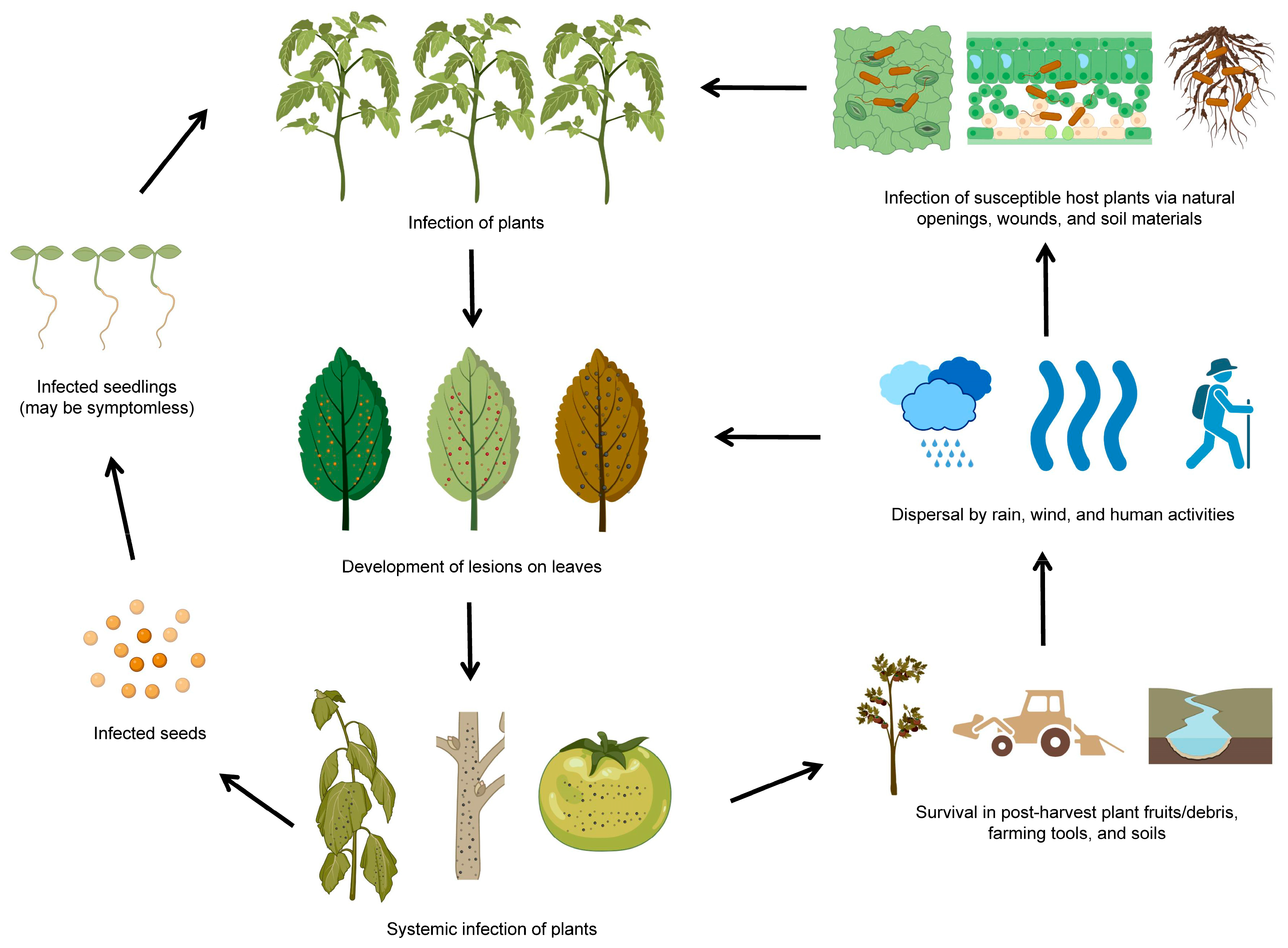
Figure 2. The life cycle of P. syringae. The diagram was adapted from [27] with some modifications and updates. The figure was created with BioRender.com, accessed on 15 February 2023.
Table 1. Documentary records of plant diseases caused by P. syringae. Data were retrieved from NCPPB (National Collection of Plant Pathogenic Bacteria, https://www.fera.co.uk/ncppb, accessed on 15 February 2023).
| Pathovar | Country | Host | Plant Symptoms | Year | References |
|---|---|---|---|---|---|
| pv. aceris | Japan | Acer buergerianum Miq. ‘Tohkaeda’(trident maple) | Necrotic leaf spots | 1990 | NCPPB, [35] |
| pv. actinidiae | Japan | Actinidia chinensis (kiwifruit) | Stem cankers | 1990 | NCPPB, [36] |
| pv. aesculi | India | Aesculus indica (horse chestnut) | Stem cankers | 1990 | NCPPB, [37] |
| pv. antirrhini | UK | Antirrhinum majus (snapdragon) | Necrotic leaf spots | 1966 | NCPPB, [38] |
| pv. apii | USA | Apium graveolens var. dulce (celery) | Necrotic leaf spots | 1964 | NCPPB, [39] |
| pv. aptata | USA | Beta vulgaris (sugar beet) | Tissue blights | 1961 | NCPPB, [40] |
| pv. atrofaciens | New Zealand | Triticum aestivum (bread wheat) | Glume rots | 1974 | NCPPB, [41] |
| pv. atropurpurea | Japan | Lolium multiflorum (ryegrass) | Shoot-tip diebacks | 1971 | NCPPB, [42] |
| pv. avellanae | Greece | Corylus avellana (hazel) | Stem cankers | 1987 | NCPPB, [43] |
| pv. avii | UK | Prunus avium (wild cherry) | Necrotic leaf spots | 1959 | NCPPB, [44] |
| pv. berberidis | New Zealand | Berberis sp. (barberry) | Necrotic leaf spots | 1975 | NCPPB, [45] |
| pv. broussonetiae | Japan | Broussonetia kazinoki (kozo) | Shoot blights | 2008 | NCPPB, [46] |
| pv. cannabina | Hungary | Cannabis sativa (hemp) | Leaf and stem rots | 1960 | NCPPB, [47] |
| pv. castaneae | Japan | Castanea crenata (chestnut) | Leaf blights | 2008 | NCPPB, [48] |
| pv. cerasicola | Japan | Prunus × yedoensis (cherry tree) | Galls on trunks and twigs. | 2008 | NCPPB, [49] |
| pv. ciccaronei | Italy | Ceratonia siliqua (carob) | Stem cankers | 1971 | NCPPB, [50] |
| pv. coriandricola | Germany | Coriandrum sativum var. micocarpur (coriander) | Necrotic leaf spots | 1991 | NCPPB, [51] |
| pv. coronafaciens | UK | Avena sativa (oat) | Leaf blights | 1958 | NCPPB, [52] |
| pv. coryli | Italy | Corylus avellena (hazel) | Stem cankers | 2001 | NCPPB, [53] |
| pv. cunninghamiae | China | Cunninghamia lanceolata (Chinese fir) | Small brown spots with yellow halos on needles (leaves) | 2008 | NCPPB, [54] |
| pv. daphniphylli | Japan | Daphniphyllum teijsmanni (himeyuzuriha) | Galls on trunks and twigs. | 1989 | NCPPB, [55] |
| pv. delphinii | New Zealand | Delphinium sp. (candle larspur) | Stem cankers | 1966 | NCPPB, [56] |
| pv. dendropanacis | Japan | Dendropanax trifidus (ivy) | Stem cankers | 1986 | NCPPB, [57] |
| pv. dysoxyli | New Zealand | Dysoxylum sp. (kohekohe) | Frost damages | 1966 | NCPPB, [58] |
| pv. eriobotryae | USA | Eriobotrya japonica (loquat) | Spots and blisters on fruit | 1970 | NCPPB, [59] |
| pv. garcae | Brazil | Coffea arabica (coffee) | Leaf and stem rots | 1958 | NCPPB, [60] |
| pv. glycinea | New Zealand | Glycine max (soybean) | Leaf blights | 1971 | NCPPB, [61] |
| pv. helianthi | Mexico | Helianthus annuus (sunflower) | Necrotic leaf spots | 1974 | NCPPB, [62] |
| pv. hibisci | USA | Hibiscus rosa seinensis (hibiscus) | Necrotic leaf spots | 1990 | NCPPB, [31] |
| pv. japonica | Japan | Hordeum vulgare (barley) | Leaf blights | 1979 | NCPPB, [63] |
| pv. lachrymans | Hungary | Cucumis sativus (cucumber) | Necrotic leaf spots | 1960 | NCPPB, [64] |
| pv. maculicola | New Zealand | Brassica oleracea var. botrytis (cauliflower) | Necrotic leaf spots | 1967 | NCPPB, [65] |
| pv. mellea | Japan | Nicotiana tabacum (tobacco) | Necrotic leaf spots | 1971 | NCPPB, [66] |
| pv. mori | Hungary | Morus alba (mulberry) | Necrotic leaf spots | 1961 | NCPPB, [67] |
| pv. morsprunorum | Switzerland | Prunus armeniaca (apricot) | Dead dormant buds | 1971 | NCPPB, [68] |
| pv. myricae | Japan | Myrica rubra (yumberry) | Necrotic leaf spots | 1981 | NCPPB, [69] |
| pv. oryzae | Japan | Oryza sativa (rice) | Sheath brown rots | 1990 | NCPPB, [70] |
| pv. papulans | Canada | Malus sylvestris (forest apple) | Blister spots | 1975 | NCPPB, [71] |
| pv. passiflorae | New Zealand | Passiflora edulis (passion fruit) | Necrotic leaf spots | 1963 | NCPPB, [72] |
| pv. persicae | France | Prunus persica (peach) | Stem cankers | 1975 | NCPPB, [73] |
| pv. phaseolicola | Canada | Phaseolus vulgaris (bean) | Necrotic leaf spots | 1941 | NCPPB, [74] |
| pv. philadelphi | UK | Philadelphus coronarius (dogwood) | Necrotic leaf spots | 1983 | NCPPB, [75] |
| pv. photiniae | Japan | Photinia glabra (Japanese photinia) | Necrotic leaf spots | 1990 | NCPPB, [76] |
| pv. pisi | New Zealand | Pisum sativum (pea) | Necrotic leaf spots | 1974 | NCPPB, [77] |
| pv. porri | France | Allium porrum (leek) | Leaf blights | 1985 | NCPPB, [78] |
| pv. primulae | USA | Primula sp. (primrose) | Necrotic leaf spots | 1939 | NCPPB, [31] |
| pv. rhaphiolepidis | Japan | Raphiolepis umbellata (yeddo hawthorne) | Necrotic leaf spots | 1989 | NCPPB, [79] |
| pv. ribicola | USA | Ribes aureum (golden currant) | Necrotic leaf spots | 1961 | NCPPB, [80] |
| pv. nerii | Spain | Nerium oleander (oleander) | Brown leaf galls | 1983 | NCPPB, [81] |
| pv. sesami | Greece | Sesamum indicum (sesame) | Necrotic leaf spots | 1961 | NCPPB, [82] |
| pv. solidagae | Japan | Solidago altissima (goldenrod) | Defoliation and terminal diebacks | 2009 | NCPPB, [83] |
| pv. striafaciens | USA | Avena sp. (oats) | Stripe blights | 1966 | NCPPB, [84] |
| pv. syringae | Japan | Hordeum vulgare (barley) | Leaf blights | 1979 | NCPPB, [85] |
| pv. tabaci | Australia | Glycine max (soybean) | Necrotic leaf spots | 1975 | NCPPB, [86] |
| pv. tagetis | Zimbabwe | Tagetes erecta (marigold) | Necrotic leaf spots | 1972 | NCPPB, [1] |
| pv. theae | Japan | Thea sinensis (tea plant) | Shoot blights | 1974 | NCPPB, [87] |
| pv. tomato | UK | Lycopersicon esculentum (tomato) | Necrotic leaf spots | 1961 | NCPPB, [32] |
| pv. tremae | Japan | Trema orientalis (charcoal-tree) | Necrotic leaf spots | 1986 | NCPPB, [88] |
| pv. ulmi | Yugoslavia | Ulmus sp. (elm) | Necrotic leaf spots | 1959 | NCPPB, [89] |
| pv. viburni | USA | Viburnum sp. (cranberry bush) | Leaf and stem spots | 1966 | NCPPB, [90] |
| pv. zizaniae | USA | Zizania aquatica (wild rice) | Leaf streaks | 1990 | NCPPB, [91] |
P. syringae has been extensively studied since the early 1980s, and it is often used as a model for understanding various aspects of bacterial pathogenicity, including molecular mechanisms of plant-microbe interactions, microbial ecology, and epidemiology [27][30]. Genomic studies have revealed specific genomic characteristics that contribute to the virulence of P. syringae. Currently, it has been found that P. syringae deploys three vital strategies to harm plants: it can survive and adapt to the surface of plants, it can suppress the plant’s immune system at different stages of infection, and it can establish a water-filled space in the plant tissues, which provides it with the access to water and nutrients [30][92][93][94].
There are various techniques available for the detection and diagnosis of P. syringae. These techniques can be broadly classified into several categories: conventional (visual examination, microscopy, culture plate or phage typing), molecular (RPA, LAMP, NGS, FISH or PCR), serological (FCM, ELISA, IF or immunoStrip), biomarker-based (plant metabolite profiling, pathogen metabolite profiling, or microbiome analysis), vision-based (hyperspectral imaging or spectroscopic imaging) and AI (artificial intelligence). Different techniques have different advantages and limitations depending on the sample type, pathovar diversity, cost-effectiveness, etc. Conventional, molecular, and serological techniques are widely used nowadays for the detection and diagnosis of P. syringae.
References
- Mohanty, S.P.; Hughes, D.P.; Salathé, M. Using Deep Learning for Image-Based Plant Disease Detection. Front. Plant Sci. 2016, 7, 1419.
- Liu, J.; Wang, X. Plant Diseases and Pests Detection Based on Deep Learning: A Review. Plant Methods 2021, 17, 22.
- Harakannanavar, S.S.; Rudagi, J.M.; Puranikmath, V.I.; Siddiqua, A.; Pramodhini, R. Plant Leaf Disease Detection Using Computer Vision and Machine Learning Algorithms. Glob. Transit. Proc. 2022, 3, 305–310.
- Plant Disease Diagnosis. Available online: https://www.apsnet.org/edcenter/disimpactmngmnt/casestudies/Pages/PlantDiseaseDiagnosis.aspx (accessed on 3 February 2023).
- Meena, A.K.; Godara, S.L.; Meena, P.N. Detection and Diagnosis of Plant Diseases; Scientific Publishers: Jodhpur, India, 2020; ISBN 978-93-89184-42-6.
- Gullino, M.L.; Bonants, P.J.M. (Eds.) Detection and Diagnostics of Plant Pathogens; Springer: Dordrecht, The Netherlands, 2014; ISBN 978-94-017-9019-2.
- FAO. The State of Food and Agriculture 2021: Making Agrifood Systems More Resilient to Shocks and Stresses; The State of Food and Agriculture (SOFA); FAO: Rome, Italy, 2021; ISBN 978-92-5-134329-6.
- Yang, P.; Chen, Y.; Wu, H.; Fang, W.; Liang, Q.; Zheng, Y.; Olsson, S.; Zhang, D.; Zhou, J.; Wang, Z.; et al. The 5-Oxoprolinase Is Required for Conidiation, Sexual Reproduction, Virulence and Deoxynivalenol Production of Fusarium graminearum. Curr. Genet. 2018, 64, 285–301.
- Yang, P.; Zhao, Z.; Fan, J.; Liang, Y.; Bernier, M.C.; Gao, Y.; Zhao, L.; Opiyo, S.O.; Xia, Y. Bacillus proteolyticus OSUB18 Triggers Induced Systemic Resistance against Bacterial and Fungal Pathogens in Arabidopsis. Front. Plant Sci. 2023, 14, 1078100.
- Zhao, L.; Yang, P.; Li, W.; Zhao, Z.; Xia, Y. First Report of Trichoderma crassum Causing Leaf Spot on Tomato (Solanum lycopersicum Cv. M82) in Ohio. Plant Dis. 2023, 107, 582.
- Cavalazzi, B.; Lemelle, L.; Simionovici, A.; Cady, S.L.; Russell, M.J.; Bailo, E.; Canteri, R.; Enrico, E.; Manceau, A.; Maris, A.; et al. Cellular Remains in a ~3.42-Billion-Year-Old Subseafloor Hydrothermal Environment. Sci. Adv. 2021, 7, eabf3963.
- Ehrlich, P. Address in Pathology, on Chemiotherapy. Br. Med. J. 1913, 2, 353–359.
- Zhao, Z.; Yang, X.; Lü, S.; Fan, J.; Opiyo, S.; Yang, P.; Mangold, J.; Mackey, D.; Xia, Y. Deciphering the Novel Role of AtMIN7 in Cuticle Formation and Defense against the Bacterial Pathogen Infection. Int. J. Mol. Sci. 2020, 21, 5547.
- Zhao, Z.; Fan, J.; Gao, Y.G.; Wang, Z.; Yang, P.; Liang, Y.; Opiyo, S.; Xia, Y. Arabidopsis Plasma Membrane ATPase AHA5 Is Negatively Involved in PAMP-Triggered Immunity. Int. J. Mol. Sci. 2022, 23, 3857.
- Zhao, Z.; Fan, J.; Yang, P.; Wang, Z.; Stephen, O.; Mackey, D.; Ye, X. Involvement of Arabidopsis Acyl Carrier Protein 1 in PAMP-Triggered Immunity. Mol. Plant-Microbe Interact. 2022, 35, 681–693.
- Plant Disease Management Strategies. Available online: https://www.apsnet.org/edcenter/disimpactmngmnt/topc/EpidemiologyTemporal/Pages/ManagementStrategies.aspx (accessed on 3 February 2023).
- Buttimer, C.; McAuliffe, O.; Ross, R.P.; Hill, C.; O’Mahony, J.; Coffey, A. Bacteriophages and Bacterial Plant Diseases. Front. Microbiol. 2017, 8, 34.
- Rapid and Accurate Disease Diagnosis as a Key Component to Successful Plant Disease Management. Available online: https://extension.unh.edu/blog/2022/02/rapid-accurate-disease-diagnosis-key-component-successful-plant-disease-management (accessed on 3 February 2023).
- Rizzo, D.M.; Lichtveld, M.; Mazet, J.A.K.; Togami, E.; Miller, S.A. Plant Health and Its Effects on Food Safety and Security in a One Health Framework: Four Case Studies. One Health Outlook 2021, 3, 6.
- Ristaino, J.B.; Anderson, P.K.; Bebber, D.P.; Brauman, K.A.; Cunniffe, N.J.; Fedoroff, N.V.; Finegold, C.; Garrett, K.A.; Gilligan, C.A.; Jones, C.M.; et al. The Persistent Threat of Emerging Plant Disease Pandemics to Global Food Security. Proc. Natl. Acad. Sci. USA 2021, 118, e2022239118.
- Mooney, H.A.; Cleland, E.E. The Evolutionary Impact of Invasive Species. Proc. Natl. Acad. Sci. USA 2001, 98, 5446–5451.
- López, M.M.; Llop, P.; Olmos, A.; Marco-Noales, E.; Cambra, M.; Bertolini, E. Are Molecular Tools Solving the Challenges Posed by Detection of Plant Pathogenic Bacteria and Viruses? Curr. Issues Mol. Biol. 2009, 11, 13–46.
- Khiyami, M.A.; Almoammar, H.; Awad, Y.M.; Alghuthaymi, M.A.; Abd-Elsalam, K.A. Plant Pathogen Nanodiagnostic Techniques: Forthcoming Changes? Biotechnol. Biotechnol. Equip. 2014, 28, 775–785.
- Buja, I.; Sabella, E.; Monteduro, A.G.; Chiriacò, M.S.; De Bellis, L.; Luvisi, A.; Maruccio, G. Advances in Plant Disease Detection and Monitoring: From Traditional Assays to In-Field Diagnostics. Sensors 2021, 21, 2129.
- Pfeilmeier, S.; Caly, D.L.; Malone, J.G. Bacterial Pathogenesis of Plants: Future Challenges from a Microbial Perspective. Mol. Plant Pathol. 2016, 17, 1298–1313.
- Katagiri, F.; Thilmony, R.; He, S.Y. The Arabidopsis thaliana-Pseudomonas syringae Interaction. In Arabidopsis Book; The American Society of Plant Biologists: Rockville, MD, USA, 2002; Volume 2002.
- Xin, X.-F.; He, S.Y. Pseudomonas syringae pv. tomato Dc3000: A Model Pathogen for Probing Disease Susceptibility and Hormone Signaling in Plants. Annu. Rev. Phytopathol. 2013, 51, 473–498.
- Gomila, M.; Busquets, A.; Mulet, M.; García-Valdés, E.; Lalucat, J. Clarification of Taxonomic Status within the Pseudomonas syringae Species Group Based on a Phylogenomic Analysis. Front. Microbiol. 2017, 8, 2422.
- Hirano, S.S.; Upper, C.D. Bacteria in the Leaf Ecosystem with Emphasis On Pseudomonas syringae—A Pathogen, Ice Nucleus, and Epiphyte. Microbiol. Mol. Biol. Rev. 2000, 64, 624–653.
- Xin, X.-F.; Kvitko, B.; He, S.Y. Pseudomonas syringae: What It Takes to Be a Pathogen. Nat. Rev. Microbiol. 2018, 16, 316–328.
- Gardan, L.; Shafik, H.; Belouin, S.; Broch, R.; Grimont, F.; Grimont, P.A.D.Y. 1999 DNA Relatedness among the Pathovars of Pseudomonas syringae and Description of Pseudomonas tremae sp. nov. and Pseudomonas cannabina sp. nov. (Ex Sutic and Dowson 1959). Int. J. Syst. Evol. Microbiol. 1999, 49, 469–478.
- Preston, G.M. Pseudomonas syringae pv. tomato: The Right Pathogen, of the Right Plant, at the Right Time. Mol. Plant Pathol. 2000, 1, 263–275.
- Morris, C.E.; Sands, D.C.; Vinatzer, B.A.; Glaux, C.; Guilbaud, C.; Buffière, A.; Yan, S.; Dominguez, H.; Thompson, B.M. The Life History of the Plant Pathogen Pseudomonas syringae Is Linked to the Water Cycle. ISME J. 2008, 2, 321–334.
- Jones, J.B.; Zitter, T.A.; Momol, T.M.; Miller, S.A. Compendium of Tomato Diseases and Pests, 2nd ed.; Jeffrey, B.J., Thomas, A.Z., Timur, M.M., Sally, A.M., Eds.; Diseases and Pests Compendium Series; The American Phytopathological Society: Saint Paul, MN, USA, 2016; ISBN 978-0-89054-434-1.
- Takikawa, Y.; Tsuyumu, S.; Goto, M. Occurrence of Bacterial Leaf Spot of Maple Incited by Pseudomonas Syrinigae pv. aceris in Japan. Jpn. J. Phytopathol. 1991, 57, 724–728.
- Takikawa, Y.; Serizawa, S.; Ichikawa, T.; Tsuyumu, S.; Goto, M. Pseudomonas syringae pv. actinidiae pv. nov.: The Causal Bacterium of Canker of Kiwifruit in Japan. Ann. Phytopathol. Soc. Jpn. 1989, 55, 437–444.
- Durgapal, J.C.; Singh, B. Taxonomy of Pseudomonads Pathogenic to Horse-Chestnut, Wild Fig and Wild Cherry in India. Indian Phytopathol. 1980, 33, 533–535.
- Hendson, M.; Hildebrand, D.C.; Schroth, M.N. Relatedness of Pseudomonas syringae pv. tomato, Pseudomonas syringae pv. maculicola and Pseudomonas syringae pv. antirrhini. J. Appl. Bacteriol. 1992, 73, 455–464.
- Gardan, L.; Cottin, S.; Bollet, C.; Hunault, G. Phenotypic Heterogeneity of Pseudomonas syringae van Hall. Res. Microbiol. 1991, 142, 995–1003.
- Maraite, H.; Weyns, J. Pseudomonas syringae pv. aptata and pv. atrofaciens, Specific Pathovars or Members of pv. Syringae? In Pseudomonas Syringae Pathovars and Related Pathogens; Rudolph, K., Burr, T.J., Mansfield, J.W., Stead, D., Vivian, A., von Kietzell, J., Eds.; Developments in Plant Pathology; Springer: Dordrecht, The Netherlands, 1997; pp. 515–520. ISBN 978-94-011-5472-7.
- Iacobellis, N.S.; Figliuolo, G.; Janse, J.; Scortichini, M.; Ciuffreda, G. Characterization of Pseudomonas syringae pv. atrofaciens. In Pseudomonas syringae Pathovars and Related Pathogens; Rudolph, K., Burr, T.J., Mansfield, J.W., Stead, D., Vivian, A., von Kietzell, J., Eds.; Developments in Plant Pathology; Springer: Dordrecht, The Netherlands, 1997; pp. 500–504. ISBN 978-94-011-5472-7.
- Sato, M.; Nishiyama, K.; Shirata, A. Involvement of Plasmid DNA in the Productivity of Coronatine by Pseudomonas syringae pv. atropurpurea. Jpn. J. Phytopathol. 1983, 49, 522–528.
- Psallidas, P.G. Pseudomonas syringae pv. avellanae pathovar nov., the Bacterium Causing Canker Disease on Corylus avellana. Plant Pathol. 1993, 42, 358–363.
- Ménard, M.; Sutra, L.; Luisetti, J.; Prunier, J.P.; Gardan, L. Pseudomonas syringae pv. avii (pv. nov.), the Causal Agent of Bacterial Canker of Wild Cherries (Prunus avium) in France. Eur. J. Plant Pathol. 2003, 109, 565–576.
- Roberts, S.j. A Note on Pseudomonas syringae pv. berberidis Infections of Berberis: Comparative Infection Studies in Attached and Detached Leaves. J. Appl. Bacteriol. 1985, 59, 369–374.
- Takahashi, K.; Nishiyama, K.; Sato, M. Pseudomonas syringae pv. broussonetiae pv. nov., the Causal Agent of Bacterial Blight of Paper Mulberry (Broussonetia kazinoki×B. papyrifera). Jpn. J. Phytopathol. 1996, 62, 17–22.
- Sato, M.; Watanabe, K.; Yazawa, M.; Takikawa, Y.; Nishiyama, K. Detection of New Ethylene-Producing Bacteria, Pseudomonas syringae pvs. cannabina and sesami, by PCR Amplification of Genes for the Ethylene-Forming Enzyme. Phytopathology 1997, 87, 1192–1196.
- Takanashi, K.; Shimizu, K. Pseudomonas syringae pv. castaneae pv. nov., causal agent of bacterial canker of chestnut (Castanea crenata Sieb. et Zucc.). Ann. Phytopathol. Soc. Jpn. 1989, 55, 397–403.
- Kamiunten, H.; Nakao, T.; Oshida, S. Pseudomonas syringae pv. cerasicola, pv. nov., the Causal Agent of Bacterial Gall of Cherry Tree. J. Gen. Plant Pathol. 2000, 66, 219–224.
- Lavermicocca, P.; Lonigro, S.L.; Evidente, A.; Andolfi, A. Bacteriocin Production by Pseudomonas syringae pv. ciccaronei NCPPB2355. Isolation and Partial Characterization of the Antimicrobial Compound. J. Appl. Microbiol. 1999, 86, 257–265.
- Toben, H.-M.; Rudoph, K. Pseudomonas syringae pv. coriandricola, Incitant of Bacterial Umbel Blight and Seed Decay of Coriander (Coriandrum sativum L.) in Germany. J. Phytopathol. 1996, 144, 169–178.
- Harper, S.; Zewdie, N.; Brown, I.R.; Mansfield, J.W. Histological, Physiological and Genetical Studies of the Responses of Leaves and Pods of Phaseolus vulgaris to Three Races of Pseudomonas syringae pv. phaseolicola and to Pseudomonas syringae pv. coronafaciens. Physiol. Mol. Plant Pathol. 1987, 31, 153–172.
- Scortichini, M.; Rossi, M.P.; Loreti, S.; Bosco, A.; Fiori, M.; Jackson, R.W.; Stead, D.E.; Aspin, A.; Marchesi, U.; Zini, M.; et al. Pseudomonas syringae pv. coryli, the Causal Agent of Bacterial Twig Dieback of Corylus avellana. Phytopathology 2005, 95, 1316–1324.
- He, X.-Y.; Goto, M. Bacterial Needle Blight of Chinese Fir (Cunninghamia lanceolate Hook) Caused by Pseudomonas syringae pv. cunninghamiae pv. nov. Jpn. J. Phytopathol. 1995, 61, 38–40.
- Oginh, C.; Kubo, Y.; Higuchi, H.; Takikawa, Y. Bacterial Gall Diseases of Himeyuzuriha (Daphniphyllum teijsmanni Z.) Caused by Pseudomonas syringae pv. daphniphylli pv. nov. J. Jpn. For. Soc. 1990, 72, 17–22.
- Zdorovenko, E.L.; Zatonsky, G.V.; Kocharova, N.A.; Shashkov, A.S.; Knirel, Y.A.; Ovod, V.V. Structure of the O-Polysaccharide of Pseudomonas syringae pv. delphinii NCPPB 1879T Having Side Chains of 3-Acetamido-3,6-Dideoxy-D-Galactose Residues. Biochemistry 2002, 67, 558–565.
- Ogimi, C.; Higuchi, H.; Takikawa, Y. Bacterial Gall Disease of Kakuremino (Dendropanax trifidus Mak.) Caused by Pseudomonas syrigae pv. dendropanacis pv. nov. Ann. Phytopathol. Soc. Jpn. 1988, 54, 296–302.
- Young, J.M.; Triggs, C.m. Evaluation of Determinative Tests for Pathovars of Pseudomonas syringae van Hall 1902. J. Appl. Bacteriol. 1994, 77, 195–207.
- Kamiunten, H. Loss of a Plasmid in Pseudomonas syringae pv. eriobotryae Is Correlated with Change of Symptoms. Jpn. J. Phytopathol. 1990, 56, 645–650.
- Kairu, G.M. Biochemical and Pathogenic Differences between Kenyan and Brazilian Isolates of Pseudomonas syringae pv. garcae. Plant Pathol. 1997, 46, 239–246.
- Osman, S.F.; Fett, W.F.; Fishman, M.L. Exopolysaccharides of the Phytopathogen Pseudomonas syringae pv. glycinea. J. Bacteriol. 1986, 166, 66–71.
- Arsenijević, M.; Vennete, R.J.; Maširević, S. Pseudomonas syringae pv. helianthi (Kawamura 1934) Dye, Wilkie et Young 1978, a Pathogen of Sunflower (Helianthus annuus L.). J. Phytopathol. 1994, 142, 199–208.
- Fukuda, T.; Azegami, K.; Tabei, H. Histological studies on bacterial black node of barley and wheat caused by Pseudomonas syringae pv. japonica. Ann. Phytopathol. Soc. Jpn. 1990, 56, 252–256.
- Yedidia, I.; Shoresh, M.; Kerem, Z.; Benhamou, N.; Kapulnik, Y.; Chet, I. Concomitant Induction of Systemic Resistance to Pseudomonas syringae pv. lachrymans in Cucumber by Trichoderma asperellum (T-203) and Accumulation of Phytoalexins. Appl. Environ. Microbiol. 2003, 69, 7343–7353.
- Cuppels, D.A.; Ainsworth, T. Molecular and Physiological Characterization of Pseudomonas syringae pv. tomato and Pseudomonas syringae pv. maculicola Strains That Produce the Phytotoxin Coronatine. Appl. Environ. Microbiol. 1995, 61, 3530–3536.
- Kiba, A.; Imanaka, Y.; Nakano, M.; Galis, I.; Hojo, Y.; Shinya, T.; Ohnishi, K.; Hikichi, Y. Silencing of Nicotiana benthamiana SEC14 Phospholipid Transfer Protein Reduced Jasmonic Acid Dependent Defense against Pseudomonas syringae. Plant Biotechnol. 2016, 33, 111–115.
- Ratanapo, S.; Ngamjunyaporn, W.; Chulavatnatol, M. Interaction of a Mulberry Leaf Lectin with a Phytopathogenic Bacterium, P. syringae Pv mori. Plant Sci. 2001, 160, 739–744.
- Sundin, G.W.; Jones, A.L.; Olson, B.D. Overwintering and Population Dynamics of Pseudomonas syringae pv. syringae and P.s. pv. morsprunorum on Sweet and Sour Cherry Trees. Can. J. Plant Pathol. 1988, 10, 281–288.
- Ogimi, C.; Higuchi, H. Bacterial gall of yamamomo (Myrica rubra S. et Z.) caused by Pseudomonas syringae pv. myricae pv. nov. Ann. Phytopathol. Soc. Jpn. 1981, 47, 443–448.
- Kuwata, H. Pseudomonas syringae pv. oryzae pv. nov., Causal Agent of Bacterial Halo Blight of Rice. Jpn. J. Phytopathol. 1985, 51, 212–218.
- Kerkoud, M.; Manceau, C.; Paulin, J.P. Rapid Diagnosis of Pseudomonas syringae pv. papulans, the Causal Agent of Blister Spot of Apple, by Polymerase Chain Reaction Using Specifically Designed HrpL Gene Primers. Phytopathology 2002, 92, 1077–1083.
- Jose, P.A.; Krishnamoorthy, R.; Kwon, S.-W.; Janahiraman, V.; Senthilkumar, M.; Gopal, N.O.; Kumutha, K.; Anandham, R. Interference in Quorum Sensing and Virulence of the Phytopathogen Pseudomonas syringae pv. passiflorae by Bacillus and Variovorax Species. BioControl 2019, 64, 423–433.
- Young, J.M. Pseudomonas syringae pv. persicae from Nectarine, Peach, and Japanese Plum in New Zealand1. EPPO Bull. 1988, 18, 141–151.
- Lindgren, P.B.; Peet, R.C.; Panopoulos, N.J. Gene Cluster of Pseudomonas syringae pv. “phaseolicola” Controls Pathogenicity of Bean Plants and Hypersensitivity of Nonhost Plants. J. Bacteriol. 1986, 168, 512–522.
- Roberts, S.J. Variation within Pseudomonas syringae pv. philadelphi, the Cause of a Leaf Spot of Philadelphus spp. J. Appl. Bacteriol. 1985, 59, 283–290.
- Goto, M. Pseudomonas syringae pv. photiniae pv. nov., the Causal Agent of Bacterial Leaf Spot of Photinia glabra Maxim. Jpn. J. Phytopathol. 1983, 49, 457–462.
- Taylor, J.D.; Bevan, J.R.; Crute, I.R.; Reader, S.L. Genetic Relationship between Races of Pseudomonas syringae pv. pisi and Cultivars of Pisum sativum. Plant Pathol. 1989, 38, 364–375.
- Samson, R.; Shafik, H.; Benjama, A.; Gardan, L. Description of the Bacterium Causing Blight of Leek as Pseudomonas syringae pv. porri (pv. nov.). Phytopathology 1998, 88, 844–850.
- Ogimi, C.; Kawano, C.; Higuchi, H.; Takikawa, Y. Bacterial Gall Disease of Sharinbai (Rhaphiolepis umbellata MARINO) Caused by Pseudomonas syringae pv. rhaphiolepidis pv. nov. J. Jpn. For. Soc. 1992, 74, 308–313.
- Ovod, V.V.; Zdorovenko, E.L.; Shashkov, A.S.; Kocharova, N.A.; Knirel, Y.A. Structure of the O Polysaccharide and Serological Classification of Pseudomonas syringae pv. ribicola NCPPB 1010. Eur. J. Biochem. 2000, 267, 2372–2379.
- Janse, J.D. Pathovar Discrimination within Pseudomonas syringae subsp. Savastanoi Using Whole Cell Fatty Acids and Pathogenicity as Criteria. Syst. Appl. Microbiol. 1991, 14, 79–84.
- Firdous, S.S.; Asghar, R.; Irfan-ul-Haque, M.; Waheed, A.; Afzal, S.N.; Mirza, M.Y. Pathogenesis of Pseudomonas syringae pv. sesami Associated with Sesame (Sesamum indicum L.) Bacterial Leaf Spot. Pak. J. Bot. 2009, 41, 927–934.
- SATO, M.; WATANABE, K.; SATO, Y. Pseudomonas syringae pv. solidagae pv. nov., the Causal Agent of Bacterial Leaf Spot of Tall Goldenrod Solidago altissima L. J. Gen. Plant Pathol. 2001, 67, 303–308.
- Barta, T.M.; Willis, D.K. Biological and Molecular Evidence That Pseudomonas syringae Pathovars coronafaciens, striafaciens and garcae Are Likely the Same Pathovar. J. Phytopathol. 2005, 153, 492–499.
- Rahimian, H. The Occurrence of Bacterial Red Streak of Sugarcane Caused by Pseudomonas syringae pv. syringae in Iran. J. Phytopathol. 1995, 143, 321–324.
- Turner, J.G.; Taha, R.R. Contribution of Tabtoxin to the Pathogenicity of Pseudomonas syringae pv. tabaci. Physiol. Plant Pathol. 1984, 25, 55–69.
- Scortichini, M.; Marchesi, U.; Di Prospero, P. Genetic Relatedness among Pseudomonas avellanae, P. syringae pv. theae and P.s. pv. Actinidiae, and Their Identification. Eur. J. Plant Pathol. 2002, 108, 269–278.
- Ogimi, C.; Higuchi, H.; Takikawa, Y. Bacterial Gall Disease of Urajiroenoki (Trema Orientalis BLUME) Caused by Pseudomonas syringae pv. Tremae pv. nov. J. Jpn. For. Soc. 1988, 70, 441–446.
- Shinohara, H.; Moriwaki, J.; Kadota, I.; Nishiyama, K. Ability of pathotype strains of Pseudomonas syringae to produce coronatine. Ann. Phytopathol. Soc. Jpn. 1999, 65, 629–634.
- Stead, D.E.; Stanford, H.; Aspin, A.; Weller, S.A. First Record of Pseudomonas syringae pv. viburni in the UK. Plant Pathol. 2006, 55, 571.
- Bowden, R.L.; Percich, J. Etiology of Bacterial Leaf Streak of Wild Rice. Phytopathology 1983, 73, 640–645.
- Xin, X.-F.; Nomura, K.; Aung, K.; Velásquez, A.C.; Yao, J.; Boutrot, F.; Chang, J.H.; Zipfel, C.; He, S.Y. Bacteria Establish an Aqueous Living Space in Plants Crucial for Virulence. Nature 2016, 539, 524–529.
- Hu, Y.; Ding, Y.; Cai, B.; Qin, X.; Wu, J.; Yuan, M.; Wan, S.; Zhao, Y.; Xin, X.-F. Bacterial Effectors Manipulate Plant Abscisic Acid Signaling for Creation of an Aqueous Apoplast. Cell Host Microbe 2022, 30, 518–529.e6.
- Yuan, M.; Jiang, Z.; Bi, G.; Nomura, K.; Liu, M.; Wang, Y.; Cai, B.; Zhou, J.-M.; He, S.Y.; Xin, X.-F. Pattern-Recognition Receptors Are Required for NLR-Mediated Plant Immunity. Nature 2021, 592, 105–109.
More
Information
Subjects:
Plant Sciences
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.3K
Revisions:
2 times
(View History)
Update Date:
22 May 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




