Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | F. David Rodriguez | -- | 2499 | 2023-05-17 10:44:51 | | | |
| 2 | Peter Tang | Meta information modification | 2499 | 2023-05-17 11:16:40 | | | | |
| 3 | F. David Rodriguez | Meta information modification | 2499 | 2023-05-17 11:22:39 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Rodríguez, F.D.; Sánchez, M.L.; Coveñas, R. Neurotensin and Neurotensin Receptors. Encyclopedia. Available online: https://encyclopedia.pub/entry/44421 (accessed on 07 February 2026).
Rodríguez FD, Sánchez ML, Coveñas R. Neurotensin and Neurotensin Receptors. Encyclopedia. Available at: https://encyclopedia.pub/entry/44421. Accessed February 07, 2026.
Rodríguez, Francisco D., Manuel Lisardo Sánchez, Rafael Coveñas. "Neurotensin and Neurotensin Receptors" Encyclopedia, https://encyclopedia.pub/entry/44421 (accessed February 07, 2026).
Rodríguez, F.D., Sánchez, M.L., & Coveñas, R. (2023, May 17). Neurotensin and Neurotensin Receptors. In Encyclopedia. https://encyclopedia.pub/entry/44421
Rodríguez, Francisco D., et al. "Neurotensin and Neurotensin Receptors." Encyclopedia. Web. 17 May, 2023.
Copy Citation
The tridecapeptide neurotensin (NT) was isolated from the bovine hypothalamus. It is involved in gut motility and belongs to a family of bioactive peptides, including contulakin, xenopsin, LANT-6, and neuromedin, showing similar amino acid sequences at the C-terminal region; this sequence is essential for the physiological effects mediated by these peptides. Neurotensin is relevant in brain reward mechanisms. Its role in substance abuse needs to be explored in depth to open new therapeutic strategies.
neurotensin
neurotensin receptors
neurotensin signaling
1. Neurotensin
The tridecapeptide NT was isolated from the bovine hypothalamus (Figure 1). It is involved in gut motility and belongs to a family of bioactive peptides, including contulakin, xenopsin, LANT-6, and neuromedin, showing similar amino acid sequences at the C-terminal region; this sequence is essential for the physiological effects mediated by these peptides [1].

NT arises from the pro-NT/neuromedin precursor (170 amino acids in humans). After its cleavage by prohormone convertases (PC, endopeptidases), the following molecules originated: large NT (140 amino acids), large neuromedin (125 amino acids), neuromedin (6 amino acids), and NT (13 amino acids). In addition, another fragment (24–140 amino acids) originated during the precursor processing; this fragment is stable in human serum and could be used as a biomarker for NT release [1]. NT is less stable than the large neuromedin in blood, and hence effective forms can exert longer-lasting biological effects due to its enhanced bloodstream stability compared to that reported for fully processed peptides [1]. It is important to note that, depending on the PC involved and expressed in the tissues (e.g., adrenal gland, intestine, brain), the peptides arising from the pro-NT/neuromedin precursor are different. Thus, PC1 originates large neuromedin and NT (intestinal pattern): PC2, neuromedin, and NT (brain pattern) and PC5A, NT, and neuromedin large forms (adrenal gland pattern). The pro-NT/neuromedin precursor is highly conserved in vertebrates. The coding region of the NT/neuromedin gene is spread over four exons and separated by three introns; it is transcribed to two transcripts (1.0 and 1.5 kb) differing in the 3′-untranslated region. In the brain, both mRNAs are equally expressed.
Experimental preclinical evidence supports the possible involvement of NT in many physiological and pathophysiological processes related to gut motility, bile acid release, glucose homeostasis, lipid metabolism, dopamine release, locomotor activity, blood pressure, angiogenesis, energy balance, body temperature, feeding, reproductive mechanisms, inflammatory processes, memory, stress, motivational and affective behaviors, antinociception, cancer, and alcohol intake [1][4][5][6][7][8][9][10][11][12]. Upregulation of NT and NTR1 has been demonstrated in patients with ulcerative colitis. The involvement of NT in irritable bowel syndrome has also been reported [5][13]. NT shows close anatomical and functional relationships with the mesocorticolimbic/nigrostriatal dopaminergic system, and it mediates some of the sensitizing and rewarding properties of drugs of abuse [14]. Moreover, NT seems to mediate the antidopaminergic action of drugs in preclinical studies, making NTR ligand candidates worth exploring for new treatments [1][15]. In this sense, NT agonists have been proposed to treat schizophrenia, drug addiction, and stress-related neuropathic pain. Neuromedin displays similar actions to those exerted by NT. Neuromedin and NT are co-released and co-localized; both peptides bind with a similar affinity to NTRs, but neuromedin is less potent on NTR1/NTR3 than NT [16][17]. Neuromedin is inactivated and degraded by aminopeptidases and NT by metalloendopeptidases [17].
2. Brain Distribution of Neurotensin
Pro-NT/neuromedin precursor mRNA expression has been reported in the caudate-putamen, hippocampus, accumbens nucleus, amygdala, bed nucleus of the stria terminalis, arcuate nucleus, paraventricular hypothalamic nucleus, medial preoptic area, cuneiform nucleus, periaqueductal gray, and dorsal raphe [1]. It has been demonstrated that steroid hormones and several neurotransmitters regulate the expression of the precursor in some of the previously mentioned regions of the central nervous system [1].
NT and its receptors are widely distributed within the mammalian central and peripheral nervous systems, and this widespread distribution confirms the many physiological actions related to the neurotensinergic system. For example, NT has been located in the bed nucleus of the stria terminalis, putamen, caudate, ventrolateral septum, subiculum, accumbens, globus pallidus, amygdaloid nuclei (e.g., central nucleus), lateral septum, thalamus (e.g., periventricular and intralaminar nuclei), hypothalamus (e.g., lateral and paraventricular nuclei), median eminence, midbrain (e.g., periaqueductal gray, midbrain limbic structures, substance nigra), pons-medulla oblongata (e.g., spinal and trigeminal substantia gelatinosa, area postrema, the nucleus of the solitary tract, nuclei parabrachialis medialis and lateralis, locus ceruleus, raphe nuclei, ventral tegmentum), and spinal cord. In some of the previous regions, NT has been involved in alcohol intake. For example, NT has been observed in the paraventricular nucleus of the thalamus, a nucleus associated with stress, motivation, and alcohol-related behaviors, and it has been suggested that in this nucleus, NT regulates alcohol/drug intake and reinstatement. Neurotensinergic neurons located in the central nucleus of the amygdala are activated after ethanol consumption, and a reduced ethanol consumption was observed after the genetic ablation of these neurons [18]. Concerning the reward circuits, NT and NTRs are mainly found in the striatum (involved in the progression from voluntary-driven to habit-driven automated and compulsive behaviors). Dopamine neurons associated with reward are primarily found in the VTA [19].
3. Neurotensin Receptors
NT binds to three different receptors: two G-protein-coupled receptors (class A), namely NTR1 and NTR2, and a single domain transmembrane protein, NTR3/Sortilin [20][21]. Class A GPCR, to which NTR1 and NTR2 belong, frequently forms homo and heterodimers with distinct properties affecting the binding affinities of ligands, signaling, and receptor endocytosis [22][23][24]. Functional heterodimers of dopamine (D2 and D3) and NTR1 receptors have been described, and the development of high-affinity bivalent ligands for these complexes opens new windows for the pharmacological control of their activity [23][24].
3.1. NTR1
The human NTR1 is a seven-transmembrane domain protein (UniProt code P30989) [25] (Figure 2A) encoded by the NTSR1 gene. The protein is made up of 418 amino acids. It exhibits typical G-protein-coupled receptors post-translational modifications, including glycosylation of asparagines 4, 37, and 41, a disulfide bridge between cysteines in positions 141 and 224, and two lipidation cysteine sites at positions 381 and 383 (Figure 2B). In the central nervous system, NTR1 regulates food intake and addiction neurotransmission. It also controls gastrointestinal and cardiovascular systems and behaves as a growth factor in different cells, affecting cell growth and survival [20][21].
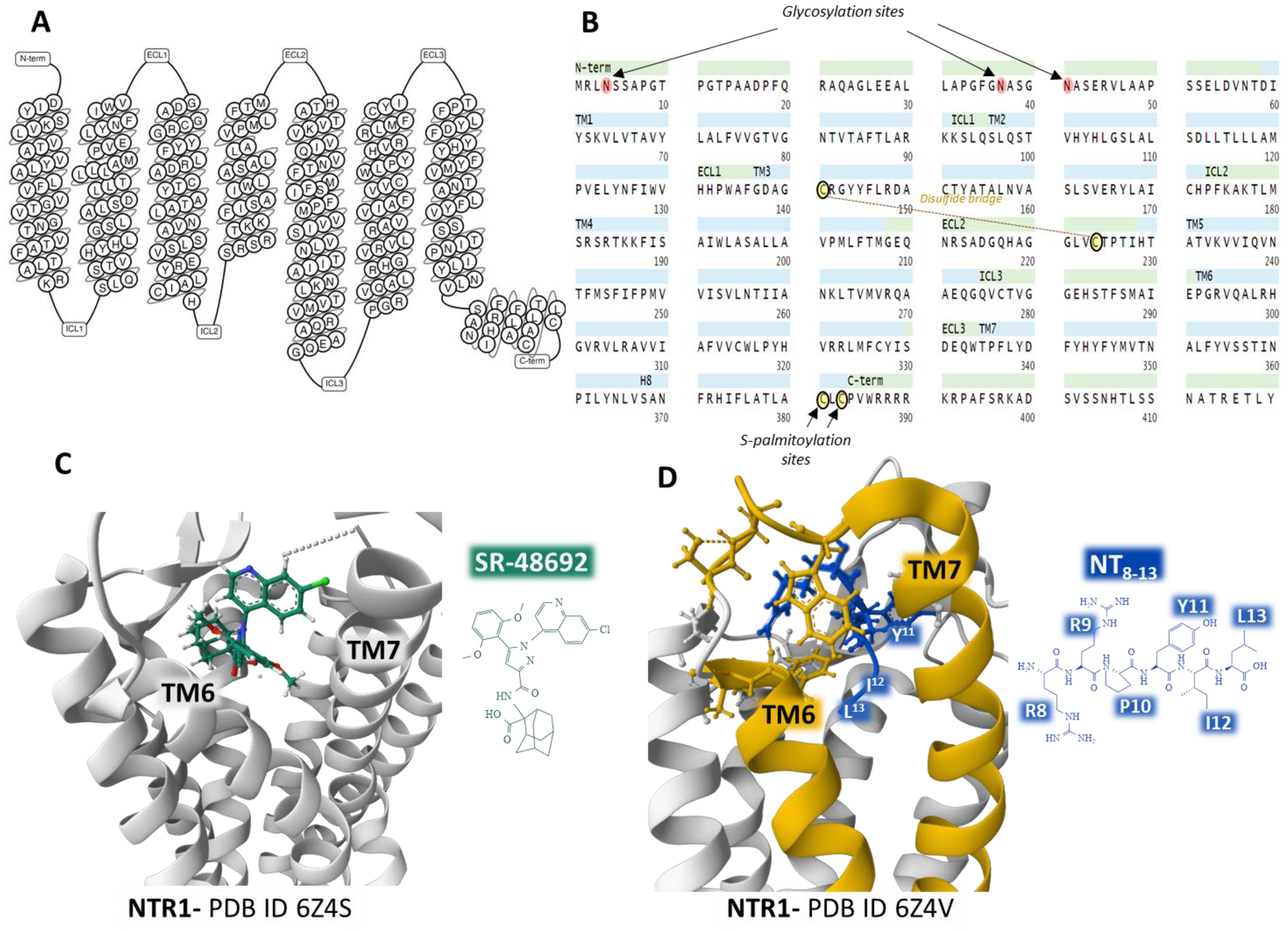
Figure 2. Structure of the seven transmembrane domains of the NTR1 (A) [26] and complete amino acid sequence of NTR1, indicating post-translational modifications (B) [25]. Structure of the NTR1 bound to the inverse agonist SR-48692 and the position of transmembrane domains TM6 and TM7 (C) [27]. Installation of the agonist peptide NT8–13 bound to NTR1 (D) [27]. (C,D) were obtained from the Protein Data Bank [3] and modified with the free-web-based software Mol* (https://molstar.org/ (accessed on 4 March 2023)) [28].
Analysis of several crystal structures of NTR1 with X-ray diffraction or electron cryomicroscopy (cryo-EM) methodologies provided essential information regarding the dynamics of the receptor. Robertson et al. determined the inactive state of the receptor bound to a nanobody [29]. Furthermore, the structure of NTR1 attached to the carboxy-terminal region of NT (NT8–13) revealed that the hexapeptide binds almost perpendicularly to the plasma membrane plane, directing the carboxy end toward the center of the receptor protein (Figure 2D) [27][30]. The structural analysis by Kato et al. [31] disclosed conformational states (canonical and non-canonical) of NTR1 in complex with heterotrimeric Gi1 protein. Cryo-EM resolution of the NTR1 structure bound to NT and G protein Gαi1β1γ1 in a lipid environment showed a tight interaction receptor-G protein in a lipid bilayer that explains the mechanism of activation of G proteins and the activated intracellular signaling [32]. These structural conformations may have relevant functional significance and help understand the transition structures that the receptors display depending on their operational situation. Cryo-EM of human NTR1 disclosed that the phosphorylated receptor (on residues on the intracellular loop three, ICL3, and the C-terminal region) builds a stable complex with β-arrestin1; also, the membrane phospholipid phosphatidylinositol-4,5-bisphosphate (PIP2) bridges transmembrane domains 1 and 4 with β-arrestin [33].
Crystal structures of NTR1 bound to different ligands (partial agonists, inverse agonists, and full agonists) have revealed many receptor structures whose function depends on the type of ligand attached [27]. Inverse agonist SR48692 provokes the opening of the binding site by forcing the aperture of the extracellular portion of the transmembrane domains TM6 and TM7, making difficult the binding of the transducer G protein (Figure 2C). In contrast, full agonist peptide NT8–13 prompts the closure of the binding pocket (Figure 2D) [27].
Accurate determination of different receptor structures and resultant functional knowledge is essential to design specific and selective nonpeptide stable compounds that fine-tune and modulate the protein signaling according to the operational necessity.
3.2. NTR2
The human NTR2 is a seven-transmembrane domain protein [26] (UniProt code P095665 [25]) (Figure 3A) encoded by the NTSR2 gene. The protein consists of a sequence of 410 amino acids. It exhibits typical G-protein-coupled receptors post-translational modifications: a disulfide bridge between cysteines in positions 108 and 194 and a lipidation site at Cys377. No glycosylation targets appear on the ensemble (Figure 3B).
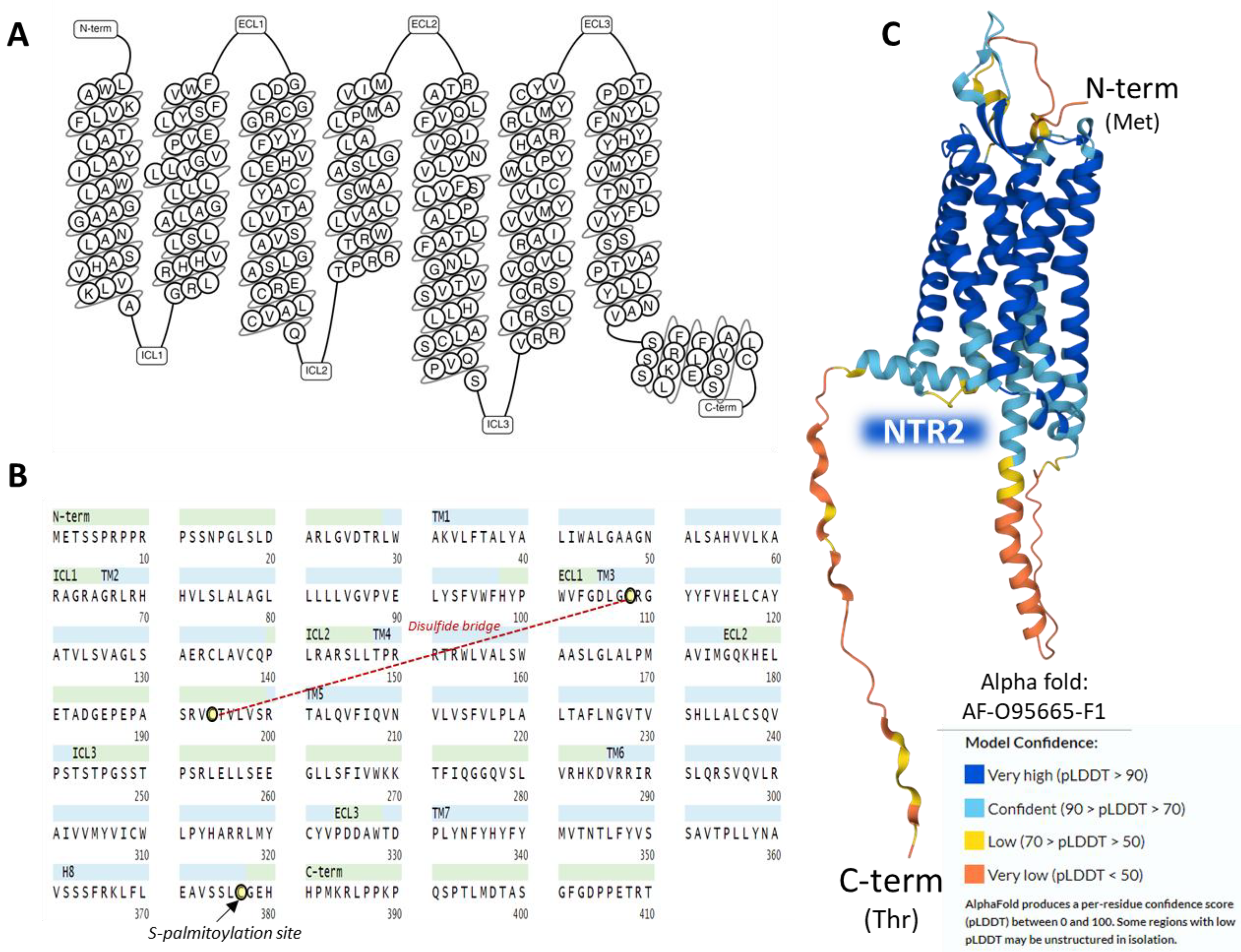
The NTR2 shows a lower affinity for NT than NTR1; it binds to the antihistaminic drug levocabastine, has low sensitivity for sodium ions, and presents no glycosylation sites on the extracellular N-terminal region [20]. NTR1 and NTR2 share approximately a 40% sequence identity as calculated by BLAST [34] and have a widespread but not identical tissue distribution [35]. The AlphaFold structure prediction shows that NTR2 outlines a seven-transmembrane serpentine structure similar to the neurokinin-1 receptor (Figure 3C).
3.3. NTR3/Sortilin
The human NTR3/sortilin is a type I 100 kD glycoprotein receptor (Uniprot code Q-99523) [25] encoded by the SORT1 gene and structurally unrelated to NTR1 and NTR2 with multifunctional tasks, including protein sorting and intracellular signaling, acting as a receptor, a co-receptor (heterodimerization), and a modulator of extracellular trafficking [36][37][38][39]. Two sortilin splice variants have been detected in human tissues (brain, heart, thyroid, or placenta, to name a few) [40]. The affinity of NT for the NTR3 stands in the low nanomolar range [20]. Because sortilin also inhabits intracellular vesicles, NT internalization stimulates its recruitment to the plasma membrane [37][40].
The protein has a signal peptide on the N-terminal end and a site for furine hydrolysis. It presents a single transmembrane domain and sequence analogies with Vps10p (on the extracellular domain) and CI-M6PR (cation-independent mannose 6-phosphate) receptor (cytoplasmic region) [20][40]. Figure 4 summarizes the complete amino acid sequence and the post-translational modifications of sortilin, as described (A). Panels B and C partially view the glycoprotein crystal structure bound to NT and resolved with X-ray diffraction.

Figure 4. Structure of NTR3/Sortilin. The complete amino acid sequence of the receptor (A). The figure highlights some domains of the protein and post-translational modifications (the furin site that cleaves the propeptide; lipidation of Cys783; glycosylation of Asn684; phosphorylation of Ser814, Ser819, and Ser825; and disulfide bridges between Cys634-Cys666, Cys668-Cys723, Cys675-Cys688, and Cys702-Cys740) [25]. Structure of the C-terminal tetrapeptide of NT bound to the ectodomain of human Sortilin (B,C). These structures corresponding to the ectodomain of human sortilin bound to an NT fragment [41] are from the Protein Data Bank [3], drawn with the free-web-based software Mol* (https://molstar.org/ (accessed on 4 March 2023)) [28].
The Protein Data Bank [3] holds a collection of structures of sortilin determined by X-ray diffraction. Figure 4C, represents the structure of the binding site of the C-terminal tetrapeptide of NT bound to the so-called ten-bladed-β-propeller domain of human sortilin. This tunnel structure also serves additional binding pockets for growth factors, thus evidencing the multifunctionality of this glycoprotein receptor [41].
4. Neurotensin Signaling Pathways
NTR1, NTR2, and NTR3 mediate the physiological effects mediated by NT through diverse intracellular signaling pathways (Figure 5).
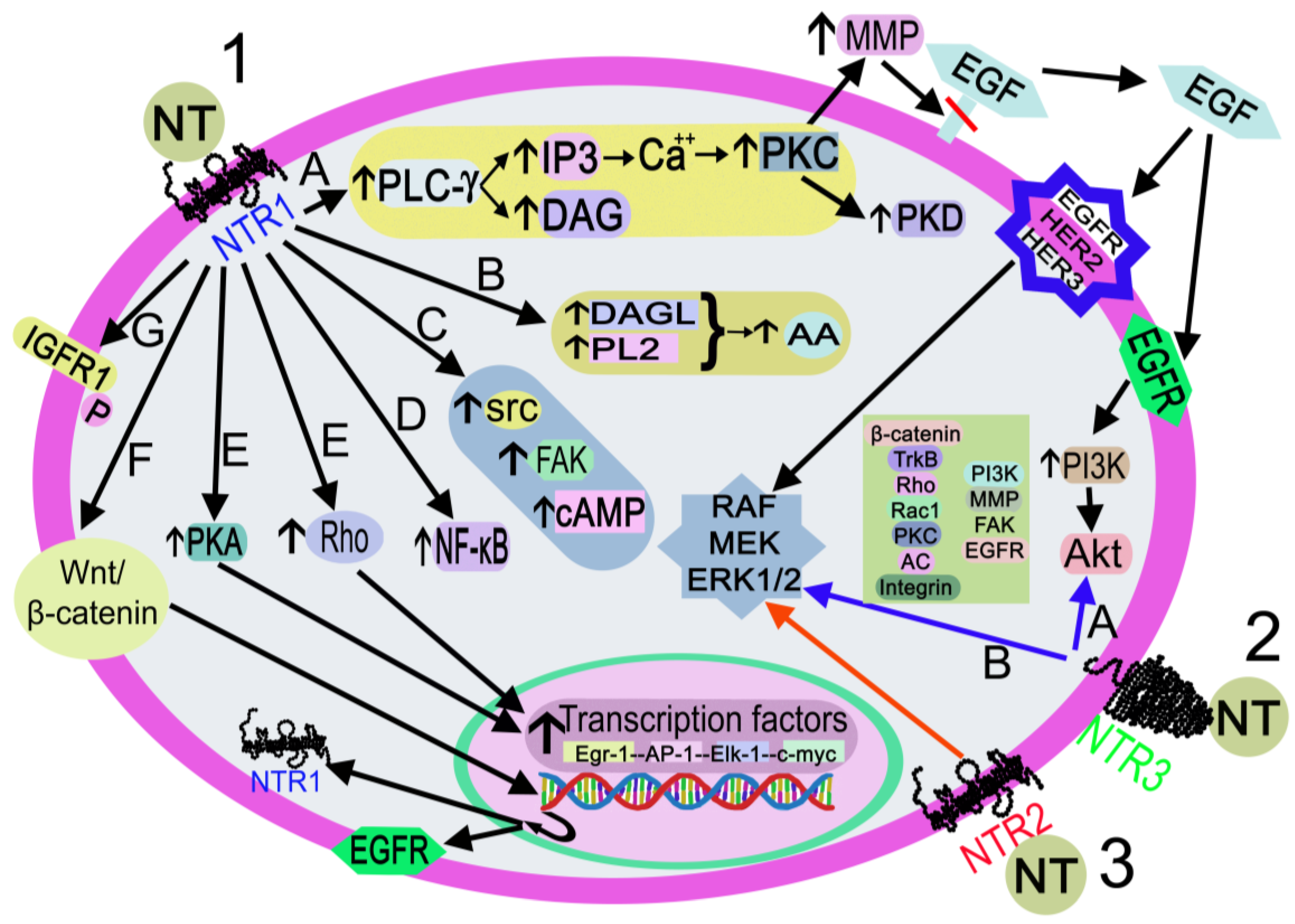
Figure 5. Activation of the signaling pathways after the binding of NT to NTR1 (black arrows), NTR2 (red arrow), or NTR3 (blue arrows). 1. NT-NTR1 binding. A: DAG/I3P synthesis after PLC-γ activation. IP3 promotes the release of Ca++, which activates PLC, PKD, and MMP. MMP cleaves and releases EGF. EGF-EGFR binding activates the PI3K-Akt pathway, and EGF-EGFR/HER2/HER3 binding activates RAF/MEK/ERK1/2. B: DAGL/PL2 activation promotes the synthesis of AA C: FAK/src/cAMP activation. D. NF-κB activation. E: Rho/PKA activation interacts with transcription factors (Egr-1, AP-1, Elk-1, c-myc). F: activation of the Wnt/beta-catenin pathway promotes the EGFR/NTR1 gene transcription. G. IGFR1 phosphorylation. 2. NT-NTR3 binding. A: Pathways involve beta-catenin, TrkB, Rho, Rac1, PKC, AC, integrin, PI3K, FAK, and EGFR B: MAPK activation. 3. NT-NTR2 binding triggers MAPK activation. Abbreviations: AA: arachidonic acid; AC, adenylyl cyclase; Akt, serine-threonine protein kinase; AP-1, activator protein 1; Ca++, calcium cation; cAMP, cyclic adenosine monophosphate; c-myc, proto-oncogene protein; DAG, diacylglycerol; DAGL, diacylglycerol lipase; EGF, epidermal growth factor; EGFR, epidermal growth factor receptor; Egr-1, early growth response protein 1; Elk-1, erythroblast transformation specific line-1 protein; ERK1/2, mitogen-activated protein kinase 1/2; FAK, focal adhesion kinase; HER2, epidermal growth factor receptor 2; HER3, epidermal growth factor receptor 3; I3P, inositol triphosphate 3; IGFR1, insulin growth factor receptor 1; MEK: mitogen-activated protein kinase kinase; MMP, metalloproteinase; NF-κB, nuclear factor kappa light chain enhancer of activated B cells; NT, neurotensin; NTR1, neurotensin receptor 1; NTR2, neurotensin receptor 2; NTR3, neurotensin receptor 3; P, phosphorylation; PI3K, phosphatidylinositol 3-kinase; PKA, protein kinase A; PKC, protein kinase C; PKD, protein kinase D; PL2, phospholipase 2; PLC-γ, phospholipase C-gamma; Rac 1, ras-related C3 botulinum toxin substrate 1; RAF, a serine/threonine-specific protein kinases (rapidly accelerated fibrosarcoma); Rho, small Rho GTPases; src, proto-oncogen tyrosine-protein kinase; TrkB, tropomyosin receptor kinase B; Wnt/β-catenin, Wnt/β-catenin pathway.
After binding to NTR1, the peptide activates the Wnt/beta-catenin pathway and the transcription of the epidermal growth factor receptor and NTR1 genes; activates diacylglycerol lipase and phospholipase 2, promoting the release of arachidonic acid; favors cyclic adenosine monophosphate synthesis and activates protein kinase A, which regulates the transcription of the early growth response protein 1, activator protein 1, erythroblast transformation specific line-1 protein, and c-myc genes; activates focal adhesion kinase, proto-oncogen tyrosine-protein kinase, and small Rho GTPases, and the latter activates transcription factors. NT also activates phospholipase C-gamma, producing inositol triphosphate 3 and diacylglycerol. The former promotes the release of Ca++ from the endoplasmic reticulum, switching on protein kinase C, which, in turn, activates metalloproteinase and protein kinase D. Metalloproteinase cleaves and releases the epidermal growth factor that binds to its receptor, epidermal growth factor receptor 2 or epidermal growth factor receptor 3, thus favoring the activation of mitogen-activated protein kinase 1/2, mitogen-activated protein kinase, and RAF (rapidly accelerated fibrosarcoma). Epidermal growth factor-epidermal growth factor receptor binding activates the phosphatidylinositol 3-kinase-serine-threonine protein kinase pathway; NT favors the phosphorylation of insulin growth factor receptor 1 and triggers nuclear factor kappa light chain enhancer of activated B cells [6]. The signaling pathways mediated by NTR2/NTR3 are not well known; the main molecules involved in these pathways, after NTR2 or NTR3 activation, are mitogen-activated protein kinase 1/2 (for NTR2) and beta-catenin, tropomyosin receptor kinase B, ras homolog family member, ras-related C3 botulinum toxin substrate 1, protein kinase C, phosphatidylinositol 3-kinase, metalloproteinase, integrin, focal adhesion kinase, mitogen-activated protein kinase 1/2, epidermal growth factor receptor, and adenylyl cyclase (for NTR3) [6][42].
References
- Carraway, R.; Dobner, P. Neurotensin/Neuromedin N. In Handbook of Biologically Active Peptides; Academic Press: Boston, MA, USA, 2013; pp. 875–882.
- Coutant, J.; Curmi, P.A.; Toma, F.; Monti, J. NMR solution structure of neurotensin in membrane-mimetic environments: Molecular basis for neurotensin receptor recognition. Biochemistry 2007, 46, 5656–5663.
- RCSB PDB: Homepage. Available online: https://www.rcsb.org/ (accessed on 30 January 2023).
- Mustain, W.C.; Rychahou, P.G.; Evers, B.M. The role of neurotensin in physiologic and pathologic processes. Curr. Opin. Endocrinol. Diabetes Obes. 2011, 18, 75–82.
- Bugni, J.; Pothoulakis, C. Neurotensin. In Handbook of Biologically Active Peptides, 2nd ed.; Academic Press: Boston, MA, USA, 2013; pp. 1265–1270.
- Sánchez, M.L.; Coveñas, R. The Neurotensinergic system: A target for cancer treatment. Curr. Med. Chem. 2022, 29, 3231–3260.
- Ma, H.; Huang, Y.; Zhang, B.; Wang, Y.; Zhao, H.; Du, H.; Cong, Z.; Li, J.; Zhu, G. Association between neurotensin receptor 1 gene polymorphisms and alcohol dependence in a male Han Chinese population. J. Mol. Neurosci. 2013, 51, 408–415.
- Lee, M.R.; Hinton, D.J.; Song, J.Y.; Lee, K.W.; Choo, C.; Johng, H.; Unal, S.S.; Richelson, E.; Choi, D. Neurotensin receptor type 1 regulates ethanol intoxication and consumption in mice. Pharmacol. Biochem. Behav. 2010, 95, 235–241.
- Rock, S.; Li, X.; Song, J.; Townsend, C.M.J.; Weiss, H.L.; Rychahou, P.; Gao, T.; Li, J.; Evers, B.M. Kinase suppressor of Ras 1 and Exo70 promote fatty acid-stimulated neurotensin secretion through ERK1/2 signaling. PLoS ONE 2019, 14, e0211134.
- Feng, Y.; Wang, J.; Dong, Y.; Wang, Y.; Li, Y. The roles of neurotensin and its analogs in pain. Curr. Pharm. Des. 2015, 21, 840–848.
- Li, H.; Namburi, P.; Olson, J.M.; Borio, M.; Lemieux, M.E.; Beyeler, A.; Calhoon, G.G.; Hitora-Imamura, N.; Coley, A.A.; Libster, A.; et al. Neurotensin orchestrates valence assignment in the amygdala. Nature 2022, 608, 586–592.
- Arbogast, P.; Gauchotte, G.; Mougel, R.; Morel, O.; Ziyyat, A.; Agopiantz, M. Neurotensin and its involvement in reproductive functions: An exhaustive review of the literature. Int. J. Mol. Sci. 2023, 24, 4594.
- Brun, P.; Mastrotto, C.; Beggiao, E.; Stefani, A.; Barzon, L.; Sturniolo, G.C.; Palu, G.; Castagliuolo, I. Neuropeptide neurotensin stimulates intestinal wound healing following chronic intestinal inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 2005, 288, 621.
- Ferraro, L.; Tiozzo Fasiolo, L.; Beggiato, S.; Borelli, A.C.; Pomierny-Chamiolo, L.; Frankowska, M.; Antonelli, T.; Tomasini, M.C.; Fuxe, K.; Filip, M. Neurotensin: A role in substance use disorder? J. Psychopharmacol. 2016, 30, 112–127.
- Cáceda, R.; Kinkead, B.; Nemeroff, C.B. Neurotensin: Role in psychiatric and neurological diseases. Peptides 2006, 27, 2385–2404.
- Cuber, J.C.; Herrmann, C.; Kitabgi, P.; Bosshard, A.; Bernard, C.; De Nadai, F.; Chayvialle, J.A. Neuromedin-N is not released with neurotensin from rat ileum. Endocrinology 1990, 126, 1584–1592.
- Kitabgi, P.; De Nadai, F.; Cuber, J.C.; Dubuc, I.; Nouel, D.; Costentin, J. Calcium-dependent release of neuromedin N and neurotensin from mouse hypothalamus. Neuropeptides 1990, 15, 111–114.
- Torruella-Suárez, M.L.; Vandenberg, J.R.; Cogan, E.S.; Tipton, G.J.; Teklezghi, A.; Dange, K.; Patel, G.K.; Mchenry, J.A.; Hardaway, J.A.; Kantak, P.A.; et al. Manipulations of central amygdala neurotensin neurons alter the consumption of ethanol and sweet fluids in mice. J. Neurosci. 2019, 40, 632.
- Torruella-Suárez, M.L.; McElligott, Z.A. Neurotensin in reward processes. Neuropharmacology 2020, 167, 108005.
- Vincent, J.; Mazella, J.; Kitabgi, P. Neurotensin, and neurotensin receptors. Trends Pharmacol. Sci. 1999, 20, 302–307.
- Vincent, J.P. Neurotensin receptors: Binding properties, transduction pathways, and structure. Cell. Mol. Neurobiol. 1995, 15, 501–512.
- Jordan, B.A.; Devi, L.A. G-protein-coupled receptor heterodimerization modulates receptor function. Nature 1999, 399, 697–700.
- Ullmann, T.; Gienger, M.; Budzinski, J.; Hellmann, J.; Hübner, H.; Gmeiner, P.; Weikert, D. Homobivalent dopamine D 2 receptor ligands modulate the dynamic equilibrium of D 2 monomers and homo- and heterodimers. ACS Chem. Biol. 2021, 16, 371–379.
- Budzinski, J.; Maschauer, S.; Kobayashi, H.; Couvineau, P.; Vogt, H.; Gmeiner, P.; Roggenhofer, A.; Prante, O.; Bouvier, M.; Weikert, D. Bivalent ligands promote endosomal trafficking of the dopamine D3 receptor-neurotensin receptor 1 heterodimer. Commun. Biol. 2021, 4, 1062.
- UniProt Database. Available online: https://www.uniprot.org/ (accessed on 26 January 2023).
- GPCRdb Database. Available online: https://gpcrdb.org/protein/ntr1_human/ (accessed on 30 January 2023).
- Deluigi, M.; Klipp, A.; Klenk, C.; Merklinger, L.; Eberle, S.A.; Morstein, L.; Heine, P.; Mittl, P.R.E.; Ernst, P.; Kamenecka, T.M.; et al. Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism. Sci. Adv. 2021, 7, eabe5504.
- Sehnal, D.; Bittrich, S.; Deshpande, M.; Svobodova, R.; Berka, K.; Bazgier, V.; Velankar, S.; Burley, S.K.; Koca, J.; Rose, A.S. Mol* Viewer: Modern web app for 3D visualization and analysis of large biomolecular structures. Nucleic Acids Res. 2021, 49, W431–W437.
- Robertson, M.J.; Papasergi-Scott, M.M.; He, F.; Seven, A.B.; Meyerowitz, J.G.; Panova, O.; Peroto, M.C.; Che, T.; Skiniotis, G. Structure determination of inactive-state GPCRs with a universal nanobody. Nat. Struct. Mol. Biol. 2022, 29, 1188.
- White, J.F.; NoinaJ, N.; Grusshammer, R.; Shibata, Y.; Love, J.; KLoss, B.; Feng, X.U.; Gvozdenovic-Jeremic, J.; Shah, P.; Shiloach, J.; et al. Structure of the agonist-bound neurotensin receptor. Nature 2012, 490, 508–513.
- Kato, H.E.; Zhang, Y.; Hu, H.; Suomivuori, C.; Kadji, F.M.N.; Aoki, J.; Krishna Kumar, K.; Fonseca, R.; Hilger, D.; Huang, W.; et al. Conformational transitions of a neurotensin receptor 1–Gi1 complex. Nature 2019, 572, 80–85.
- Zhang, M.; Gui, M.; Wang, Z.; Gorgulla, C.; Yu, J.J.; Wu, H.; Sun, Z.J.; Klenk, C.; Merklinger, L.; Morstein, L.; et al. Cryo-EM structure of an activated GPCR–G protein complex in lipid nanodiscs. Nat. Struct. Mol. Biol. 2021, 28, 258.
- Huang, W.; Masureel, M.; Qu, Q.; Janetzko, J.; Inoue, A.; Kato, H.E.; Robertson, M.J.; Nguyen, K.C.; Glenn, J.S.; Skiniotis, G.; et al. Structure of the neurotensin receptor 1 in complex with β-arrestin 1. Nature 2020, 579, 303.
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402.
- Labbé-Jullié, C.; Barroso, S.; Nicolas-Etève, D.; Reversat, J.L.; Botto, J.M.; Mazella, J.; Bernassau, J.M.; Kitabgi, P. Mutagenesis and modeling of the neurotensin receptor NTR1. Identification of residues that are critical for binding SR 48692, a nonpeptide neurotensin antagonist. J. Biol. Chem. 1998, 273, 16351–16357.
- Talbot, H.; Saada, S.; Naves, T.; Gallet, P.; Fauchais, A.; Jauberteau, M. Regulatory Roles of Sortilin and SorLA in Immune-Related Processes. Front. Pharmacol. 2019, 9, 1507.
- Leloup, N.; Lössl, P.; Meijer, D.H.; Brennich, M.; Heck, A.J.R.; Thies-Weesie, D.M.E.; Janssen, B.J. C Low pH-induced conformational change and dimerization of sortilin triggers endocytosed ligand release. Nat. Commun. 2017, 8, 1708–1716.
- Ghaemimanesh, F.; Mehravar, M.; Milani, S.; Poursani, E.M.; Saliminejad, K. The multifaceted role of sortilin/neurotensin receptor 3 in human cancer development. J. Cell. Physiol. 2021, 236, 6271–6281.
- Blondeau, N.; Beraud-Dufour, S.; Lebrun, P.; Hivelin, C.; Coppola, T. Sortilin in glucose homeostasis: From accessory protein to key player? Front. Pharmacol. 2019, 9, 1561.
- Petersen, C.M.; Nielsen, M.S.; Nykjær, A.; Jacobsen, L.; Tommerup, N.; Rasmussen, H.H.; RØigaard, H.; Gliemann, J.; Madsen, P.; Moestrup, S.K. Molecular identification of a novel candidate sorting receptor purified from the human brain by receptor-associated protein affinity chromatography. J. Biol. Chem. 1997, 272, 3599–3605.
- Petersen, C.M.; Thirup, S.S.; Quistgaard, E.M.; Madsen, P.; Grøftehauge, M.K.; Nissen, P. Ligands bind to Sortilin in the tunnel of a ten-bladed β-propeller domain. Nat. Struct. Mol. Biol. 2009, 16, 96–98.
- Moody, T.W.; Ramos-Alvarez, I.; Jensen, R.T. Bombesin, endothelin, neurotensin, and pituitary adenylate cyclase-activating polypeptide cause tyrosine phosphorylation of receptor tyrosine kinases. Peptides 2021, 137, 170480.
More
Information
Subjects:
Pharmacology & Pharmacy
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.4K
Revisions:
3 times
(View History)
Update Date:
17 May 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




