
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jianhua Wang | -- | 2061 | 2023-05-12 02:22:25 | | | |
| 2 | Beatrix Zheng | Meta information modification | 2061 | 2023-05-16 10:25:10 | | | | |
| 3 | Beatrix Zheng | -12 word(s) | 2049 | 2023-05-16 10:32:42 | | | | |
| 4 | Beatrix Zheng | + 2 word(s) | 2051 | 2023-05-16 10:33:40 | | |
Video Upload Options
With the accelerating growth of antimicrobial resistance (AMR), there is an urgent need for new antimicrobial agents with low or no AMR. Antimicrobial peptides (AMPs) have been extensively studied as alternatives to antibiotics (ATAs). Coupled with the new generation of high-throughput technology for AMP mining, the number of derivatives has increased dramatically, but manual running is time-consuming and laborious. Therefore, it is necessary to establish databases that combine computer algorithms to summarize, analyze, and design new AMPs. A number of AMP databases have already been established, such as the Antimicrobial Peptides Database (APD), the Collection of Antimicrobial Peptides (CAMP), the Database of Antimicrobial Activity and Structure of Peptides (DBAASP), and the Database of Antimicrobial Peptides (dbAMPs). These four AMP databases are comprehensive and are widely used.
1. Introduction
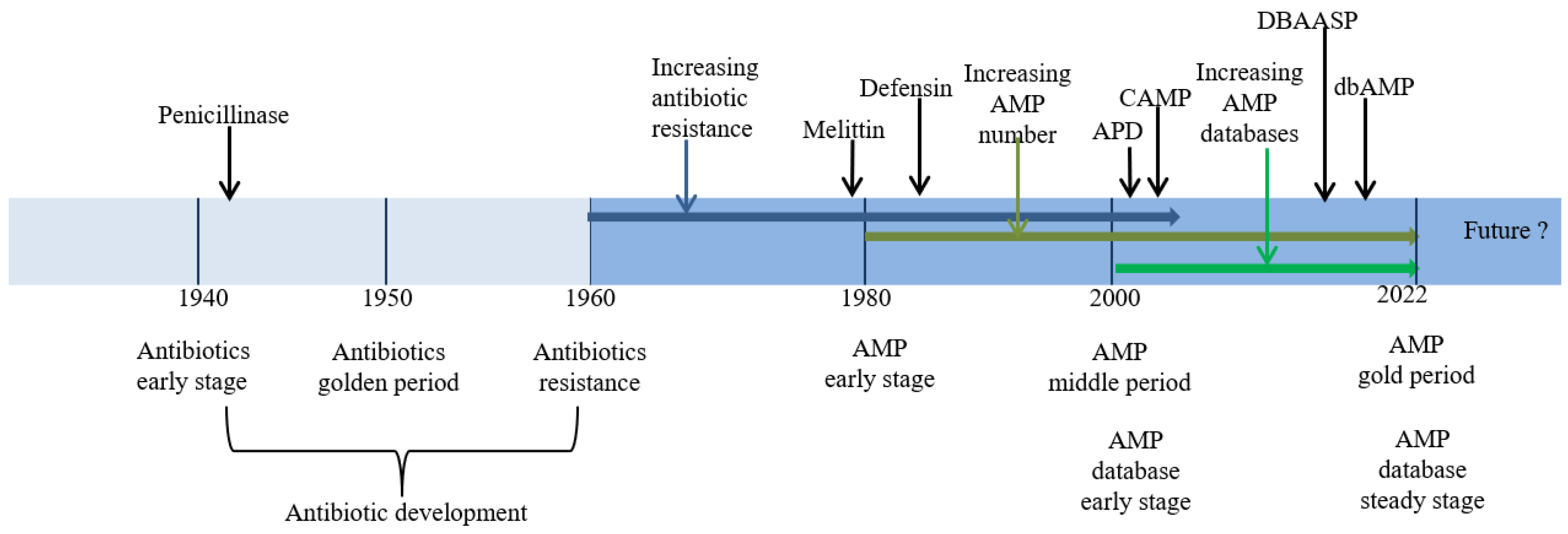
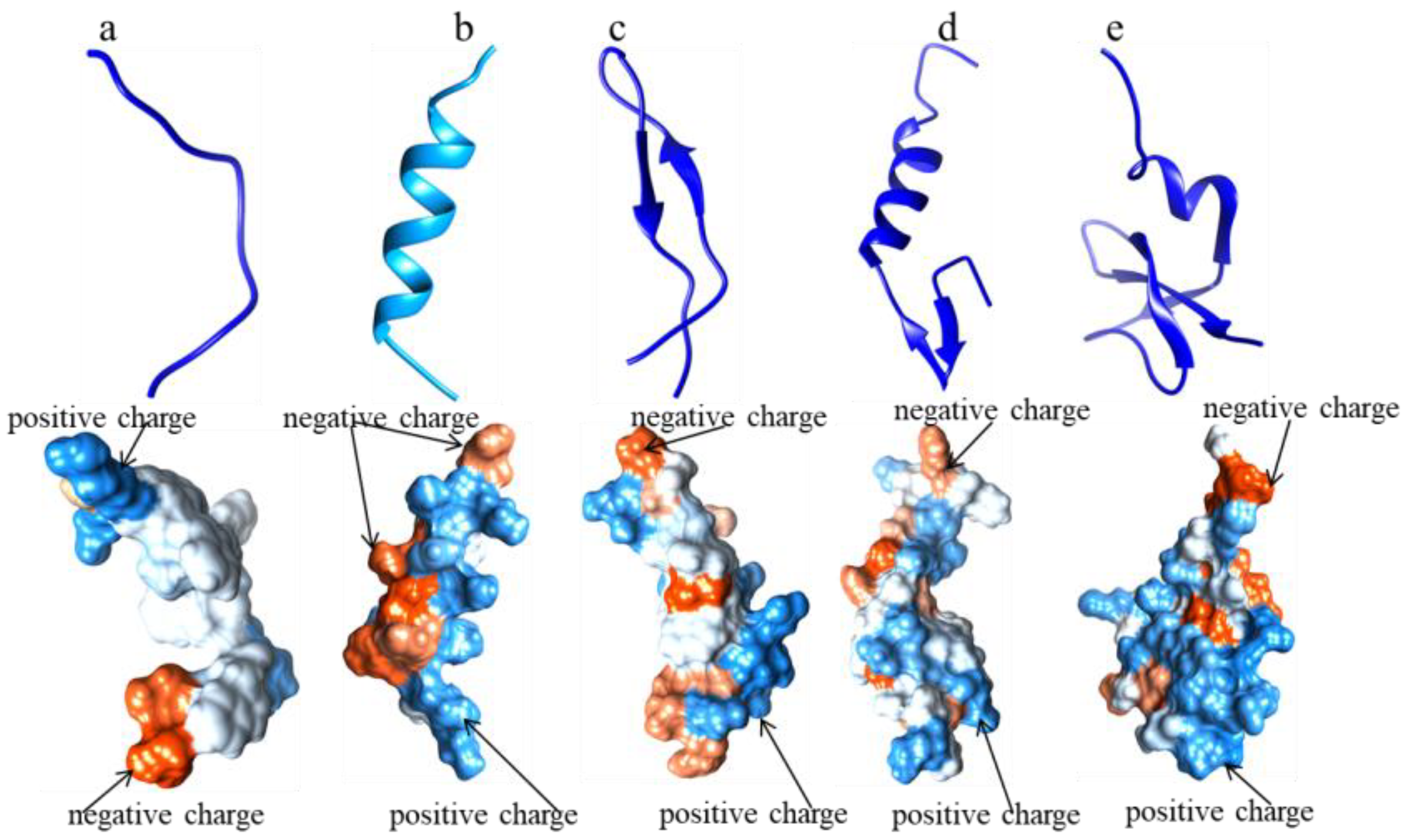
2. Four Typical AMP Databases
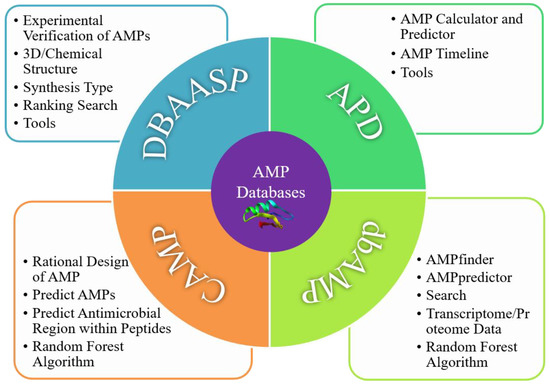
2.1. DBAASP
2.2. APD
2.3. CAMP
2.4. dbAMP
References
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433.
- Gale, E.F. Correlation between penicillin resistance and assimilation affinity in Staphylococcus aureus. Nature 1947, 160, 407.
- Shehreen, S.; Chyou, T.Y.; Fineran, P.C.; Brown, C.M. Genome-wide correlation analysis suggests different roles of CRISPR-Cas systems in the acquisition of antibiotic resistance genes in diverse species. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019, 374, 20180384.
- Huemer, M.; Shambat, S.M.; Brugger, S.D.; Zinkernagel, A.S. Antibiotic resistance and persistence-Implications for human health and treatment perspectives. EMBO Rep. 2020, 21, e51034.
- Karadag, A.S.; Kayıran, M.A.; Wu, C.Y.; Chen, W.; Parish, L.C. Antibiotic resistance in acne: Changes, consequences and concerns. J. Eur. Acad. Dermatol. Venereol. 2021, 35, 73–78.
- Luo, Y.; Song, Y. Mechanism of Antimicrobial Peptides: Antimicrobial, Anti-inflammatory and antibiofilm activities. Int. J. Mol. Sci. 2021, 22, 11401.
- Kaschnitz, R.; Kreil, G. Processing of prepromelittin by subcellular fractions from rat liver. Biochem. Biophys. Res. Commun. 1978, 83, 901–907.
- Fennell, J.F.; Shipman, W.H.; Cole, L.J. Antibacterial Action of a Bee Venom Fraction (Melittin) against a Penicillin-Resistant Staphylococcus and Other Microorganisms; USNRDL-TR-67-101; Research and Development Technical Report; United States Naval Radiological Defense Laboratory: San Francisco, CA, USA, 1967; Volume 5, pp. 1–13.
- Boman, H.G.; Nilsson, I.; Rasmuson, B. Inducible antibacterial defence system in Drosophila. Nature 1972, 237, 232–235.
- Hultmark, D.; Engstrom, A.; Bennich, H.; Kapur, R.; Boman, H.G. Insect immunity: Isolation and structure of cecropin D and four minor antibacterial components from Cecropiapupae. Eur. J. Biochem. 1982, 127, 207–217.
- Qu, Z.; Steiner, H.; Engstrom, A.; Bennich, H.; Boman, H.G. Insect immunity: Isolation and structure of cecropins B and D from pupae of the Chinese oak silk moth, Antheraeapernyi. Eur. J. Biochem. 1982, 127, 219–224.
- Boman, H.G.; Faye, I.; von Hofsten, P.; Kockum, K.; Lee, J.Y.; Xanthopoulos, K.G.; Bennich, H.; Engstrom, A.; Merrifield, R.B.; Andreu, D. On the primary structures of lysozyme, cecropins and attacins from Hyalophoracecropia. Dev. Comp. Immunol. 1985, 9, 551–558.
- Lidholm, D.A.; Gudmundsson, G.H.; Xanthopoulos, K.G.; Boman, H.G. Insect immunity: cDNA clones coding for the precursor forms of cecropins A and D, antibacterial proteins from Hyalophoracecropia. FEBS Lett. 1987, 226, 8–12.
- Hou, F.; Li, J.; Pan, P.; Xu, J.; Liu, L.; Liu, W.; Song, B.; Li, N.; Wan, J.; Gao, H. Isolation and characterisation of a new antimicrobial peptide from the skin of Xenopuslaevis. Int. J. Antimicrob. Agents 2011, 38, 510–515.
- Kumar, N.S.S.; Nazeer, R.A.; Jaiganesh, R. Purification and identification of antioxidant peptides from the skin protein hydrolysate of two marine fishes, horse mackerel (Magalaspiscordyla) and croaker (Otolithesruber). Amino Acids 2012, 42, 1641–1649.
- Elsbach, P.; Weiss, J.; Levy, O. Integration of antimicrobial host defenses: Role of the bactericidal/permeability-increasing protein. Trends Microbiol. 1994, 2, 324–328.
- Ramamoorthy, A.; Thennarasu, S.; Lee, D.K.; Tan, A.; Maloy, L. Solid-state NMR investigation of the membrane-disrupting mechanism of antimicrobial peptides MSI-78 and MSI-594 derived from magainin 2 and melittin. Biophys. J. 2006, 91, 206–216.
- Mor, A.; Nicolas, P. Isolation and structure of novel defensive peptides from frog skin. Eur. J. Biochem. 1994, 219, 145–154.
- Levy, O. Antimicrobial proteins and peptides of blood: Templates for novel antimicrobial agents. Blood 2000, 96, 2664–2672.
- Gong, Z.; Pei, X.; Ren, S.; Chen, X.; Wang, L.; Ma, C.; Xi, X.; Chen, T.; Shaw, C.; Zhou, M. Identification and rational design of a novel antibacterial peptide dermaseptin-AC from the skin secretion of the red-eyed tree frog Agalychnis callidryas. Antibiotics 2020, 9, 243.
- Pasupuleti, M.; Schmidtchen, A.; Malmsten, M. Antimicrobial peptides: Key components of the innate immune system. Crit. Rev. Biotechnol. 2012, 32, 143–171.
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial Peptides: Diversity, mechanism of action and strategies to improve the activity and biocompatibility in vivo. Biomolecules 2018, 8, 4.
- Lai, R.; Zheng, Y.T.; Shen, J.H.; Liu, G.J.; Liu, H.; Lee, W.H.; Tang, S.Z.; Zhang, Y. Antimicrobial peptides from skin secretions of Chinese red belly toad Bombina maxima. Peptides 2002, 23, 427–435.
- Cao, X.; Zhang, Y.; Mao, R.; Teng, D.; Wang, X.; Wang, J. Design and recombination expression of a novel plectasin-derived peptide MP1106 and its properties against Staphylococcus aureus. Appl. Microbiol. Biotechnol. 2015, 99, 2649–2662.
- Slaninová, J.; Mlsová, V.; Kroupová, H.; Alán, L.; Tůmová, T.; Monincová, L.; Borovičková, L.; Fučík, V.; Ceřovský, V. Toxicity study of antimicrobial peptides from wild bee venom and their analogs toward mammalian normal and cancer cells. Peptides 2012, 33, 18–26.
- Zheng, X.; Yang, N.; Mao, R.; Hao, Y.; Teng, D.; Wang, J. Pharmacokinetics and pharmacodynamics of fungal defensin NZX against Staphylococcus aureus-Induced mouse peritonitis model. Front. Microbiol. 2022, 13, 865774.
- Liu, H.; Yang, N.; Teng, D.; Mao, R.; Hao, Y.; Ma, X.; Wang, J. Design and pharmacodynamics of recombinant fungus defensin NZL with improved activity against Staphylococcus hyicus In Vitro and In Vivo. Int. J. Mol. Sci. 2021, 22, 5435.
- Hao, Y.; Wang, J.; de la Fuente-Nunez, C.; Franco, O.L. Editorial: Antimicrobial Peptides: Molecular design, structure-function relationship, and biosynthesis optimization. Front. Microbiol. 2022, 13, 888540.
- Wu, Y.; Yang, N.; Mao, R.; Hao, Y.; Teng, D.; Wang, J. In vitro pharmacodynamics and bactericidal mechanism of fungal defensin-derived peptides NZX and P2 against Streptococcus agalactiae. Microorganisms 2022, 10, 881.
- Ma, X.; Yang, N.; Mao, R.; Hao, Y.; Yan, X.; Teng, D.; Wang, J. The pharmacodynamics study of insect defensin DLP4 against toxigenic Staphylococcus hyicus ACCC 61734 in Vitro and Vivo. Front. Cell. Infect. Microbiol. 2021, 11, 638598.
- Yang, N.; Teng, D.; Mao, R.; Hao, Y.; Wang, X.; Wang, Z.; Wang, X.; Wang, J. A recombinant fungal defensin-like peptide-P2 combats multidrug-resistant Staphylococcus aureus and biofilms. Appl. Microbiol. Biotechnol. 2019, 103, 5193–5213.
- Lee, S.B.; Li, B.; Jin, S.; Daniell, H. Expression and characterization of antimicrobial peptides Retrocyclin-101 and Protegrin-1 in chloroplasts to control viral and bacterial infections. Plant Biotechnol. J. 2011, 9, 100–115.
- Bellamy, W.; Takase, M.; Wakabayashi, H.; Kawase, K.; Tomita, M. Antibacterial spectrum of lactoferricin B, a potent bactericidal peptide derived from the N-terminal region of bovine lactoferrin. J. Appl. Bacteriol. 1992, 73, 472–479.
- Hastings, J.W.; Sailer, M.; Johnson, K.; Roy, K.L.; Vederas, J.C.; Stiles, M.E. Characterization of leucocin A-UAL 187 and cloning of the bacteriocin gene from Leuconostoc gelidum. J. Bacteriol. 1991, 173, 7491–7500.
- Bals, R. Epithelial antimicrobial peptides in host defense against infection. Respir. Res. 2000, 1, 141–150.
- Selsted, M.E.; Novotny, M.J.; Morris, W.L.; Tang, Y.Q.; Smith, W.; Cullor, J.S. Indolicidin, a novel bactericidal tridecapeptide amide from neutrophils. J. Biol. Chem. 1992, 267, 4292–4295.
- Nguyen, L.T.; Haney, E.F.; Vogel, H.J. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 2011, 29, 464–472.
- Silverstein, K.A.; Moskal, W.A., Jr.; Wu, H.C.; Underwood, B.A.; Graham, M.A.; Town, C.D.; VandenBosch, K.A. Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J. 2007, 51, 262–280.
- Resende, J.M.; Moraes, C.M.; Prates, M.V.; Cesar, A.; Almeida, F.C.; Mundim, N.C.; Valente, A.P.; Bemquerer, M.P.; Piló-Veloso, D.; Bechinger, B. Solution NMR structures of the antimicrobial peptides phylloseptin-1, -2, and -3 and biological activity: The role of charges and hydrogen bonding interactions in stabilizing helix conformations. Peptides 2008, 29, 1633–1644.
- Sakagami-Yasui, Y.; Shirafuji, Y.; Yamasaki, O.; Morizane, S.; Hamada, T.; Umemura, H.; Iwatsuki, K. Two arginine residues in the COOH-terminal of human β-defensin-3 constitute an essential motif for antimicrobial activity and IL-6 production. Exp. Dermatol. 2017, 26, 1026–1032.
- Zhu, Y.; Akhtar, M.U.; Li, B.; Chou, S.; Shao, C.; Li, J.; Shan, A. The design of cell-selective tryptophan and arginine-rich antimicrobial peptides by introducing hydrophilic uncharged residues. Acta Biomater. 2022, 153, 557–572.
- Jin, L.; Bai, X.; Luan, N.; Yao, H.; Zhang, Z.; Liu, W.; Chen, Y.; Yan, X.; Rong, M.; Lai, R.; et al. A designed tryptophan- and lysine/arginine-rich antimicrobial peptide with therapeutic potential for clinical antibiotic-resistant Candida albicans vaginitis. J. Med. Chem. 2016, 59, 1791–1799.
- Saravanan, R.; Li, X.; Lim, K.; Mohanram, H.; Peng, L.; Mishra, B.; Basu, A.; Lee, J.M.; Bhattacharjya, S.; Leong, S.S. Design of short membrane selective antimicrobial peptides containing tryptophan and arginine residues for improved activity, salt-resistance, and biocompatibility. Biotechnol. Bioeng. 2014, 111, 37–49.
- Svenson, J.; Karstad, R.; Flaten, G.E.; Brandsdal, B.O.; Brandl, M.; Svendsen, J.S. Altered activity and physicochemical properties of short cationic antimicrobial peptides by incorporation of arginine analogues. Mol. Pharm. 2009, 6, 996–1005.
- Panteleev, P.V.; Bolosov, I.A.; Balandin, S.V.; Ovchinnikova, T.V. Design of antimicrobial peptide arenicin analogs with improved therapeutic indices. J. Pept. Sci. 2015, 21, 105–113.
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250.
- Rončević, T.; Gerdol, M.; Mardirossian, M.; Maleš, M.; Cvjetan, S.; Benincasa, M.; Maravić, A.; Gajski, G.; Krce, L.; Aviani, I.; et al. Anisaxins, helical antimicrobial peptides from marine parasites, kill resistant bacteria by lipid extraction and membrane disruption. Acta Biomater. 2022, 146, 131–144.
- Sychev, S.V.; Balandin, S.V.; Panteleev, P.V.; Barsukov, L.I.; Ovchinnikova, T.V. Lipid-dependent pore formation by antimicrobial peptides arenicin-2 and melittin demonstrated by their proton transfer activity. J. Pept. Sci. 2015, 21, 71–76.
- Teixeira, V.; Feio, M.J.; Bastos, M. Role of lipids in the interaction of antimicrobial peptides with membranes. Prog. Lipid Res. 2012, 51, 149–177.
- Mygind, P.H.; Fischer, R.L.; Schnorr, K.M.; Hansen, M.T.; Sönksen, C.P.; Ludvigsen, S.; Raventós, D.; Buskov, S.; Christensen, B.; De Maria, L.; et al. Plectasin is a peptide antibiotic with therapeutic potential from a saprophytic fungus. Nature 2005, 437, 975–980.
- Lazzaro, B.P.; Zasloff, M.; Rolff, J. Antimicrobial peptides: Application informed by evolution. Science 2020, 368, eaau5480.
- Nagarajan, D.; Nagarajan, T.; Roy, N.; Kulkarni, O.; Ravichandran, S.; Mishra, M.; Chakravortty, D.; Chandra, N. Computational antimicrobial peptide design and evaluation against multidrug-resistant clinical isolates of bacteria. J. Biol. Chem. 2018, 293, 3492–3509.
- Porto, W.F.; Pires, A.S.; Franco, O.L. Computational tools for exploring sequence databases as a resource for antimicrobial peptides. Biotechnol. Adv. 2017, 35, 337–349.
- Wang, G.; Zietz, C.M.; Mudgapalli, A.; Wang, S.; Wang, Z. The evolution of the antimicrobial peptide database over 18 years: Milestones and new features. Protein Sci. 2022, 31, 92–106.
- Chakraborty, S.; Phu, M.; de Morais, T.P.; Nascimento, R.; Goulart, L.R.; Rao, B.J.; Asgeirsson, B.; Dandekar, A.M. The PDB database is a rich source of alpha-helical anti-microbial peptides to combat disease causing pathogens. F1000 Res. 2014, 3, 295.
- Plisson, F.; Ramírez-Sánchez, O.; Martínez-Hernández, C. Machine learning-guided discovery and design of non-hemolytic peptides. Sci Rep. 2020, 10, 16581.
- Brahmachary, M.; Krishnan, S.P.; Koh, J.L.; Khan, A.M.; Seah, S.H.; Tan, T.W.; Brusic, V.; Bajic, V.B. ANTIMIC: A database of antimicrobial sequences. Nucleic Acids Res. 2004, 32, D586–D589.
- Hammami, R.; Ben Hamida, J.; Vergoten, G.; Fliss, I. PhytAMP: A database dedicated to antimicrobial plant peptides. Nucleic Acids Res. 2009, 37, D963–D968.
- Shi, G.; Kang, X.; Dong, F.; Liu, Y.; Zhu, N.; Hu, Y.; Xu, H.; Lao, X.; Zheng, H. DRAMP 3.0: An enhanced comprehensive data repository of antimicrobial peptides. Nucleic Acids Res. 2022, 50, D488–D496.
- Gabere, M.N.; Noble, W.S. Empirical comparison of web-based antimicrobial peptide prediction tools. Bioinformatics 2017, 33, 1921–1929.
- Wang, G. Improved methods for classification, prediction, and design of antimicrobial peptides. Methods Mol. Biol. 2015, 1268, 43–66.
- Thomas, S.; Karnik, S.; Barai, R.S.; Jayaraman, V.K.; Idicula-Thomas, S. CAMP: A useful resource for research on antimicrobial peptides. Nucleic Acids Res. 2010, 38, D774–D780.
- Pirtskhalava, M.; Gabrielian, A.; Cruz, P.; Griggs, H.L.; Squires, R.B.; Hurt, D.E. DBAASP v.2: An enhanced database of structure and antimicrobial/cytotoxic activity of natural and synthetic peptides. Nucleic Acids Res. 2016, 44, D1104–D1112, Erratum in Nucleic Acids Res. 2016, 44, 6503.
- Jhong, J.H.; Chi, Y.H.; Li, W.C.; Lin, T.H.; Huang, K.Y.; Lee, T.Y. dbAMP: An integrated resource for exploring antimicrobial peptides with functional activities and physicochemical properties on transcriptome and proteome data. Nucleic Acids Res. 2019, 47, D285–D297.
- Jhong, J.H.; Yao, L.; Pang, Y.; Li, Z.; Chung, C.R.; Wang, R.; Li, S.; Li, W.; Luo, M.; Ma, R.; et al. dbAMP 2.0: Updated resource for antimicrobial peptides with an enhanced scanning method for genomic and proteomic data. Nucleic Acids Res. 2022, 50, D460–D470.
- Pirtskhalava, M.; Amstrong, A.A.; Grigolava, M.; Chubinidze, M.; Alimbarashvili, E.; Vishnepolsky, B.; Gabrielian, A.; Rosenthal, A.; Hurt, D.E.; Tartakovsky, M. DBAASP v3: Database of antimicrobial/cytotoxic activity and structure of peptides as a resource for development of new therapeutics. Nucleic Acids Res. 2021, 49, D288–D297.
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093.
- Lee, J.; Kang, H.K.; Cheong, H.; Park, Y. A novel antimicrobial peptides from pine needles of Pinus densiflora Sieb. Et Zucc. against foodborne bacteria. Front. Microbiol. 2021, 12, 662462.
- Dean, S.N.; Alvarez, J.A.E.; Zabetakis, D.; Walper, S.A.; Malanoski, A.P. PepVAE: Variational autoencoder framework for antimicrobial peptide generation and activity prediction. Front. Microbiol. 2021, 12, 725727.
- Gu, J.; Isozumi, N.; Yuan, S.; Jin, L.; Gao, B.; Ohki, S.; Zhu, S. Evolution-based protein engineering for antifungal peptide improvement. Mol. Biol. Evol. 2021, 38, 5175–5189.
- Yang, Y.; Gao, H.; Liu, W.; Liu, X.; Jiang, X.; Li, X.; Wu, Q.; Xu, Z.; Zhao, Q. Arctiumlappa L. roots ameliorates cerebral ischemia through inhibiting neuronal apoptosis and suppressing AMPK/mTOR-mediated autophagy. Phytomedicine 2021, 85, 153526.
- Li, Y.; Li, G.X.; Yu, M.L.; Liu, C.L.; Qu, Y.T.; Wu, H. Association between anxiety symptoms and problematic smartphone use among chinese university students: The mediating/moderating role of self-efficacy. Front. Psychiatry 2021, 12, 581367.
- Mao, R.; Teng, D.; Wang, X.; Xi, D.; Zhang, Y.; Hu, X.; Yang, Y.; Wang, J. Design, expression, and characterization of a novel targeted plectasin against methicillin-resistant Staphylococcus aureus. Appl. Microbiol. Biotechnol. 2013, 97, 3991–4002, Erratum in Appl. Microbiol. Biotechnol. 2019, 97, 4233–4234.
- Zavala-Soto, J.O.; Hernandez-Rivero, L.; Tapia-Fonllem, C. Pro-lactation cesarean section: Immediate skin-to-skin contact and its influence on prolonged breastfeeding. Front. Sociol. 2022, 7, 908811.
- Moreno-Barahona, M.; Fraijo-Sing, B.; Fleury-Bahi, G.; Navarro-Carrascal, O.; Tapia-Fonllem, C. Conceptual integration and empirical validation of a unified taxonomy: Quantitative data analysis for virtual learning environments. Front. Psychol. 2022, 13, 814592.
- Wang, G.; Li, X.; Wang, Z. APD2: The updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 2009, 37, D933–D937.
- Waghu, F.H.; Gopi, L.; Barai, R.S.; Ramteke, P.; Nizami, B.; Idicula-Thomas, S. CAMP: Collection of sequences and structures of antimicrobial peptides. Nucleic Acids Res. 2014, 42, D1154–D1158.
- Waghu, F.H.; Idicula-Thomas, S. Collection of antimicrobial peptides database and its derivatives: Applications and beyond. Protein Sci. 2020, 29, 36–42.
- Fan, L.; Sun, J.; Zhou, M.; Zhou, J.; Lao, X.; Zheng, H.; Xu, H. DRAMP: A comprehensive data repository of antimicrobial peptides. Sci. Rep. 2016, 6, 24482.




