Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Lorena Urbanelli | -- | 1849 | 2023-05-09 11:32:18 | | | |
| 2 | Sirius Huang | Meta information modification | 1849 | 2023-05-10 02:40:23 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Sagini, K.; Urbanelli, L.; Buratta, S.; Emiliani, C.; Llorente, A. Lipidic Metabolism in Cancer. Encyclopedia. Available online: https://encyclopedia.pub/entry/44028 (accessed on 07 February 2026).
Sagini K, Urbanelli L, Buratta S, Emiliani C, Llorente A. Lipidic Metabolism in Cancer. Encyclopedia. Available at: https://encyclopedia.pub/entry/44028. Accessed February 07, 2026.
Sagini, Krizia, Lorena Urbanelli, Sandra Buratta, Carla Emiliani, Alicia Llorente. "Lipidic Metabolism in Cancer" Encyclopedia, https://encyclopedia.pub/entry/44028 (accessed February 07, 2026).
Sagini, K., Urbanelli, L., Buratta, S., Emiliani, C., & Llorente, A. (2023, May 09). Lipidic Metabolism in Cancer. In Encyclopedia. https://encyclopedia.pub/entry/44028
Sagini, Krizia, et al. "Lipidic Metabolism in Cancer." Encyclopedia. Web. 09 May, 2023.
Copy Citation
Altered cellular metabolism is a well-established hallmark of cancer. Although most studies have focused on the metabolism of glucose and glutamine, the upregulation of lipid metabolism is also frequent in cells undergoing oncogenic transformation. In fact, cancer cells need to meet the enhanced demand of plasma membrane synthesis and energy production to support their proliferation. Moreover, lipids are precursors of signaling molecules, termed lipid mediators, which play a role in shaping the tumor microenvironment.
biofluids
biomarkers
cancer
extracellular vesicles
lipid metabolism
lipidomics
liquid biopsy
mass spectrometry
1. Introduction
Altered cellular metabolism is a well-established hallmark of cancer [1]. The first observation of cancer metabolic alterations was made in the 1920s by Otto Warburg. He noticed that cancer cells use large amounts of glucose to generate lactate, even in the presence of oxygen, a phenomenon termed aerobic glycolysis or the Warburg effect [2]. Since then, our understanding of tumor-associated metabolic alterations, underlying molecular mechanisms and functional consequences in tumorigenesis has expanded. A variety of well-known oncogenes, such as c-myc, AKT, mTOR, hypoxia-inducible factors and RAS, have been shown to contribute to the metabolic adaptations of cancer cells [3]. Multiple studies have uncovered how glycolysis and the tricarboxylic acid cycle generate metabolic intermediates that sustain the de novo synthesis of nucleotides, lipids and amino acids, supporting cancer cell proliferation [4]. Moreover, some metabolites, known as oncometabolites, accumulate in cancer cells in response to an altered expression of metabolic enzymes and have been directly linked to tumor growth. A canonical example is the cancer-associated mutations in isocitrate dehydrogenase 1 and 2, which result in the aberrant production of the oncometabolite 2-hydroxyglutarate (2HG). The cellular accumulation of 2HG inhibits enzymes controlling histone and DNA demethylation, thus altering the chromatin landscape and gene expression [5]. The majority of studies so far have focused on the metabolism of glucose and glutamine, but it is becoming clear that lipid metabolism is also frequently altered in cells undergoing oncogenic transformation [6]. Cancer cells often up-regulate de novo lipogenesis, fatty acid (FA) uptake, FA oxidation (FAO) and lipid accumulation to support cellular proliferation and the consequent higher demand of plasma membrane synthesis and energy production [7]. Improved lipid analysis techniques based on liquid/gas chromatography coupled with mass spectrometry (MS) have enabled quantitative profiling of lipids as individual molecular species from minute amounts of samples, allowing the in-depth characterization of the lipidome [8]. These technologies allow for obtaining cancer-specific lipid profiles, which reflect, at a phenotypic level, molecular alterations and environmental factors (such as exposure to high-fat diets). These profiles may help in unraveling novel therapeutic targets and biomarkers for the early detection of cancer, possibly leading to improvements in the clinical strategy to treat the disease.
2. Lipidic Metabolism in Cancer
Lipids represent a complex group of biomolecules that vary in structure and perform three main functions: energy storage, membrane formation and signaling. The majority of lipids derivates from FAs, molecules consisting of hydrocarbon chains varying in length and saturation.
Triglycerides (TAGs) are formed by a glycerol moiety esterified with three FAs and, in cells, are localized within lipid droplets. They represent the main storage form of lipids for energetic purposes, as TAG-derived FAs in mitochondria can be catabolized by FAO for ATP production. Overexpression of FAO enzymes, such as the rate-limiting enzyme carnitine palmitoyltransferase 1 and acyl-CoA synthetase long-chain 3 (ACSL3), has been found in numerous malignancies and has been correlated to tumor growth, especially in adverse environmental conditions, such as glucose deprivation [9]. In some cancers, FAO is activated by specific oncogenes, such as c-Myc or mutant K-Ras [7].
Lipids organized in lamellar bilayers form the architecture of biological membranes. The main membrane constituents in mammals are phospholipids and cholesterol. Phospholipids can be further classified according to their structure into glycerophospholipids (consisting of glycerol bound to two fatty acyl chains and a polar head formed by a phosphate group linked to a polar group) and sphingolipids (consisting of ceramide and a polar head formed by a phosphate group linked to choline or ethanolamine) (Figure 1). Glycerophospholipids carry different polar groups, giving rise to various lipid classes: phosphatidic acid (PA), phosphatidylcholine (PC), phosphatidylserine (PS), phosphatidylethanolamine (PE), phosphatidylglycerol (PG) and phosphatidylinositol (PI). Although most glycerophospholipids contain ester-bound FAs, ether-linked fatty acyl chains are also common [10]. Sphingolipids also can have different head groups and are therefore classified into ceramide (Cer, consisting of a long-chain base, often sphingosine, linked to a FA via the amino group) and complex sphingolipids such as sphingomyelin (SM) and glycosphingolipids (glucosylceramides, GlcCer; galactosylceramides, GalCer; sulfatides, Sulf; lactosylceramides, LacCer; gangliosides, GM). The lipid bilayer of the plasma membrane has an asymmetric distribution of lipids. Its outer leaflet contains mostly PC and sphingolipids, and the inner leaflet contains PE, PS, and PI, whereas cholesterol is more evenly distributed between the two leaflets. Similar asymmetry is also often found in other organelle membranes. The fatty acyl chains in glycerophospholipids mostly contain 16 or 18 carbon atoms and zero or few double bonds in the cis configuration. Nevertheless, longer polyunsaturated FAs (PUFAs), such as arachidonic acid (AA, C20:4), eicosapentaenoic acid (EPA, C22:5) and docosahexanoic acid (DHA, C22:6), are frequently bound to the sn-2 position [9]. In sphingolipids, instead, a marked difference in acyl chain length, often with C16:0 as the shortest species and C22-C24 as the longest species, is common [11]. The saturation degree of membrane lipids regulates membrane fluidity and cell homeostasis. Consistently, the accumulation of lipids containing saturated FAs can lead to endoplasmic reticulum stress and apoptosis [12], while high amounts of PUFAs sensitize cells to lipid peroxidation and ferroptosis, a non-apoptotic iron-dependent form of cell death [13]. The relative abundance of saturated and unsaturated FAs in membrane phospholipids primarily depends on FA availability and can be regulated by FA remodeling. In particular, the Lands’ cycle consists of a series of deacylation at the sn-2 position of glycerophospholipids by phospholipase A2 followed by reacylation by lysophospholipid acyltransferases (LPLATs) [14]. As different LPLAT isoforms differ in their affinity for FAs and lysophospholipids, this process generates diverse phospholipid species.
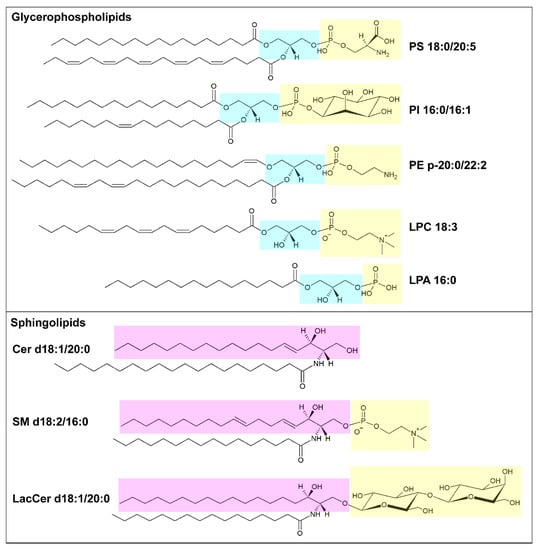
Figure 1. Schematic illustration of membrane lipids. In the upper box, example of glycerophospholipids: phosphatidylserine (PS) 18:0/20:5, phosphatidylinositol (PI) 16:0/16:1, alkenyl ether (plasmalogen) phosphatidylethanolamine (PE) p20:0/22:2, lysophosphatidylcholine (LPC) 18:3 and lysophosphatidic acid (LPA) 16:0. The glycerol moiety is marked in light blue, and the polar head is marked in yellow. In the lower box, examples of sphingolipids: ceramide (Cer) d18:1/20:0, sphingomyelin (SM) d18:2/16:0 and lactosylceramide (LacCer) d18:1/20:0. The pink box highlights the long-chain base (LCB), sphingosine, and the yellow one highlights the polar head. Structures have been made using the Structure Drawing Tools available at Lipid Maps [15].
The de novo synthesis of FAs is low in normal adult cells (except for lipogenic tissues, such as liver and adipose tissue). In contrast, increased lipogenesis is well documented in cancer cells, making them less dependent on the availability of nutrients and enabling the building of a cell membrane enriched in phospholipids containing oxidative damage-resistant saturated FAs [16]. The biosynthesis of FAs begins with the carboxylation of cytosolic acetyl-CoA by acetyl-CoA carboxylase (ACC) to produce malonyl-CoA, which becomes the substrate of FA synthase (FASN), forming palmitic acid (C16:0). Palmitate can be further elongated by FA elongases (ELOVL1-7) and desaturated by stearoyl-CoA desaturases (SCD) and FA desaturases (FADS1-3), to generate a cellular pool of non-essential FAs, such as palmitoleate (C16:1), stearate (C18:0) and oleate (C18:1). All three enzyme families involved in FA synthesis (ACC, FASN, SCD) are frequently up-regulated in several cancer types, including glioblastoma, breast, ovarian, lung, prostate and liver cancer; furthermore, their expression correlates with poorer prognosis and high cancer grade [7]. To be incorporated into membrane lipids, free FAs need to be activated to their corresponding acyl-CoA by acyl-CoA synthetases (ACS). Among ACS family members, characterized by different lengths of fatty acyl chains used as substrates, a relevant role in cancer cells has been assigned to long-chain ACS (ACSL), which activates FAs formed by 12 to 20 carbon atoms. ACSLs exhibit distinct substrate preferences, e.g., ACSL4 has a strong preference for PUFAs, and ACSL3 conjugates both PUFAs and monounsaturated FAs (MUFAs). Therefore, by regulating the activity of ACSLs, cancer cells can control the saturation degree of their membrane [17]. Various oncogenic signals, such as the PI3K/AKT/mTORC1 axis, BRAF and c-Myc, have been implicated in the upregulation of lipogenesis by activating sterol regulatory element-binding protein 1 (SREBP1), the major regulator of genes involved in FA synthesis [18][19][20]. Moreover, in some cases, cancer cells may increase the availability of free FAs by increasing TAG degradation. Hence, a higher level of monoacylglycerol lipase has been detected across various aggressive cancer types [21].
Mammals can only produce certain FAs, whereas others, such as linoleic acid (C18:2 n6) and alpha-linolenic acid (C18:3 n3), are essential and must be taken up from the diet. Cells can uptake FAs through multiple routes, including receptor-mediated endocytosis of low-density lipoproteins (LDLs) or free FA import via membrane FA transporters, such as the FA translocase CD36. In particular, elevated CD36 levels and the increased uptake of free FAs have been correlated with enhanced metastasis formation and aggressiveness in oral cancer [22]. Moreover, tumor cells can increase the availability of Fas by inducing, in neighboring adipocytes, lipolysis and mobilization of Fas via the overexpression on their surface of the FA-binding protein FABP4 [23]. Metastatic cancers, including ovarian, breast and colorectal cancer [7], are also able to promote systemic adipose tissue atrophy in a TNF-α- and IL-6-dependent manner, which may result in cancer-associated cachexia [24].
Apart from their structural and energetic functions, lipids are important for cell signaling and protein trafficking. For instance, the peculiar composition of lipid rafts (i.e., sphingolipid- and cholesterol-rich plasma membrane microdomains) can aid the clustering of receptors, such as tyrosine kinase receptors, and their downstream signaling [25][26]. Moreover, membrane lipids can act as precursors for signaling molecules. Phosphoinositides (PIPs) are phosphorylated derivates of PI that help in specifying organelle identity and intracellular transport by recruiting cytosolic proteins containing specific recognition domains [27]. Phospholipase C-mediated hydrolysis of PIPs generates two important second messengers, diacylglycerol (DAG) and inositol trisphosphate, which are implicated in multiple signal cascades [28]. PLAs release free FAs from the sn-1 and sn-2 position of phospholipids, creating lysophospholipids that can be converted into lysoPA (LPA) via cleavage of the polar group by lysophospholipase D. LPAs might bind to G protein-coupled receptors, triggering the activation of different signaling axes, such as AKT signaling, to promote cell migration and survival [29]. Interestingly, the lysophospholipase D autotaxin has recently been shown to activate a stroma-cancer signaling axis that promotes tumor progression in pancreatic cancer [30]. Moreover, phospholipid-derived free FAs may serve as precursors for lipid mediators themselves, such as in the case of AA, the substrate for the synthesis of proinflammatory eicosanoids via the cyclooxygenase pathway. One of these, prostaglandin E2 has been implicated in the establishment of a tumor-promoting microenvironment by inducing cancer cell proliferation, migration and angiogenesis [31].
In summary, lipids play an important role in tumor development, sustaining cell proliferation and metastasis formation. Dysregulated lipid metabolism is not only a key component in cancer metabolic adaptation but also creates an altered lipid profile that can distinguish tumors from normal tissues. For this reason, lipidomics may add an additional layer of information to proteomics and genomics, expanding our knowledge of lipid functions and opening the path to new opportunities for drug and biomarker development.
References
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674.
- Warburg, O.; Wind, F.; Negelein, E. The metabolism of tumors in the body. J. Gen. Physiol. 1927, 8, 519–530.
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47.
- Vander Heiden, M.G.; DeBerardinis, R.J. Understanding the Intersections between Metabolism and Cancer Biology. Cell 2017, 168, 657–669.
- Liu, J.Y.; Wellen, K.E. Advances into understanding metabolites as signaling molecules in cancer progression. Curr. Opin. Cell Biol. 2020, 63, 144–153.
- Snaebjornsson, M.T.; Janaki-Raman, S.; Schulze, A. Greasing the Wheels of the Cancer Machine: The Role of Lipid Metabolism in Cancer. Cell Metab. 2020, 31, 62–76.
- Broadfield, L.A.; Pane, A.A.; Talebi, A.; Swinnen, J.V.; Fendt, S.-M. Lipid metabolism in cancer: New perspectives and emerging mechanisms. Dev. Cell 2021, 56, 1363–1393.
- Shevchenko, A.; Simons, K. Lipidomics: Coming to grips with lipid diversity. Nat. Rev. Mol. Cell Biol. 2010, 11, 593–598.
- Ma, Y.; Temkin, S.M.; Hawkridge, A.M.; Guo, C.; Wang, W.; Wang, X.-Y.; Fang, X. Fatty acid oxidation: An emerging facet of metabolic transformation in cancer. Cancer Lett. 2018, 435, 92–100.
- Nagan, N.; Zoeller, R.A. Plasmalogens: Biosynthesis and functions. Prog. Lipid Res. 2001, 40, 199–229.
- Skotland, T.; Sagini, K.; Sandvig, K.; Llorente, A. An emerging focus on lipids in extracellular vesicles. Adv. Drug Deliv. Rev. 2020, 159, 308–321.
- Ackerman, D.; Simon, M.C. Hypoxia, lipids, and cancer: Surviving the harsh tumor microenvironment. Trends Cell Biol. 2014, 24, 472–478.
- Yang, W.S.; Stockwell, B.R. Ferroptosis: Death by Lipid Peroxidation. Trends Cell Biol. 2016, 26, 165–176.
- Wang, B.; Tontonoz, P. Phospholipid Remodeling in Physiology and Disease. Annu. Rev. Physiol. 2019, 81, 165–188.
- LIPID MAPS®. Available online: https://lipidmaps.org/resources/tools/structure (accessed on 10 October 2022).
- Rysman, E.; Brusselmans, K.; Scheys, K.; Timmermans, L.; Derua, R.; Munck, S.; Van Veldhoven, P.P.; Waltregny, D.; Daniëls, V.W.; Machiels, J.; et al. De novo Lipogenesis Protects Cancer Cells from Free Radicals and Chemotherapeutics by Promoting Membrane Lipid Saturation. Cancer Res. 2010, 70, 8117–8126.
- Klett, E.L.; Chen, S.; Yechoor, A.; Lih, F.B.; Coleman, R.A. Long-chain acyl-CoA synthetase isoforms differ in preferences for eicosanoid species and long-chain fatty acids. J. Lipid Res. 2017, 58, 884–894.
- Porstmann, T.; Santos, C.R.; Griffiths, B.; Cully, M.; Wu, M.; Leevers, S.; Griffiths, J.R.; Chung, Y.-L.; Schulze, A. SREBP Activity Is Regulated by mTORC1 and Contributes to Akt-Dependent Cell Growth. Cell Metab. 2008, 8, 224–236.
- Carroll, P.; Diolaiti, D.; McFerrin, L.; Gu, H.; Djukovic, D.; Du, J.; Cheng, P.F.; Anderson, S.; Ulrich, M.; Hurley, J.B.; et al. Deregulated Myc Requires MondoA/Mlx for Metabolic Reprogramming and Tumorigenesis. Cancer Cell 2015, 27, 271–285.
- Talebi, A.; Dehairs, J.; Rambow, F.; Rogiers, A.; Nittner, D.; Derua, R.; Vanderhoydonc, F.; Duarte, J.A.G.; Bosisio, F.; Eynde, K.V.D.; et al. Sustained SREBP-1-dependent lipogenesis as a key mediator of resistance to BRAF-targeted therapy. Nat. Commun. 2018, 9, 2500.
- Nomura, D.K.; Long, J.Z.; Niessen, S.; Hoover, H.S.; Ng, S.-W.; Cravatt, B.F. Monoacylglycerol Lipase Regulates a Fatty Acid Network that Promotes Cancer Pathogenesis. Cell 2010, 140, 49–61.
- Pascual, G.; Avgustinova, A.; Mejetta, S.; Martín, M.; Castellanos, A.; Attolini, C.S.-O.; Berenguer, A.; Prats, N.; Toll, A.; Hueto, J.A.; et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature 2017, 541, 41–45.
- Nieman, K.M.; Kenny, H.A.; Penicka, C.V.; Ladanyi, A.; Buell-Gutbrod, R.; Zillhardt, M.R.; Romero, I.L.; Carey, M.S.; Mills, G.B.; Hotamisligil, G.S.; et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat. Med. 2011, 17, 1498–1503.
- Argilés, J.M.; Busquets, S.; Stemmler, B.; López-Soriano, F.J. Cancer cachexia: Understanding the molecular basis. Nat. Rev. Cancer 2014, 14, 754–762.
- Lingwood, D.; Simons, K. Lipid Rafts As a Membrane-Organizing Principle. Science 2010, 327, 46–50.
- Irwin, M.E.; Mueller, K.L.; Bohin, N.; Ge, Y.; Boerner, J.L. Lipid raft localization of EGFR alters the response of cancer cells to the EGFR tyrosine kinase inhibitor gefitinib. J. Cell. Physiol. 2011, 226, 2316–2328.
- Schink, K.O.; Tan, K.-W.; Stenmark, H. Phosphoinositides in Control of Membrane Dynamics. Annu. Rev. Cell Dev. Biol. 2016, 32, 143–171.
- Cantley, L.; Fleischman, L.; Whitman, M. The role of lipid-derived second messengers in cell growth and transformation. Anti Cancer Drug Des. 1987, 2, 129–138.
- Moolenaar, W.H.; Perrakis, A. Insights into autotaxin: How to produce and present a lipid mediator. Nat. Rev. Mol. Cell Biol. 2011, 12, 674–679.
- Auciello, F.R.; Bulusu, V.; Oon, C.; Tait-Mulder, J.; Berry, M.; Bhattacharyya, S.; Tumanov, S.; Allen-Petersen, B.L.; Link, J.; Kendsersky, N.D.; et al. A Stromal Lysolipid–Autotaxin Signaling Axis Promotes Pancreatic Tumor Progression. Cancer Discov. 2019, 9, 617–627.
- Wang, D.; DuBois, R.N. Eicosanoids and cancer. Nat. Rev. Cancer 2010, 10, 181–193.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
741
Revisions:
2 times
(View History)
Update Date:
10 May 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




