You're using an outdated browser. Please upgrade to a modern browser for the best experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Hideo Tsubouchi | -- | 2531 | 2023-05-08 04:29:23 | | | |
| 2 | Sirius Huang | Meta information modification | 2531 | 2023-05-09 02:47:43 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Tsubouchi, H. The Hop2-Mnd1 Complex. Encyclopedia. Available online: https://encyclopedia.pub/entry/43952 (accessed on 24 December 2025).
Tsubouchi H. The Hop2-Mnd1 Complex. Encyclopedia. Available at: https://encyclopedia.pub/entry/43952. Accessed December 24, 2025.
Tsubouchi, Hideo. "The Hop2-Mnd1 Complex" Encyclopedia, https://encyclopedia.pub/entry/43952 (accessed December 24, 2025).
Tsubouchi, H. (2023, May 08). The Hop2-Mnd1 Complex. In Encyclopedia. https://encyclopedia.pub/entry/43952
Tsubouchi, Hideo. "The Hop2-Mnd1 Complex." Encyclopedia. Web. 08 May, 2023.
Copy Citation
The Hop2-Mnd1 complex was originally identified as a meiosis-specific factor that is indispensable for successful meiosis in budding yeast. Later, it was found that Hop2-Mnd1 is conserved from yeasts to humans, playing essential roles in meiosis. Accumulating evidence suggests that Hop2-Mnd1 promotes RecA-like recombinases towards homology search/strand exchange.
Dmc1
homologous recombination
meiosis
Rad51
RecA homologs
synaptonemal complex
1. Meiotic Role of the Hop2-Mnd1 Complex
1.1. Mutant Phenotypes
HOP2 was originally identified in S. cerevisiae as a meiosis-specific gene whose absence causes a complete cell cycle arrest at meiotic prophase [1]. Notably in hop2, many chromosomes form SC between nonhomologous partners. The hop2 mutant arrests at meiotic prophase with an aberrant accumulation of Dmc1 at unrepaired DSBs, indicating that HR is blocked. Homologous pairing is severely reduced in the hop2 mutant, which is more severe than that in dmc1. Accumulation of nonhomologous synapsis in the hop2 mutant lead to the proposal that Hop2 prevents synapsis between nonhomologous chromosomes [1]. MND1 was identified from a whole-genome expression screen for genes required for meiosis and spore formation [2][3]. MND1 was also identified as a multicopy suppressor of the temperature-sensitive allele of HOP2 [4]. The hop2 and mnd1 mutants essentially share the same phenotypes [1][3][4][5][6][7][8]. Both show a tight cell cycle arrest at meiotic prophase. Meiotic DSBs are poorly repaired, and homolog paring is substantially reduced. Both Rad51 and Dmc1 accumulate aberrantly on meiotic chromosomes in these mutants.
Genetic analysis in S. cerevisiae suggests that Hop2-Mnd1 functions in the same pathway as Dmc1. Although meiotic defects of the dmc1 mutant are very severe, those observed in the hop2/mnd1 mutants tend to be more severe. In a strain background where the dmc1 mutant slows down but does not completely arrest, the hop2 and mnd1 mutants show an almost complete prophase arrest [9]. Interestingly, a very small fraction of cells form spores in hop2 and mnd1, and their viability is ~10%, which is around the same level as that of the dmc1 mutant. This is in contrast to the rad51 mutant where sporulation is much higher (~30%) but spores become mostly inviable. Similarly, DSB repair defect in the hop2/mnd1 mutants is more severe than that of dmc1 [9]. By introducing the dmc1 mutation, however, the meiotic defects observed in the hop2/mnd1 mutants are suppressed to the level of the dmc1 single mutant, indicating that the dmc1 mutant is epistatic to hop2 and mnd1; ~10% of spores become viable in both the dmc1 and hop2 dmc1 mutants. Defects caused by the absence of Dmc1, or its meiosis-specific auxiliary factors Mei5 and Sae3, are partially suppressed by the overproduction of Rad51 [5]. The meiotic defects of the hop2 and mnd1 mutants are also suppressed by Rad51 overproduction. These observations strongly suggest the specific involvement of Hop2 and Mnd1 in the Dmc1 function. Homologous pairing defects in the double mutants are partially alleviated by introducing rad51 and/or dmc1 mutations, leading to the proposal that the meiotic defects in the hop2/mnd1 mutants are caused by RecA homolog-mediated aberrant activities [9].
In fission yeast S. pombe, homologues for Hop2 and Mnd1 were identified: Meu13 and Mcp7, respectively [10][11]. Although, unlike S. cerevisiae, S. pombe cells do not form the SC, the basic phenotypes of these S. pombe mutants are analogous to those found in S. cerevisiae. Both mutants show a substantial cell cycle delay during meiosis, which is suppressed by the absence of meiotic DSBs [12]. Homologous pairing and crossing over are heavily compromised in these mutants. The dmc1 mutant is epistatic to mcp7 with respect to meiotic cell cycle, spore viability, and crossing over [11].
In mice, Hop2 knockout mice are infertile in both male and female, consistent with its primary role in meiosis [13]. Spermatocytes from knockout mice arrest prior to the pachytene stage with aberrant accumulation of RAD51 and DMC1, eventually leading to apoptosis. They also accumulate eukaryotic ssDNA binding protein RPA and show persistent γ-H2AX signals. Homologous pairing and formation of the SC are greatly reduced, while formation of chromosome axes is barely affected. Chromosome synapsis is limited, and some advanced cells have SC formed between nonhomologous chromosomes. All these phenotypes are in line with those found in the results obtained from yeast. One striking difference is in the relationship between DMC1 and HOP2. The absence of HOP2 causes a more severe defect in chromosome synapsis than the absence of Dmc1 in mice. The double Hop2−/− Dmc1−/− mice, however, show a similar synapsis defect to the Hop2−/− mice. This is in sharp contrast to the observations in budding and fission yeast in which the dmc1 mutant is epistatic to hop2 (i.e., the dmc1 hop2 mutant behaves like dmc1). Perhaps Hop2 has another function in mice, possibly at an earlier stage, before the loading of RecA homologs onto ssDNA.
Hop2-Mnd1 roles have been extensively studied in a plant model Arabidopsis thaliana. The hop2 (the HOP2 homolog is called AHP2 in Arabidopsis) or mnd1 mutant plants are sterile [14][15][16][17][18]. They are defective in homologous pairing, and chromosomes look entangled, likely reflecting synapsis between nonhomologous chromosomes. At the stage of prophase corresponding to pachytene and later, chromosome fragmentation becomes evident, indicating their defects in DSB repair. The Hop2-Mnd1 complex likely plays an important role in stabilizing homolog association and preventing connections between nonhomologous chromosomes [19][20]. Unlike the case with S. cerevisiae or mice, where hop2/mnd1 mutants arrest the cell cycle or undergo associated apoptosis, meiosis of hop2/mnd1 mutants is not arrested in Arabidopsis. This is likely because the DNA damage checkpoint seems less stringent in Arabidopsis meiosis, which is reminiscent of S. pombe meiosis. Meiosis is completed in dmc1 mutants in both S. pombe and A. thaliana [21][22]. It is likely that Hop2-Mnd1 functions exclusively with Dmc1 because the hop2 mnd1 dmc1 triple mutant shows the same phenotype as that of dmc1 single in which DSBs are repaired through Rad51-mediated intersister HR [23].
In the ciliated protist Tetrahymena thermophila, there are two homologs for Hop2 and Mnd1, named Hop2A, Hop2B, Mnd1A, and Mnd1B [24]. Hop2A and Mnd1A are meiosis-specific, while Hop2B and Mnd1B are also expressed in somatic cells. The attempt to knockout HOP2B was not successful, suggesting its essential role in this organism. In general, the meiotic defects of the hop2A mutant are mild. Meiotic DSBs are likely formed and repaired because γ-H2AX and Rad51 signals appear transiently along the course of meiotic prophase. However, mostly univalents are found at anaphase I, suggesting a defect in crossing over. Homologous pairing is reduced only mildly. The presence of HOP2B may account for these relatively mild phenotypes of the hop2A mutant. Tetrahymena chromosomes at mid meiosis exhibit a unique arrangement where telomeres are clustered at one pole and this bouquet-like configuration is constrained inside the meiotically elongated micronucleus. This unique meiotic configuration may account for the relatively high homologous pairing observed in the absence of Hop2A or Spo11 (and thus no meiotic DSBs).
1.2. Localization of Hop2-Mnd1
One unique feature of Hop2 and Mnd1 is their meiotic localization pattern. The meiotic localization of Hop2 and Mnd1, in S. cerevisiae, appears to punctate at early prophase, which becomes more intense as chromosomes condense towards the pachytene stage [1][4][6]. Hop2/Mnd1 foci do not overlap with Rad51. Importantly, this localization is neither abolished by the absence of meiotic DSBs nor enriched at recombination hot spots [1][4][6]. This is in sharp contrast to the typical localization pattern of RecA homologs and their auxiliary factors. Rad51 and Dmc1 appear as chromosome-associated foci that transiently appear and disappear during the meiotic prophase. This localization pattern coincides with the kinetics of HR maturation. These signals are believed to represent the binding of Rad51 and Dmc1 to the ssDNA produced at DSB ends. This idea is supported by their localization dependency on meiotic DSB formation. Similar localization patterns of Hop2/Mnd1 are observed in S. pombe [10][11], Arabidopsis [15][16], and the rice Oryza sativa [25]. In Arabidopsis, although the spatial distribution of Hop2 and Mnd1 shows substantial overlap as foci, the more diffusive Mnd1 signal beyond foci also appears all along the chromosomes, covering a wider area than Hop2. This trend is most prominent at the leptotene and at the nucleolus [15][16]. Super-resolution imaging was used to closely examine Hop2 localization using O. sativa [25]. OsHop2 is localized to mostly chromatin loops, some of which overlap with chromosome axes and the central region of the SC. Interaction between OsHop2 and Zep1, the transverse filament protein of the SC in rice, is detected on the yeast two-hybrid system, suggesting a possible interplay between Hop2 and the SC [25].
2. Structure and Biochemical Properties of the Hop2-Mnd1 Complex
2.1. Structure
Hop2 interacts with Mnd1 and forms the Hop2-Mnd1 complex. A crystallographic structure of the Giardia lamblia Hop2-Mnd1 complex has been elucidated [26] (Figure 1A,B). Overall, the complex forms a curved, rod-like structure with one end linked to two juxtaposed winged-helix domains (WHD) consisting of the N-terminal residues of each protein, while the other is capped by extra alpha-helices that form a helical bundle structure. Their association is supported by a pair of parallelly arranged alpha-helices. Each helix domain carries three consecutive leucine zippers (LZ1, 2, 3) interrupted by two non-helical regions that form kinked junctions in the complex. The C-terminal leucine zipper of Hop2 and Mnd1 folds back onto LZ3, forming a cap made of alpha-helical bundles, thus designated as LZ3wCH (leucine zipper 3 with capping helices). The WHD domain has a positively charged patch, likely responsible for specific binding to dsDNA binding [27][28][29][30]. On the other hand, the LZ3wCH domain, located on the opposite side, is responsible for binding the Dmc1 nucleoprotein filament.
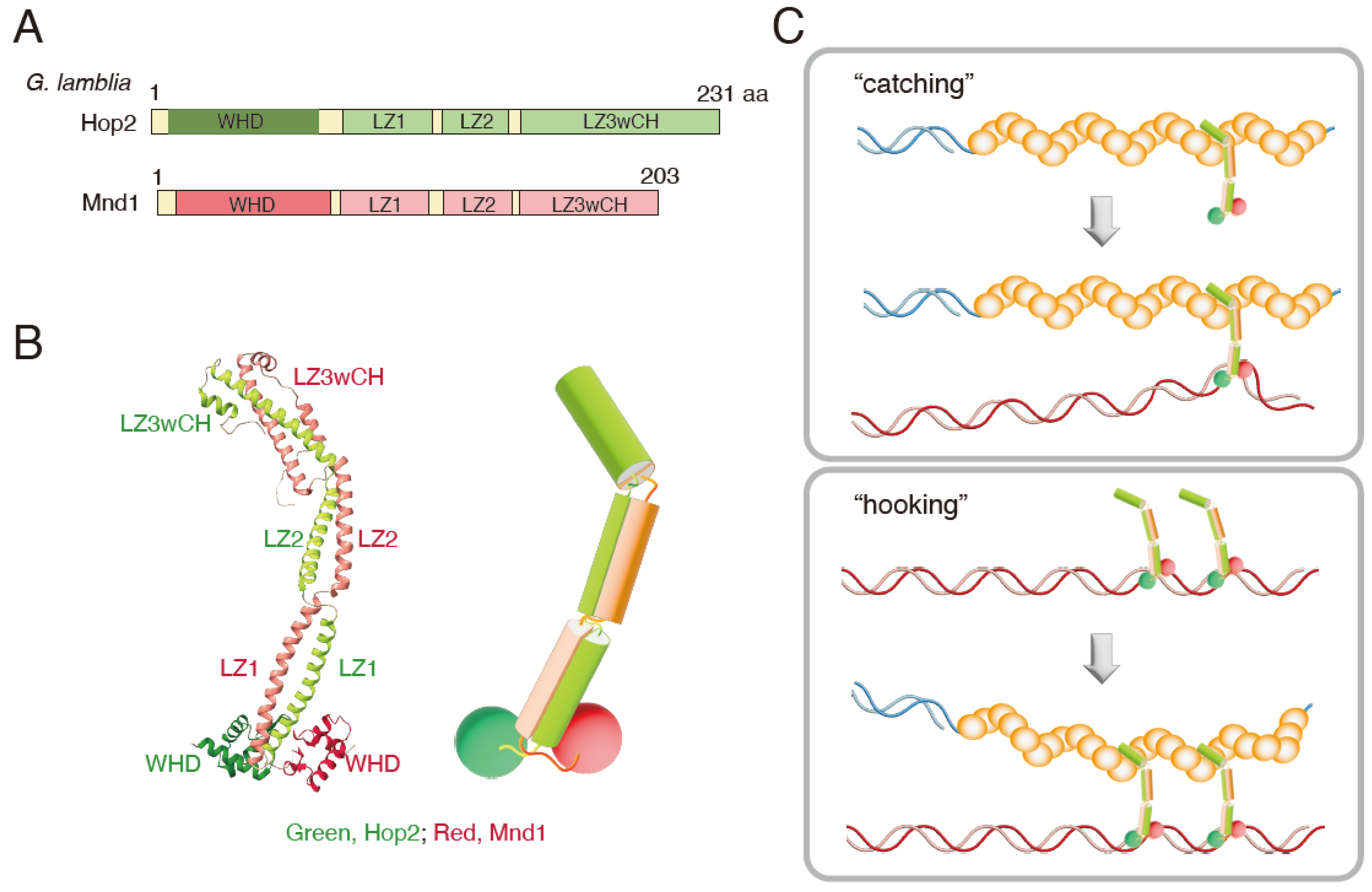
Figure 1. (A) Domain organizations of G. lamblia Hop2 and Mnd1. WHD, winged-helix domain; LZ, leucine zipper; LZwCH, leucine zipper with capping helices. (B) Left, crystal structure of the G. lamblia Hop2-Mnd1 complex (4Y66, chains D and C). Right, schematic drawing of the Hop2-Mnd1 complex. (C) Two models of the function of Hop2-Mnd1 in homologous recombination. See the main text for details.
So far, G. lamblia Hop2-Mnd1 is the only complex whose structure has been determined. A combination of cross-linking and mass spectrometry reveals that the Arabidopsis Hop2-Mnd1 complex takes a parallel configuration, like that of G. lamblia [31]. At the same time, the results suggest the possible coexistence of at least two configurations, open and elongated, and closed and folded. The results also suggest the highly flexible nature of the complex. Interestingly, the mouse HOP2-MND1 complex was proposed to take a V-shape configuration in solution, based on the analysis using a combination of small-angle X-ray scattering and electron microscopy. Two domains for dsDNA-binding (two WHD domains, one for HOP2 and the other for MND1) are separately located on the arms of the V shape. The third DNA binding domain, which preferentially binds ssDNA, is at the C-terminus of HOP2. This is located at the bundled region of the V shape that plays a critical role in interacting with RAD51/DMC1 [29][32]. Variations in the configuration of Hop2-Mnd1 from different species could be due to the rather flexible nature of this complex.
2.2. Biochemical Activity
The biochemical properties of mammalian HOP2-MND1 have been extensively studied. Mouse and human HOP2-MND1 preferentially bind dsDNA over ssDNA, interact with both human RAD51 and DMC1, and promote RAD51- and DMC1-driven D-loop formation and strand exchange [27][33][34][35][36][37]. Mouse HOP2-MND1 promotes at least two critical steps of RAD51- and DMC1-mediated reactions: one at the presynaptic filament formation step and the other at the step where the presynaptic filament catches dsDNA to assemble the synaptic complex [35][36]. In the latter step, dsDNA capture was assayed by pulling down presynaptic filaments pre-mixed with dsDNA. Importantly, homology between ssDNA in the presynaptic filament and dsDNA is not a prerequisite for efficient dsDNA capture. Combined with the above-mentioned G. lamblia Hop2-Mnd1 structure, LZ3wCH is likely involved in promoting presynaptic filament formation while WHD is responsible for dsDNA binding [26]. HOP2-MND1 stimulates DNA network formation where nucleoprotein filaments containing either Rad51 or Dmc1 associate with multiple dsDNA molecules simultaneously [38]. Single molecular analysis shows that HOP2-MND1 condenses dsDNA [38]. It is interesting to note that E. coli RecA can condense DNA into networks while Dmc1 or Rad51, by itself, cannot. Hop2-Mnd1 might provide such an activity to eukaryotic RecA homologs to achieve an efficient homology search. HOP2-MND1 also induces conformational changes of RAD51 that modulate RAD51 binding to nucleotide cofactors and DNA [39]. HOP2-MND1 circumvents the metal ion requirement of RAD51 for ATP binding as well as increases RAD51 binding preference for ssDNA.
Mouse HOP2 promotes D-loop formation by itself, while HOP2-MND1 does not. This raises the possibility that HOP2 can function independently from MND1, and that the intrinsic activity of HOP2 is quenched by MND1 in the complex [33][40]. According to another report, mouse HOP2 showed little D-loop formation activity on its own, but promoted DMC1-driven D-loop formation without MND1 [41]. Indeed, in the Mnd1 knockout mouse where Hop2 is intact, spermatocytes show a much higher level of chromosome synapsis, with most meiotically formed DSBs repaired. This raises the possibility that HOP2, either by itself or together with DMC1, supports homologous pairing and synapsis [40]. Further investigation will be necessary to address whether and how HOP2 functions without MND1 in wild type meiosis.
The characterization of the S. cerevisiae Hop2-Mnd1 complex initially lagged because the original annotation of the HOP2 coding sequence (CDS) turned out to be erroneous [8][42]. Nonetheless, the Hop2-Mnd1 complex, using correctly annotated CDS, robustly promotes Dmc1-driven D-loop formation, while it had no effect on Rad51 [42]. Interaction between presynaptic filaments and Hop2-Mnd1 was examined using DNA curtains and single-molecule imaging [43]. Hop2-Mnd1 specifically binds Dmc1-ssDNA filaments, not Rad51 filaments. The association is characterized as quick in binding and slow in dissociation. Such interaction specificity might contribute to the preferential usage of Dmc1 in S. cerevisiae meiosis.
S. pombe Hop2-Mnd1 promotes Dmc1-driven D-loop formation and strand exchange [37][44]. Interestingly, Hop2-Mnd1 promotes the production of hyper joint molecules, likely a DNA network consisting of associated ss and dsDNA. This is reminiscent of what was observed with mouse Hop2-Mnd1 [38]. S. pombe Hop2-Mnd1 interacts with both Dmc1 and Rad51, but it stimulates neither D-loop formation nor strand exchange driven by Rad51. A minor activation of Rad51 was detected when Hop2-Mnd1 was combined with Swi5-Sfr1, a Rad51 auxiliary factor [44][45]. Just like mouse, S. pombe Hop2-Mnd1 promotes dsDNA binding by the Dmc1 presynaptic filaments in a homology-independent manner. Hop2-Mnd1 does not stabilize Dmc1 presynaptic filaments in a sense that it does not reduce dissociation of Dmc1 from ssDNA. Similarly, Hop2-Mnd1 is not an efficient mediator of Dmc1 either (i.e., it does not promote the replacement of RPA preassembled on ssDNA with Dmc1). Efficient strand exchange requires Hop2-Mnd1, especially when homology is embedded in the middle of the donor dsDNA, suggesting its critical role in initiating strand exchange [44].
In other organisms, A. thaliana Hop2-Mnd1 stimulates Dmc1-mediated D-loop formation [23]. The Entamoeba histolytica Hop2-Mnd1 interacts with Dmc1 and also promotes Dmc1-driven D-loop formation and strand exchange [46]. These observations are essentially in line with the reported biochemical properties of Hop2-Mnd1 from other organisms.
References
- Leu, J.-Y.; Chua, P.R.; Roeder, G.S. The Meiosis-Specific Hop2 Protein of S. Cerevisiae Ensures Synapsis between Homologous Chromosomes. Cell 1998, 94, 375–386.
- Rabitsch, K.P.; Tóth, A.; Gálová, M.; Schleiffer, A.; Schaffner, G.; Aigner, E.; Rupp, C.; Penkner, A.M.; Moreno-Borchart, A.C.; Primig, M.; et al. A Screen for Genes Required for Meiosis and Spore Formation Based on Whole-Genome Expression. Curr. Biol. 2001, 11, 1001–1009.
- Gerton, J.L.; DeRisi, J.L. Mnd1p: An Evolutionarily Conserved Protein Required for Meiotic Recombination. Proc. Natl. Acad. Sci. USA 2002, 99, 6895–6900.
- Tsubouchi, H.; Roeder, G.S. The Mnd1 Protein Forms a Complex with Hop2 To Promote Homologous Chromosome Pairing and Meiotic Double-Strand Break Repair. Mol. Cell Biol. 2002, 22, 3078–3088.
- Tsubouchi, H.; Roeder, G.S. The Budding Yeast Mei5 and Sae3 Proteins Act Together with Dmc1 during Meiotic Recombination. Genetics 2004, 168, 1219–1230.
- Zierhut, C.; Berlinger, M.; Rupp, C.; Shinohara, A.; Klein, F. Mnd1 Is Required for Meiotic Interhomolog Repair. Curr. Biol. 2004, 14, 752–762.
- Henry, J.M.; Camahort, R.; Rice, D.A.; Florens, L.; Swanson, S.K.; Washburn, M.P.; Gerton, J.L. Mnd1/Hop2 Facilitates Dmc1-Dependent Interhomolog Crossover Formation in Meiosis of Budding Yeast. Mol. Cell Biol. 2006, 26, 2913–2923.
- Chen, Y.-K.; Leng, C.-H.; Olivares, H.; Lee, M.-H.; Chang, Y.-C.; Kung, W.-M.; Ti, S.-C.; Lo, Y.-H.; Wang, A.H.-J.; Chang, C.-S.; et al. Heterodimeric Complexes of Hop2 and Mnd1 Function with Dmc1 to Promote Meiotic Homolog Juxtaposition and Strand Assimilation. Proc. Natl. Acad. Sci. USA 2004, 101, 10572–10577.
- Tsubouchi, H.; Roeder, G.S. The Importance of Genetic Recombination for Fidelity of Chromosome Pairing in Meiosis. Dev. Cell 2003, 5, 915–925.
- Nabeshima, K.; Kakihara, Y.; Hiraoka, Y.; Nojima, H. A Novel Meiosis-Specific Protein of Fission Yeast, Meu13p, Promotes Homologous Pairing Independently of Homologous Recombination. EMBO J. 2001, 20, 3871–3881.
- Saito, T.T.; Tougan, T.; Kasama, T.; Okuzaki, D.; Nojima, H. Mcp7, a Meiosis-Specific Coiled-Coil Protein of Fission Yeast, Associates with Meu13 and Is Required for Meiotic Recombination. Nucleic Acids Res. 2004, 32, 3325–3339.
- Shimada, M.; Nabeshima, K.; Tougan, T.; Nojima, H. The Meiotic Recombination Checkpoint Is Regulated by Checkpoint Rad+genes in Fission Yeast. EMBO J. 2002, 21, 2807–2818.
- Petukhova, G.; Romanienko, P.J.; Camerini-Otero, R.D. The Hop2 Protein Has a Direct Role in Promoting Interhomolog Interactions during Mouse Meiosis. Dev. Cell 2003, 5, 927–936.
- Schommer, C.; Beven, A.; Lawrenson, T.; Shaw, P.; Sablowski, R. AHP2 Is Required for Bivalent Formation and for Segregation of Homologous Chromosomes in Arabidopsis Meiosis. Plant J. 2003, 36, 1–11.
- Stronghill, P.; Pathan, N.; Ha, H.; Supijono, E.; Hasenkampf, C. Ahp2 (Hop2) Function in Arabidopsis Thaliana (Ler) Is Required for Stabilization of Close Alignment and Synaptonemal Complex Formation except for the Two Short Arms That Contain Nucleolus Organizer Regions. Chromosoma 2010, 119, 443–458.
- Vignard, J.; Siwiec, T.; Chelysheva, L.; Vrielynck, N.; Gonord, F.; Armstrong, S.J.; Schlögelhofer, P.; Mercier, R. The Interplay of RecA-Related Proteins and the MND1-HOP2 Complex during Meiosis in Arabidopsis Thaliana. PLoS Genet. 2007, 3, 1894–1906.
- Panoli, A.P.; Ravi, M.; Sebastian, J.; Nishal, B.; Reddy, T.; Marimuthu, M.P.; Subbiah, V.; Vijaybhaskar, V.; Siddiqi, I. AtMND1 Is Required for Homologous Pairing during Meiosis in Arabidopsis. BMC Mol. Biol. 2006, 7, 24.
- Kerzendorfer, C.; Vignard, J.; Pedrosa-Harand, A.; Siwiec, T.; Akimcheva, S.; Jolivet, S.; Sablowski, R.; Armstrong, S.; Schweizer, D.; Mercier, R.; et al. The Arabidopsis Thaliana MND1 Homologue Plays a Key Role in Meiotic Homologous Pairing, Synapsis and Recombination. J. Cell Sci. 2006, 119, 2486–2496.
- Pathan, N.; Stronghill, P.; Hasenkampf, C. Transmission Electron Microscopy and Serial Reconstructions Reveal Novel Meiotic Phenotypes for the Ahp2 Mutant of Arabidopsis Thaliana. Genome 2013, 56, 139–145.
- Farahani-Tafreshi, Y.; Wei, C.; Gan, P.; Daradur, J.; Riggs, C.D.; Hasenkampf, C.A. The Arabidopsis HOP2 Gene Has a Role in Preventing Illegitimate Connections between Nonhomologous Chromosome Regions. Chromosome Res. 2022, 30, 59–75.
- Fukushima, K.; Tanaka, Y.; Nabeshima, K.; Yoneki, T.; Tougan, T.; Tanaka, S.; Nojima, H. Dmc1 of Schizosaccharomyces Pombe Plays a Role in Meiotic Recombination. Nucleic Acids Res. 2000, 28, 2709–2716.
- Couteau, F.; Belzile, F.; Horlow, C.; Grandjean, O.; Vezon, D.; Doutriaux, M.P. Random Chromosome Segregation without Meiotic Arrest in Both Male and Female Meiocytes of a Dmc1 Mutant of Arabidopsis. Plant Cell 1999, 11, 1623–1634.
- Uanschou, C.; Ronceret, A.; von Harder, M.; de Muyt, A.; Vezon, D.; Pereira, L.; Chelysheva, L.; Kobayashi, W.; Kurumizaka, H.; Schlögelhofer, P.; et al. Sufficient Amounts of Functional HOP2/MND1 Complex Promote Interhomolog DNA Repair but Are Dispensable for Intersister DNA Repair during Meiosis in Arabidopsis. Plant Cell 2013, 25, 4924–4940.
- Mochizuki, K.; Novatchkova, M.; Loidl, J. DNA Double-Strand Breaks, but Not Crossovers, Are Required for the Reorganization of Meiotic Nuclei in Tetrahymena. J. Cell Sci. 2008, 121, 2148–2158.
- Shi, W.; Tang, D.; Shen, Y.; Xue, Z.; Zhang, F.; Zhang, C.; Ren, L.; Liu, C.; Du, G.; Li, Y.; et al. OsHOP2 Regulates the Maturation of Crossovers by Promoting Homologous Pairing and Synapsis in Rice Meiosis. New Phytol. 2019, 222, 805–819.
- Kang, H.A.; Shin, H.C.; Kalantzi, A.S.; Toseland, C.P.; Kim, H.M.; Gruber, S.; Peraro, M.D.; Oh, B.H. Crystal Structure of Hop2-Mnd1 and Mechanistic Insights into Its Role in Meiotic Recombination. Nucleic Acids Res. 2015, 43, 3841–3856.
- Pezza, R.J.; Petukhova, G.; Ghirlando, R.; Camerini-Otero, R.D. Molecular Activities of Meiosis-Specific Proteins Hop2, Mnd1, and the Hop2-Mnd1 Complex. J. Biol. Chem. 2006, 281, 18426–18434.
- Moktan, H.; Guiraldelli, M.F.; Eyster, C.A.; Zhao, W.; Lee, C.Y.; Mather, T.; Camerini-Otero, R.D.; Sung, P.; Zhou, D.H.; Pezza, R.J. Solution Structure and DNA-Binding Properties of the Winged Helix Domain of the Meiotic Recombination HOP2 Protein. J. Biol. Chem. 2014, 289, 14682–14691.
- Zhao, W.; Saro, D.; Hammel, M.; Kwon, Y.; Xu, Y.; Rambo, R.P.; Williams, G.J.; Chi, P.; Lu, L.; Pezza, R.J.; et al. Mechanistic Insights into the Role of Hop2-Mnd1 in Meiotic Homologous DNA Pairing. Nucleic Acids Res. 2014, 42, 906–917.
- Moktan, H.; Zhou, D.H. Wing 1 of Protein HOP2 Is as Important as Helix 3 in DNA Binding by MD Simulation. J. Biomol. Struct. Dyn. 2018, 36, 1853–1866.
- Rampler, E.; Stranzl, T.; Orban-Nemeth, Z.; Hollenstein, D.M.; Hudecz, O.; Schloegelhofer, P.; Mechtler, K. Comprehensive Cross-Linking Mass Spectrometry Reveals Parallel Orientation and Flexible Conformations of Plant HOP2-MND1. J. Proteome Res. 2015, 14, 5048–5062.
- Zhao, W.; Sung, P. Significance of Ligand Interactions Involving Hop2-Mnd1 and the RAD51 and DMC1 Recombinases in Homologous DNA Repair and XX Ovarian Dysgenesis. Nucleic Acids Res. 2015, 43, 4055–4066.
- Petukhova, G.; Pezza, R.J.; Vanevski, F.; Ploquin, M.; Masson, J.Y.; Camerini-Otero, R.D. The Hop2 and Mnd1 Proteins Act in Concert with Rad51 and Dmc1 in Meiotic Recombination. Nat. Struct. Mol. Biol. 2005, 12, 449–453.
- Enomoto, R.; Kinebuchi, T.; Sato, M.; Yagi, H.; Kurumizaka, H.; Yokoyama, S. Stimulation of DNA Strand Exchange by the Human TBPIP/Hop2-Mnd1 Complex. J. Biol. Chem. 2006, 281, 5575–5581.
- Pezza, R.J.; Voloshin, O.N.; Vanevski, F.; Camerini-Otero, R.D. Hop2/Mnd1 Acts on Two Critical Steps in Dmc1-Promoted Homologous Pairing. Genes Dev. 2007, 21, 1758–1766.
- Chi, P.; San Filippo, J.; Sehorn, M.G.; Petukhova, G.; Sung, P. Bipartite Stimulatory Action of the Hop2-Mnd1 Complex on the Rad51 Recombinase. Genes Dev. 2007, 21, 1747–1757.
- Ploquin, M.; Petukhova, G.; Morneau, D.; Déry, U.; Bransi, A.; Stasiak, A.; Camerini-Otero, R.D.; Masson, J.Y. Stimulation of Fission Yeast and Mouse Hop2-Mnd1 of the Dmc1 and Rad51 Recombinases. Nucleic Acids Res. 2007, 35, 2719–2733.
- Pezza, R.J.; Camerini-Otero, R.D.; Bianco, P.R. Hop2-Mnd1 Condenses DNA to Stimulate the Synapsis Phase of DNA Strand Exchange. Biophys. J. 2010, 99, 3763–3772.
- Bugreev, D.; Huang, F.; Mazina, O.M.; Pezza, R.J.; Voloshin, O.N.; Daniel Camerini-Otero, R.; Mazin, A. HOP2-MND1 Modulates RAD51 Binding to Nucleotides and DNA. Nat. Commun. 2014, 5, 4198.
- Pezza, R.J.; Voloshin, O.N.; Volodin, A.A.; Boateng, K.A.; Bellani, M.A.; Mazin, A.; Camerini-Otero, R.D. The Dual Role of HOP2 in Mammalian Meiotic Homologous Recombination. Nucleic Acids Res. 2014, 42, 2346–2357.
- Enomoto, R.; Kinebuchi, T.; Sato, M.; Yagi, H.; Shibata, T.; Kurumizaka, H.; Yokoyama, S. Positive Role of the Mammalian TBPIP/HOP2 Protein in DMC1-Mediated Homologous Pairing. J. Biol. Chem. 2004, 279, 35263–35272.
- Chan, Y.L.; Brown, M.S.; Qin, D.; Handa, N.; Bishop, D.K. The Third Exon of the Budding Yeast Meiotic Recombination Gene HOP2 Is Required for Calcium-Dependent and Recombinase Dmc1-Specific Stimulation of Homologous Strand Assimilation. J. Biol. Chem. 2014, 289, 18076–18086.
- Crickard, J.B.; Kwon, Y.; Sung, P.; Greene, E.C. Dynamic Interactions of the Homologous Pairing 2 (Hop2)–Meiotic Nuclear Divisions 1 (Mnd1) Protein Complex with Meiotic Presynaptic Filaments in Budding Yeast. J. Biol. Chem. 2019, 294, 490–501.
- Tsubouchi, H.; Argunhan, B.; Ito, K.; Takahashi, M.; Iwasaki, H. Two Auxiliary Factors Promote Dmc1-Driven DNA Strand Exchange via Stepwise Mechanisms. Proc. Natl. Acad. Sci. USA 2020, 117, 12062–12070.
- Haruta, N.; Kurokawa, Y.; Murayama, Y.; Akamatsu, Y.; Unzai, S.; Tsutsui, Y.; Iwasaki, H. The Swi5-Sfr1 Complex Stimulates Rhp51/Rad51—and Dmc1-Mediated DNA Strand Exchange in Vitro. Nat. Struct. Mol. Biol. 2006, 13, 823–830.
- Kelso, A.A.; Say, A.F.; Sharma, D.; Ledford, L.A.L.; Turchick, A.; Saski, C.A.; King, A.; Attaway, C.C.; Temesvari, L.A.; Sehorn, M.G.; et al. Entamoeba Histolytica Dmc1 Catalyzes Homologous DNA Pairing and Strand Exchange That Is Stimulated by Calcium and Hop2-Mnd1. PLoS ONE 2015, 10, e0139399.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
764
Revisions:
2 times
(View History)
Update Date:
09 May 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




