Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Anne Galinier | -- | 4389 | 2023-05-05 17:33:08 | | | |
| 2 | Sirius Huang | + 117 word(s) | 4506 | 2023-05-08 11:51:58 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Galinier, A.; Delan-Forino, C.; Foulquier, E.; Lakhal, H.; Pompeo, F. Peptidoglycan Synthesis and Regulation in Bacteria. Encyclopedia. Available online: https://encyclopedia.pub/entry/43888 (accessed on 14 January 2026).
Galinier A, Delan-Forino C, Foulquier E, Lakhal H, Pompeo F. Peptidoglycan Synthesis and Regulation in Bacteria. Encyclopedia. Available at: https://encyclopedia.pub/entry/43888. Accessed January 14, 2026.
Galinier, Anne, Clémentine Delan-Forino, Elodie Foulquier, Hakima Lakhal, Frédérique Pompeo. "Peptidoglycan Synthesis and Regulation in Bacteria" Encyclopedia, https://encyclopedia.pub/entry/43888 (accessed January 14, 2026).
Galinier, A., Delan-Forino, C., Foulquier, E., Lakhal, H., & Pompeo, F. (2023, May 05). Peptidoglycan Synthesis and Regulation in Bacteria. In Encyclopedia. https://encyclopedia.pub/entry/43888
Galinier, Anne, et al. "Peptidoglycan Synthesis and Regulation in Bacteria." Encyclopedia. Web. 05 May, 2023.
Copy Citation
Peptidoglycan is a three-dimensional polymer that enables bacteria to resist cytoplasmic osmotic pressure, maintain their cell shape and protect themselves from environmental threats. Numerous antibiotics target enzymes involved in the synthesis of the cell wall, particularly peptidoglycan synthases.
peptidoglycan
PBP
SEDS
hydrolases
1. Introduction
Peptidoglycan (PG) plays a vital role in protecting bacteria from external stress and cytoplasmic pressure while maintaining its morphology [1][2]. This polymer comprises glycan chains crosslinked by short peptides and surrounds the lipid bilayer cytoplasmic membrane [2]. In Gram-negative bacteria with an outer membrane, such as Escherichia coli, PG forms a thin layer, while in Gram-positive bacteria without an external membrane, such as Bacillus subtilis, it forms a thick layer. Throughout a bacterium’s life, PG is constantly synthesized, remodeled and repaired to enable cell elongation and division [1][3][4]. The newly synthesized PG is integrated into the existing layer, while the old PG is simultaneously released. These processes are mediated by enzymes such as PG synthases and hydrolases, which belong to dynamic complexes that are spatially and temporally regulated [3][5].
The various steps Involved in PG synthesis have been described in numerous studies and reviews, and this section highlights the main ones (Figure 1) [1][6]. The initial stages of PG synthesis occur in the cytoplasm [7]. Uridine diphosphate N-acetylglucosamine (UDP-GlcNAc), a precursor of PG, is synthesized from fructose-6-phosphate via the hexosamine pathway [8][9]. MurA and MurB enzymes catalyze the formation of UDP-N-acetylmuramic acid (MurNAc) from UDP-GlcNAc and phosphoenolpyruvate [10][11]. Five amino acids, including D-amino acids, are then successively added to form UDP-MurNAc-pentapeptide. The composition of the pentapeptide depends on bacteria but is similar in E. coli [12] and in B. subtilis [13] and is L-Ala–D-Glu–m-A2pm–D-Ala–D-Ala.
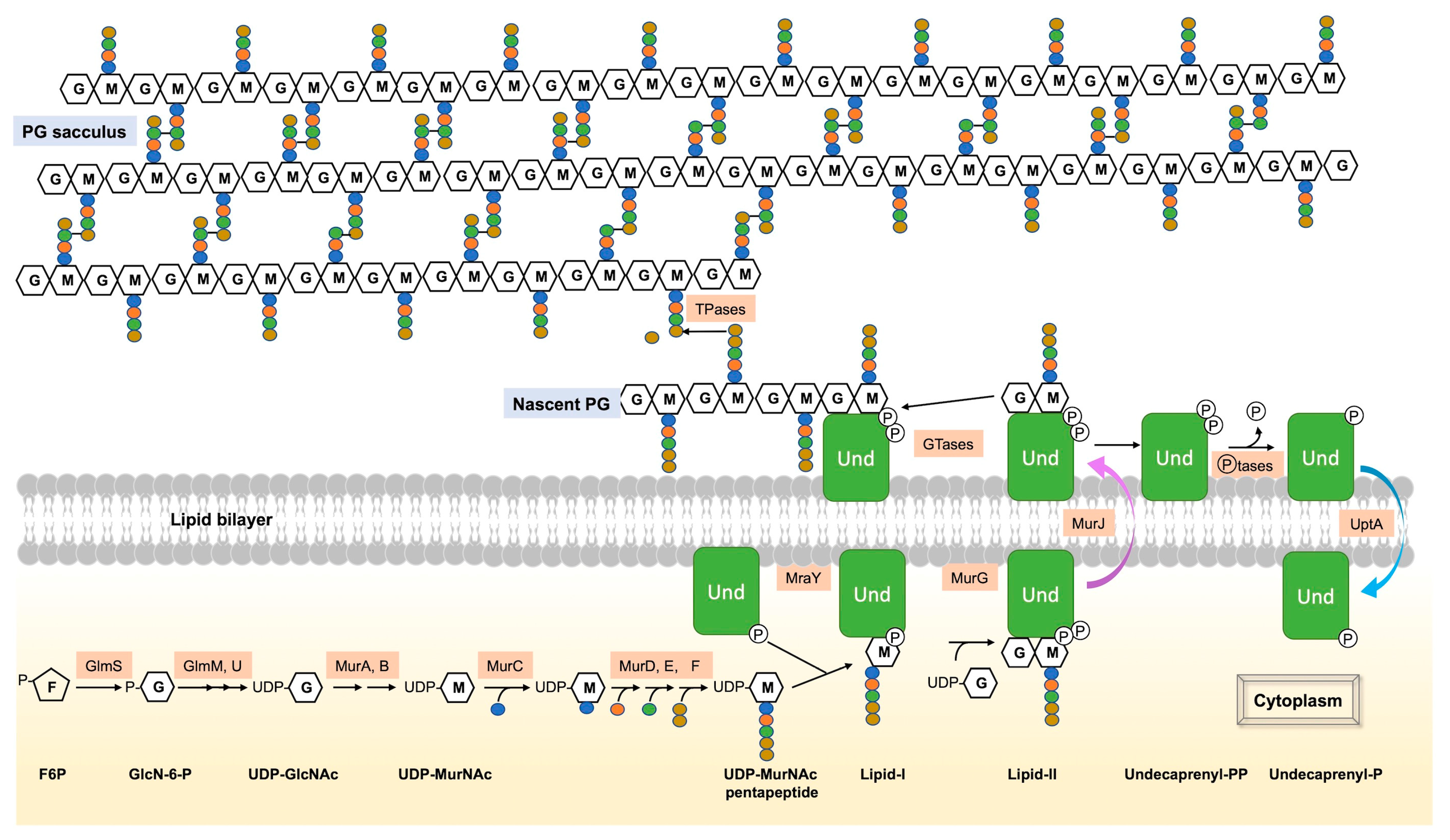
Figure 1. Summary of the main steps of PG synthesis. PG synthesis begins with a series of steps in the cytoplasm. The initial PG precursor, UDP-GlcNAc, is formed from F6P via the hexosamine pathway. UDP-MurNAc is then synthesized from UDP-GlcNAc by the enzymes MurA and MurB. Successive addition of five amino acids forms UDP-MurNAc-pentapeptide. The subsequent steps of PG synthesis take place on the inner face of the cytoplasmic membrane. MraY catalyzes the combination of UDP-MurNAc-pentapeptide with UndP to form Lipid-I, which is then modified by the transfer of UDP-GlcNAc to the MurNAc unit to form Lipid-II by MurG. Flippase MurJ facilitates the translocation of Lipid-II across the cytoplasmic membrane. On the external face of the membrane, GTases and TPases utilize the disaccharide pentapeptide of Lipid-II as a substrate for PG polymerization and synthesis of the PG layer (sacculus) in the periplasm (Gram-negative) or exterior (Gram-positive) of the cell. UndPP, released during PG synthesis, is dephosphorylated by membrane phosphatases (such as BacA, YbjG, PgpB, and LpxT in E. coli or UppP and BcrC in B. subtilis), and the resulting UndP is recycled to the cytoplasm by the UndP transporter UptA of the DedA superfamily, found in both B. subtilis and E. coli.
The subsequent steps of PG assembly take place on the inner face of the cytoplasmic membrane with enzymes bound to the membrane. In both Gram-positive and Gram-negative bacteria, MraY catalyzes the initial membrane step, forming undecaprenyl-phosphate-N-acetylmuramyl-pentapeptide (Lipid-I) from UDP-MurNAc-pentapeptide and undecaprenyl-phosphate (UndP), a C55 polyisoprenoid lipid phosphate, [14][15][16]. The final intracellular step involves the transfer of a molecule of UDP-GlcNAc to the MurNAc of Lipid-I under the action of MurG, forming Lipid-II (UndPP-GlcNAc-MurNAc-pentapeptide) [17]. Lipid-II is then translocated across the cytoplasmic membrane by the flippase MurJ to the periplasm (Gram-negative) or the exterior (Gram-positive) [18]. The disaccharide-pentapeptide part of Lipid-II serves as a substrate for PG synthases, i.e., PBPs and Shape, Elongation, Division and Sporulation (SEDS) proteins, for polymerization and cross-linking to the PG layer, also named sacculus. Finally, the remaining undecaprenyl pyrophosphate (UndPP) is then dephosphorylated by membrane phosphatases to be transported and recycled as UndP to the cytoplasm [19][20].
2. Regulation of GlcN-6-P Synthesis, the Initial Cytoplasmic Precursor of UDP-GlcNAc
The synthesis of PG begins in the cytoplasm with the formation of UDP-GlcNAc (Figure 1). The UDP-GlcNAc biosynthesis pathway involves four steps and three enzymes: GlmS, GlmM and GlmU, first identified in E. coli [21]. GlmM and GlmU are essential. In contrast, GlmS is required only in the absence of amino sugars in the environment. These sugars can be incorporated and converted into glucosamine-6-phosphate (GlcN-6-P), thus bypassing the GlmS-catalyzed reaction. The first, rate-limiting step is the conversion of fructose-6-phosphate (F6P) into GlcN-6-P in the presence of glutamine by GlmS. This step was shown to be highly feedback-regulated in both E. coli and B. subtilis, although the studies used different molecular mechanisms [22][23][24][25].
2.1. Regulation of GlmS Synthesis in E. coli
In E. coli, GlmS synthesis is regulated by the GlcN-6-P intracellular level via a mechanism based on four main actors: the two small RNAs (sRNAs), GlmZ and GlmY, the RNase adaptor RapZ and the endoribonuclease RNaseE [26] (Figure 2A,B).
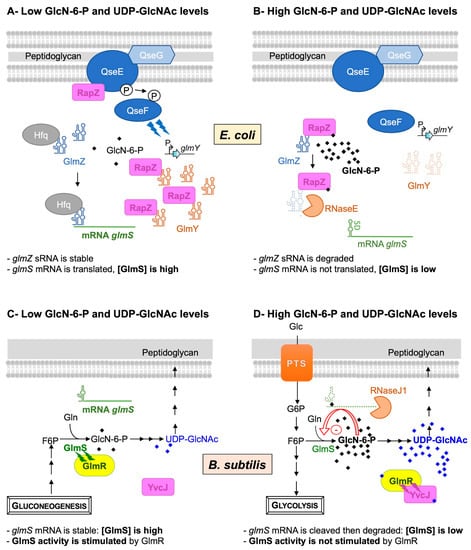
Figure 2. Feedback regulation of GlmS in E. coli (A,B) and in B. subtilis (C,D). In E. coli, when the intracellular GlcN6P concentration is low (A), the two-component system QseE/QseF, associated with the lipoprotein QseG, boosts the expression of the sRNA GlmY that protects the second sRNA GlmZ from degradation and thus indirectly activates glmS. The sRNA GlmZ accumulates and interacts with the glmS mRNA through a base-pairing interaction, stabilized by Hfq, to stimulate its translation and increase the GlmS enzyme level. When the intracellular level of GlcN6P is high (B), RapZ binds to GlcN6P, thereby interfering with sRNA binding and leading to the stimulation of QseE/QseF. RapZ is released from complexes with GlmY, and the sRNA is rapidly degraded. Once free, RapZ binds and targets GlmZ sRNA to the RNase E endoribonuclease, which cleaves the sRNA at the base-pairing site, thus preventing the stimulation of glmS translation. In B. subtilis, when intracellular GlcN6P and UDP-GlcNAc concentrations are low (C), the ribozyme is not complexed to GlcN-6-P; the glmS transcript is stable and translated to increase the GlmS enzyme level. In addition, GlmR interacts with GlmS to stimulate its activity. When the intracellular GlcN6P and UDP-GlcNAc concentrations are high (D), GlcN6P binds to the ribozyme of the glmS transcript and stimulates its self-cleavage. No longer protected by a 5′ triphosphate end, the glmS transcript undergoes rapid exonucleolytic degradation by RNase J to decrease the GlmS enzyme level. In addition, GlmR binds UDP-GlcNAc and no longer interacts with GlmS to stimulate its activity. However, it binds to YvcJ, a protein homologous to RapZ.
2.2. Regulation of Synthesis and Activity of GlmS in B. subtilis
In B. subtilis, both GlmS synthesis and activity are feedback-regulated by the GlcN-6-P and UDP-GlcNAc intracellular pools, respectively (Figure 2C,D).
In this Gram-positive bacterium, the level of glmS transcript is also controlled according to the GlcN-6-P intracellular concentration. The discovery of this well-studied regulation highlighted a new class of ribozymes [22][27][28]. In fact, the glmS transcript contains a unique type of riboswitch in its 5′ UTR: a self-cleaving ribozyme activated by GlcN-6-P. At a low GlcN-6-P concentration (Figure 2C), the already fully folded ribozyme is not complexed to GlcN-6-P; the glmS transcript is stable and translated. At high GlcN-6-P concentrations (Figure 2D), this amino sugar binds to the ribozyme, acting as a coenzyme to stimulate RNA self-cleavage [29][30]. No longer protected by a 5′ triphosphate, the glmS transcript undergoes rapid exonucleolytic degradation by RNase J1; thus, the GlmS concentration is reduced.
An additional level of regulation of GlcN-6-P synthesis was demonstrated via the feedback regulation of GlmS activity by UDP-GlcNAc [24][31]. In fact, GlmS activity was shown to be stimulated by GlmR, a UDP-GlcNAc-binding protein required for growth on non-glycolytic carbon sources [32][33]. At a low UDP-GlcNAc concentration, i.e., growth on non-glycolytic carbon sources, GlmR is not complexed with UDP-GlcNAc and binds to GlmS in order to stimulate its activity (Figure 2C). In such conditions, GlmR is essential. This stimulation is prevented by the presence of UDP-GlcNAc in vitro or when growth conditions are favorable in vivo, i.e., glycolytic carbon sources (Figure 2D) [24][31]. In such conditions, GlmR is dispensable; it does not interact with GlmS but is complexed with UDP-GlcNAc to bind to YvcJ, a protein homologous to RapZ [24]. In B. subtilis, the glmS transcript level is not modified by the deletion of yvcJ, and YvcJ does not regulate the synthesis of GlmS; its molecular role is not characterized [25]. There is no evidence to show that YvcJ is an RNA-binding protein, with the RNA-binding domain of E. coli RapZ being weakly conserved in B. subtilis YvcJ [34]. However, YvcJ function is directly or indirectly related to natural competence. Indeed, competence efficiency is affected in yvcJ mutant strains in comparison to WT strains, and the expression of ComK regulon is also affected [25][35]. A recent publication confirmed the central role of GlmR in PG synthesis and antibiotic sensitivity [36]. In fact, mutations of rpoB, encoding the β-subunit of the RNA polymerase, can alter B. subtilis sensitivity to antibiotics such as rifampicin and/or β-lactam cefuroxime. The cefuroxime induces the accumulation of UDP-GlcNAc, which feedback regulates GlmS activity in a GlmR-dependent manner and thus PG synthesis.
3. Flipping over the Cytoplasmic Membrane
For decades, the mechanism behind the flipping and recycling of Lipid-II across the cytoplasmic membrane remained unknown. However, two recent publications shed light on this mystery through the simultaneous discovery of two protein families capable of recycling UndP [19][20].
3.1. The Lipid-II Flippases
Lipid-II flippases, such as MurJ, are responsible for flipping Lipid-II to the outer leaf of the cytoplasmic membrane (Figure 1) [18]. This protein requires proton motive force (PMF) in order to drive conformational changes for the flip of Lipid-II [37][38]. In E. coli, MurJ is the sole Lipid-II flippase. It is essential, and the depletion of this protein results in the inhibition of PG biosynthesis and accumulation of lipid-linked PG precursors [18]. By contrast, B. subtilis has four MurJ homologs. One of them (SpoVB) is required for PG synthesis during sporulation [39]. E. coli MurJ has been shown to be able to complement the sporulation defect of a spoVB mutant. In turn, both SpoVB and YtgP, another MurJ from B. subtilis, were shown to complement the growth defect of an E. coli strain depleted of murJ, thus confirming their status as Lipid-II flippases. Unexpectedly, a B. subtilis strain lacking these four MurJ homologs did not exhibit a growth defect, suggesting the existence of an alternative Lipid-II flippase family. Indeed, an additional Lipid-II flippase, Amj (alternate to MurJ), was discovered in B. subtilis a few years ago [40]. Cells lacking both Amj and the four MurJ homologs exhibited cell shape defects and lysis. Furthermore, the expression of amj or murJ from B. subtilis in an E. coli murJ mutant restored Lipid-II flipping and, consequently, the viability of this mutant. In addition, the SEDS protein, FtsW [41], an essential protein carrying GTase activity required for glycan strand polymerization during septum formation in cell division, has also been shown to transport Lipid-II in vitro [42]. It may thus be an alternative Lipid-II flippase, but this assumption is controversial. This proposition was based on an in vitro FtsW reconstitution assay that reported both a Lipid-II flippase activity and an ability of FtsW to translocate various phospholipids [42]. However, no FtsW-dependent flippase activity could be detected when MurJ was incorporated into liposomes. In addition, no genetic evidence supports the argument that FtsW flips Lipid-II. In fact, it was shown that FtsW activity and Lipid-II synthesis are required for the recruitment of MurJ to the mid-cell in E. coli [43]. The mid-cell is the place where septal PG is synthesized and thus where the Lipid-II is flipped during cell division. This result strongly suggests that MurJ and FtsW work together in vivo for the flipping of Lipid-II.
3.2. The UndP Flippases
The step following the flipping of Lipid-II to the outer face of the cytoplasmic membrane, where the muropeptide is polymerized and crosslinked to the existing PG meshwork, is the dephosphorylation of UndPP by UndPP membrane phosphatases (BacA, YbjG, PgpB and LpxT in E. coli [44] or UppP and BcrC in B. subtilis [45]) for its recycling (Figure 1). The resulting UndP is flipped back to the inner side of the cytoplasmic membrane to be recycled for the production of novel Lipid-II. For years, the transporters involved in this recycling were unidentified. It was only very recently that, using genetic screens in B. subtilis, Staphylococcus aureus [19] and Vibrio cholerae [20], two broadly conserved families of flippases were shown to be responsible for UndP transport across the membrane. Genetic, cytological and syntenic analyses support the idea that these two UndP transporter families (corresponding to UptA and PopT proteins) are indeed UndP flippases.
UptA (for UndP transporter A) is a member of the DedA superfamily, a broadly conserved but poorly characterized membrane protein family [19][20]. The UptA structural model resembles the structure of membrane transporters. Bioinformatic analysis revealed that B. subtilis possesses six paralogs of DedA, whereas the E. coli genome encodes eight of them [19]. In addition to UptA, S. aureus and V. cholerae possess another protein involved in UndP recycling, named PopT (polyprenyl-phosphate transporter). This protein is absent in B. subtilis and in E. coli [19][20]. It carries a DUF368 domain and belongs to a protein family distinct from DedA. Its structural model resembles the structure of canonical membrane transporters, with a two-fold inverted symmetry. The two types of transporters, UptA and PopT, possess two membrane re-entrant loops into the lipid bilayer. The importance and respective roles of UptA- and PopT-like proteins have not yet been clearly characterized in bacteria that possess these two types of transporters, such as S. aureus and V. cholerae. However, it was proposed that both transport and recycle UndP. To support this proposition, it was proposed that UndP recycling is reduced in a B. subtilis mutant strain lacking YngC, one of the six DedA proteins [19], and in an S. aureus mutant strain lacking DUF368-containing proteins [20]. In addition, an S. aureus mutant with both uptA and popT deleted for is highly susceptible to tunicamycin, consistent with reduced levels of inward-facing UndP due to an accumulation of outward-facing UndP in this double mutant [19]. Moreover, members of these two protein families were found in several Gram-positive and Gram-negative bacteria, indicating that the UndP recycling mediated by these flippases is broadly conserved [19][20]. Nevertheless, the mechanisms of UndP transport by these proteins remain unknown.
4. Regulation of PG Expansion
After Lipid-II flips to the outer side of the cytoplasmic membrane, GTases polymerize GlcNAc-MurNAc-pentapeptide moieties into glycan strands (Figure 1). TPases then crosslink these strands to form the newly synthesized PG. PBPs (from classes A and B) and SEDS proteins catalyze these reactions [3]. Widely used antibiotics target PG synthesis, highlighting its essential function in bacteria. Rod-shaped bacteria have two modes of PG synthesis during the cell cycle: the elongasome and the divisome. The elongasome catalyzes lateral PG synthesis during cell elongation, while the divisome catalyzes septal PG synthesis during cell division [1][2][46]. The composition of these cell wall machineries varies according to the bacteria. In rod-shaped bacteria, the elongasome is associated with the actin-like protein MreB. This cytoskeletal protein polymerizes into small filaments that localize in patches on the inner face of the lateral cell wall of bacteria in order to drive the elongasome and thus the insertion of new PG during elongation [47][48]. In addition, B. subtilis encodes three actin-like proteins of the MreB family (MreB, Mbl and MreBH), MreBH being essential for the activation of the major autolysin LytE and for its localization to the sites of new PG insertion [49][50]. The elongasome, also called Rod-complex, is composed of a variety of enzymes, some of which are found in the composition of the core of this machinery in E. coli and B. subtilis, such as MreB, MreC, MreD, RodA, RodZ and bPBPs [1][2][50][51]. RodZ physically links the cytoplasmic MreB to the elongasome. aPBPs are not considered as components of this elongasome.
Cell division is controlled by another cytoskeletal protein, FtsZ, a tubulin-like protein present in almost all bacteria. FtsZ polymerizes in the mid-cell to form a circumferential ring, called the Z-ring, that defines the site of division, named the septum [52][53]. FtsZ also recruits numerous proteins to form the divisome [2][54]. The divisome is more complex than the elongasome. In E. coli, approximately 40 proteins were identified in its composition. Among them, a dozen are essential or conditionally essential and highly conserved in bacteria. The proteins FtsZ, FtsA, ZipA, FtsE, FtsX, FtsK, FtsQ, FtsL, FtsB, FtsW, FtsI and FtsN constitute the basic components of the divisome [55]. Some of these proteins are also part of the divisome of B. subtilis, which is assembled in at least two distinct steps [54]. The first step involves the polymerization of FtsZ and the concomitant recruitment of “early” divisome proteins including FtsA, SepF, ZapA and EzrA. Then, a second step takes place with the recruitment of the “late” proteins that have extracellular or membrane domains and the regulatory proteins, such as GpsB, DivIVA, FtsL, DivIB, FtsW, PBP2b, MinJ, MinD and MinC. The divisome is responsible for both the constriction of the inner (and outer) membrane(s) and PG synthesis at the division site [46] and is almost ubiquitous.
Because PG plays a fundamental role in bacteria, the enzymes involved in its synthesis, remodeling and repair are expected to be highly controlled in a cell-cycle-dependent manner, particularly their synthesis, localization and activity. For example, cell wall elongation and cell division machineries are in competition, and the rates of side wall and septal PG synthesis are inversely correlated [56]. This observation strongly supports the notion of antagonist activities of the elongasome and the divisome, as well as the spatiotemporal regulation of these machineries. Although the way in which SEDS proteins are regulated remains unknown, many regulatory proteins have been found to regulate the localization or activity of PBPs and hydrolases.
4.1. Regulation of PBPs
E. coli possesses 12 PBPs, which include 3 aPBPs (PBP1a, PBP1b and PBP1c), 2 bPBPs (PBP2 and PBP3) and 7 low-MW PBPs. PBP1a and PBP1b are the major aPBPs, whereas PBP2 is involved in cell elongation and PBP3 belongs to the divisome [57]. The seven low-MW PBPs are involved in cell separation, PG maturation and recycling. B. subtilis encodes 16 PBPs, including 4 aPBPs (PBP1a, PBP2c, PBP4 and PBP1d) and 6 bPBPs (PBP2a, PBP2b, PBP3, SpoVD, PBPH and YrrR) [58][59]. The main aPBP is PBP1a. PBP2a and PBPH are involved in cell elongation, with the simultaneous deletion of the two corresponding genes being lethal [60]. PBP2b belongs to the divisome [61]. Some of these PBPs are functionally redundant [59][62]. The production and the properties of these enzymes are relatively well-characterized, but the regulation of their activity and their dynamic localization are not entirely understood. For example, the cell division protein GpsB, which was first proposed to be involved in the shuttling of PBP1a, the main B. subtilis aPBP, through the facilitation of its removal from the cell pole [63], was recently shown to have a more complex role. It was proposed to mediate the interaction between PBP1a and various proteins to form larger protein complexes at specific sites, probably to drive PG synthesis in a bacterial cell-cycle-dependent manner [64]. In E. coli, two outer-membrane lipoproteins, LpoA and LpoB, were shown to regulate PBP activities and be essential aPBP cofactors [65][66]. LpoA interacts with its cognate PBP, PBP1a, whereas LpoB specifically interacts with PBP1b. LpoA stimulates TPase activity of PBP1a in vitro [67]. In fact, LpoA not only modulates PG crosslinking but is also required for PBP1a to form PG glycan strands. Concerning LpoB, it regulates the PG synthesis rate by spanning the periplasm and reaching its cognate PG synthase, PBP1b [68]. In B. subtilis, no essential regulator of PBPs has been highlighted thus far, but the non-essential TseB protein was proposed to regulate PBP2a, a bPBP [69]. Indeed, this membrane protein was shown to be required for efficient cell wall elongation and to specifically interact with PBP2a. However, TseB is not required for PBP2a activity or localization, although PBP2a overproduction is deleterious in the absence of TseB. It was proposed to be a component of the elongasome, regulating PBP2a and cell elongation. In Streptococcus pneumoniae and S. aureus, it was proposed that MacP and the CozE proteins can act as aPBP regulators [70][71][72] and that EloR can regulate the lytic pneumococcus GTase MltG in the mid-cell [73][74].
4.2. Regulation of PG Hydrolases
In addition to PG synthase, PG hydrolases are essential for cell wall elongation and daughter cell separation during division [1]. In the rod-shaped bacteria E. coli and B. subtilis, cell elongation requires the cleavage of peptide crosslinks within the existing PG via endopeptidases to create space for the incorporation of newly synthetized PG into the cell cylinder. These enzymes are thus required for PG expansion during cell growth. Despite their importance, the regulation of these enzymes is not well-understood (see [5] for a recent review).
B. subtilis encodes two functionally redundant D,L-endopeptidases (CwlO and LytE) that, together, are essential [75]. They act on the lateral PG, cleaving peptide crosslinks. Their activity is highly regulated. One aspect of this regulation involves the essential WalR–WalK two-component system. It was shown that WalR–WalK senses the cleavage products generated by CwlO and LytE and, in response, modulates their hydrolase activity during cell wall elongation [76]. The hydrolase activity of CwlO is also regulated by FtsEX, a conserved ABC transporter, with its essential co-factors SweD and SweC, required for cell wall elongation [77].
In E. coli, FtsEX plays a role in cell division [78], specifically in the assembly and activation of the divisome so as to connect cell wall synthesis to cell wall hydrolysis at the septum. The FtsEX complex is recruited to the Z-ring through FtsE interaction with FtsZ and then interacts with FtsA to initiate the assembly of the divisome complex while halting septal PG synthesis until the divisome assembly is complete [78][79].
5. Respective Role of PBPs and SEDS
It was long believed that aPBPs, which are essential in most bacteria [80], are the only GTases involved in PG synthesis. However, a B. subtilis mutant lacking the four aPBPs was shown to be viable and to produce a PG exhibiting an almost normal structure [81], which contradicted this assumption. The discovery of the SEDS proteins RodA and FtsW [41][82] explained the viability of this quadruple mutant. It was proposed that in bacteria with a cell wall, aPBPs are essential, excepting cases where SEDS proteins can replace them [83]. Indeed, RodA and FtsW were shown to be the main PG synthases and to serve as primary GTases [41]. FtsW seems to be universally essential, but some bacteria do not require RodA for growth. In E. coli and B. subtilis, RodA and FtsW are both essential [84][85]. They interact with their bPBP partners and are the core constituents of the elongasome and the divisome, respectively [82][84]. RodA is necessary for lateral PG synthesis during elongation and for the maintenance of the rod shape of B. subtilis. Conditional mutants depleted of either RodA or its two associated bPBPs, PBP2a and PBPH, have a spherical morphology [60][86]. RodA serves as a GTase, while the two redundant bPBPs, PBP2a and PBPH, and possibly aPBPs, provide TPase activity [41][82]. In addition, structural data suggest that these associated bPBPs act as allosteric activators of RodA, permitting the SEDS/bPBP complex to coordinate its double enzymatic activities of PG polymerization and crosslinking in order to build the cell wall [87].
Concerning FtsW, it is an essential component of the divisome and is responsible for septal PG synthesis during cell division, together with its cognate class bPBP (PBP3 in E. coli) [88][89]. The precise role of aPBPs had to be reinvestigated in light of the essentiality of SEDS. It was proposed that the SEDS/bPBPs complex builds the foundational PG structure, while the aPBPs fill in the gaps and play a role in the repair of PG defects [90].
In a B. subtilis mutant strain that lacks the main aPBP, PBP1a, the overexpression of rodA occurs [50]. Moreover, an artificial increase in rodA expression can rescue a mutant lacking PBP1a [82]. These observations suggest that the overproduction of RodA can compensate for the absence of aPBP, triggering alternative mechanisms to restore the normal function of the cell wall in B. subtilis [50]. For instance, the alternative RNA polymerase sigma-54 factor, σI, was shown to be vital in the absence of aPBPs. In this mutant, an upregulation of MreBH, a homolog of MreB that localizes the LytE autolysin to the elongasome, occurs in a σI-dependent manner. An equilibrium in MreBH–LytE activity is critical for optimal elongasome function. In the absence of aPBPs, increased levels of RodA, MreBH and LytE stimulate the elongasome to enhance lateral PG synthesis, compensating for the loss of mechanical stability of the PG conferred by aPBPs [50]. Thus, aPBPs do not contribute to cell shape determination but are responsible for the mechanical stability of PG, which can be restored through the overproduction or stimulation of the elongasome (particularly RodA).
A study on the respective roles of RodA, the elongasome and aPBPs in cell growth and width demonstrated that increased elongasome activity is correlated with an increased density of directional MreB filaments [91]. The width of B. subtilis and E. coli rods depends on the balance between the activities of the elongasome, which reduces the cell diameter, and aPBPs, which increase it, indicating a mechanical or reparatory function of aPBPs (especially PBP1a). This hypothesis was reinforced by a recent study proposing that the intrinsically disordered domain (IDR) of some membrane proteins can sense gaps in the PG meshwork and play a role in maintaining cell wall homeostasis in B. subtilis [92]. Specifically, PBP1a exhibits an extracytoplasmic IDR that is critical for its function. The IDR of PBP1a directs it to gaps in the PG meshwork in order to repair and strengthen it. In these conditions, PBP1a, whose role is to repair the PG meshwork, complements the elongasome, whose role is to build the foundational PG structure [92].
Based on a model proposed for S. pneumoniae, in which the mature PG is synthesized by three functional entities, the divisome (with FtsW), the elongasome (with RodA) and bifunctional PBPs (aPBPs) [93], we can suggest that a similar model exists in E. coli and B. subtilis, with distinct and specific roles for each of these entities in cell division, cell wall elongation, and cell wall repair, respectively (Figure 3).
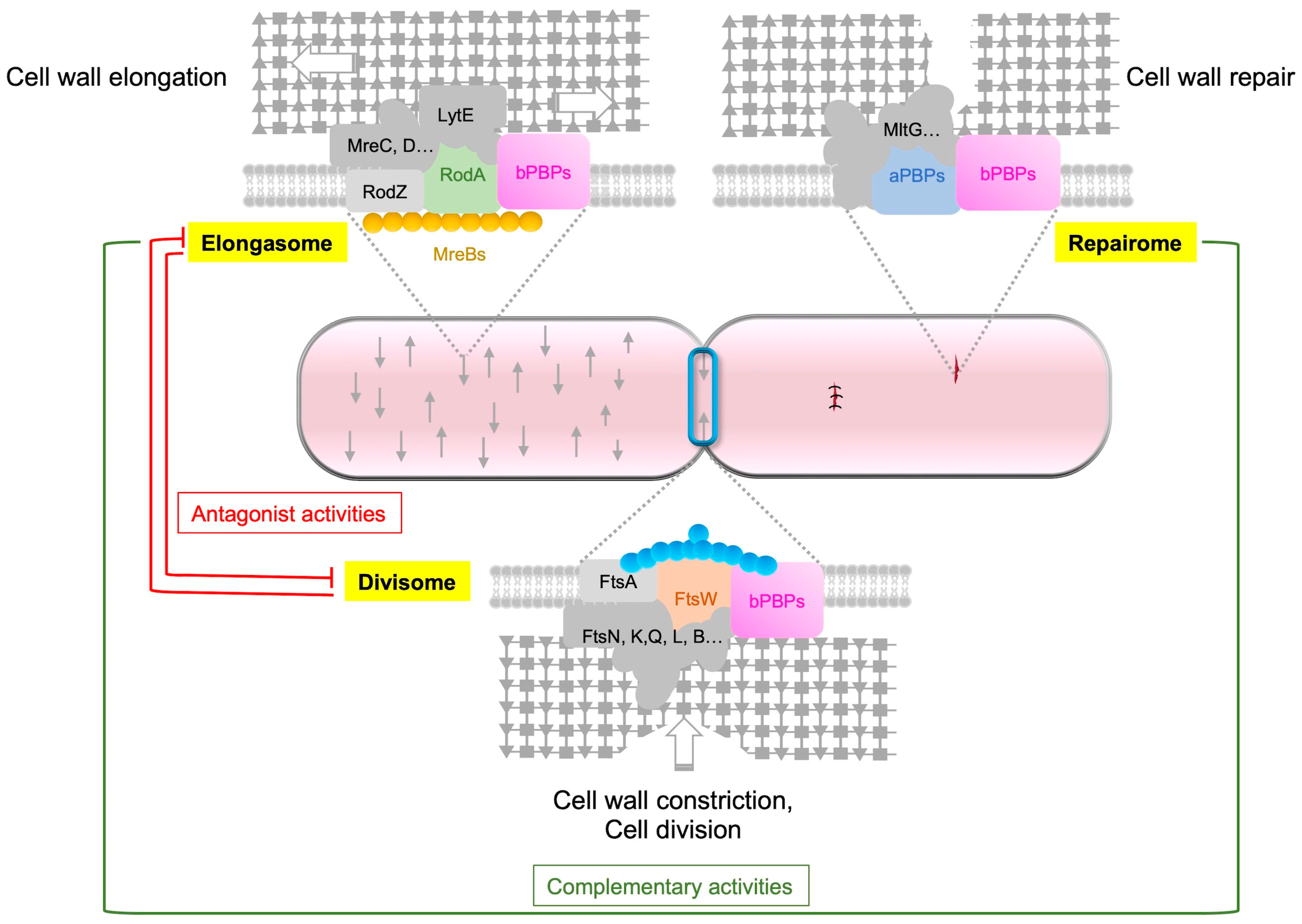
Figure 3. Respective roles of aPBP, the elongasome and the divisome. The synthesis of PG is carried out by three distinct complexes, namely, the elongasome, the divisome and the repairome. The elongasome is responsible for constructing the foundational lateral PG structure, and the GTase RodA plays a key role in this process. The repairome, which includes aPBPs, is involved in repairing and strengthening the PG meshwork in collaboration with the elongasome complex. In B. subtilis, PBP1a, the main aPBP, has an extracytoplasmic IDR that guides the GTase to gaps in the PG meshwork for repair. When aPBPs are absent, cells increase the expression of rodA, mreBH and lytE genes to upregulate elongasome activity. The divisome, on the other hand, is responsible for building the septal PG, and during cell division, the GTase FtsW plays a key role. Antagonistic activities are observed between the elongasome and the divisome, with the activity of one being repressed during the activity of the other.
References
- Egan, A.J.F.; Errington, J.; Vollmer, W. Regulation of peptidoglycan synthesis and remodelling. Nat. Rev. Microbiol. 2020, 18, 446–460.
- Errington, J.; Wu, L.J. Cell Cycle Machinery in Bacillus subtilis. Subcell. Biochem. 2017, 84, 67–101.
- Sassine, J.; Sousa, J.; Lalk, M.; Daniel, R.A.; Vollmer, W. Cell morphology maintenance in Bacillus subtilis through balanced peptidoglycan synthesis and hydrolysis. Sci. Rep. 2020, 10, 17910.
- Weaver, A.; Taguchi, A.; Dörr, T. Masters of Misdirection: Peptidoglycan Glycosidases in Bacterial Growth. J. Bacteriol. 2023, 205, e0042822.
- Brogan, A.P.; Rudner, D.Z. Regulation of peptidoglycan hydrolases: Localization, abundance, and activity. Curr. Opin. Microbiol. 2023, 72, 102279.
- Kumar, S.; Mollo, A.; Kahne, D.; Ruiz, N. The Bacterial Cell Wall: From Lipid II Flipping to Polymerization. Chem. Rev. 2022, 122, 8884–8910.
- Barreteau, H.; Kovac, A.; Boniface, A.; Sova, M.; Gobec, S.; Blanot, D. Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008, 32, 168–207.
- Mengin-Lecreulx, D.; Flouret, B.; van Heijenoort, J. Pool levels of UDP N-acetylglucosamine and UDP N-acetylglucosamine-enolpyruvate in Escherichia coli and correlation with peptidoglycan synthesis. J. Bacteriol. 1983, 154, 1284–1290.
- Mengin-Lecreulx, D.; Flouret, B.; van Heijenoort, J. Cytoplasmic steps of peptidoglycan synthesis in Escherichia coli. J. Bacteriol. 1982, 151, 1109–1117.
- Benson, T.E.; Walsh, C.T.; Hogle, J. The structure of the substrate-free form of MurB, an essential enzyme for the synthesis of bacterial cell walls. Structure 1996, 4, 47–54.
- Brown, E.D.; Vivas, E.I.; Walsh, C.T.; Kolter, R. MurA (MurZ), the enzyme that catalyzes the first committed step in peptidoglycan biosynthesis, is essential in Escherichia coli. J. Bacteriol. 1995, 177, 4194–4197.
- Vollmer, W.; Bertsche, U. Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim. Biophys. Acta (BBA)-Biomembr. 2008, 1778, 1714–1734.
- Morales Angeles, D.; Liu, Y.; Hartman, A.M.; Borisova, M.; de Sousa Borges, A.; de Kok, N.; Beilharz, K.; Veening, J.W.; Mayer, C.; Hirsch, A.K.; et al. Pentapeptide-rich peptidoglycan at the Bacillus subtilis cell-division site. Mol. Microbiol. 2017, 104, 319–333.
- Bouhss, A.; Mengin-Lecreulx, D.; Le Beller, D.; Van Heijenoort, J. Topological analysis of the MraY protein catalysing the first membrane step of peptidoglycan synthesis. Mol. Microbiol. 1999, 34, 576–585.
- Ikeda, M.; Wachi, M.; Jung, H.K.; Ishino, F.; Matsuhashi, M. The Escherichia coli mraY gene encoding UDP-N-acetylmuramoyl-pentapeptide: Undecaprenyl-phosphate phospho-N-acetylmuramoyl-pentapeptide transferase. J. Bacteriol. 1991, 173, 1021–1026.
- Lloyd, A.J.; Brandish, P.E.; Gilbey, A.M.; Bugg, T.D. Phospho-N-acetyl-muramyl-pentapeptide translocase from Escherichia coli: Catalytic role of conserved aspartic acid residues. J. Bacteriol. 2004, 186, 1747–1757.
- Mengin-Lecreulx, D.; Texier, L.; Rousseau, M.; van Heijenoort, J. The murG gene of Escherichia coli codes for the UDP-N-acetylglucosamine: N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase involved in the membrane steps of peptidoglycan synthesis. J. Bacteriol. 1991, 173, 4625–4636.
- Ruiz, N. Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proc. Natl. Acad. Sci. USA 2008, 105, 15553–15557.
- Roney, I.J.; Rudner, D.Z. Two broadly conserved families of polyprenyl-phosphate transporters. Nature 2023, 613, 729–734.
- Sit, B.; Srisuknimit, V.; Bueno, E.; Zingl, F.G.; Hullahalli, K.; Cava, F.; Waldor, M.K. Undecaprenyl phosphate translocases confer conditional microbial fitness. Nature 2023, 613, 721–728.
- Mengin-Lecreulx, D.; van Heijenoort, J. Identification of the glmU gene encoding N-acetylglucosamine-1-phosphate uridyltransferase in Escherichia coli. J. Bacteriol. 1993, 175, 6150–6157.
- Winkler, W.C.; Nahvi, A.; Roth, A.; Collins, J.A.; Breaker, R.R. Control of gene expression by a natural metabolite-responsive ribozyme. Nature 2004, 428, 281–286.
- Kalamorz, F.; Reichenbach, B.; März, W.; Rak, B.; Görke, B. Feedback control of glucosamine-6-phosphate synthase GlmS expression depends on the small RNA GlmZ and involves the novel protein YhbJ in Escherichia coli. Mol. Microbiol. 2007, 65, 1518–1533.
- Foulquier, E.; Pompeo, F.; Byrne, D.; Fierobe, H.P.; Galinier, A. Uridine diphosphate N-acetylglucosamine orchestrates the interaction of GlmR with either YvcJ or GlmS in Bacillus subtilis. Sci. Rep. 2020, 10, 15938.
- Luciano, J.; Foulquier, E.; Fantino, J.R.; Galinier, A.; Pompeo, F. Characterization of YvcJ, a conserved P-loop-containing protein, and its implication in competence in Bacillus subtilis. J. Bacteriol. 2009, 191, 1556–1564.
- Durica-Mitic, S.; Görke, B. Feedback regulation of small RNA processing by the cleavage product. RNA Biol. 2019, 16, 1055–1065.
- McCown, P.J.; Winkler, W.C.; Breaker, R.R. Mechanism and distribution of glmS ribozymes. Methods Mol. Biol. 2012, 848, 113–129.
- Winkler, W.C.; Breaker, R.R. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 2005, 59, 487–517.
- Cochrane, J.C.; Lipchock, S.V.; Smith, K.D.; Strobel, S.A. Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme. Biochemistry 2009, 48, 3239–3246.
- Cochrane, J.C.; Strobel, S.A. Catalytic strategies of self-cleaving ribozymes. Acc. Chem. Res. 2008, 41, 1027–1035.
- Patel, V.; Wu, Q.; Chandrangsu, P.; Helmann, J.D. A metabolic checkpoint protein GlmR is important for diverting carbon into peptidoglycan biosynthesis in Bacillus subtilis. PLoS Genet. 2018, 14, e1007689.
- Görke, B.; Foulquier, E.; Galinier, A. YvcK of Bacillus subtilis is required for a normal cell shape and for growth on Krebs cycle intermediates and substrates of the pentose phosphate pathway. Microbiology 2005, 151 Pt 11, 3777–3791.
- Foulquier, E.; Galinier, A. YvcK, a protein required for cell wall integrity and optimal carbon source utilization, binds uridine diphosphate-sugars. Sci. Rep. 2017, 7, 4139.
- Islam, M.S.; Hardwick, S.W.; Quell, L.; Durica-Mitic, S.; Chirgadze, D.Y.; Görke, B.; Luisi, B.F. Structure of a bacterial ribonucleoprotein complex central to the control of cell envelope biogenesis. EMBO J. 2022, e112574.
- Pompeo, F.; Luciano, J.; Brochier-Armanet, C.; Galinier, A. The GTPase function of YvcJ and its subcellular relocalization are dependent on growth conditions in Bacillus subtilis. J. Mol. Microbiol. Biotechnol. 2011, 20, 156–167.
- Patel, Y.; Soni, V.; Rhee, K.Y.; Helmann, J.D. Mutations in rpoB That Confer Rifampicin Resistance Can Alter Levels of Peptidoglycan Precursors and Affect β-Lactam Susceptibility. mBio 2023, e0316822.
- Kumar, S.; Rubino, F.A.; Mendoza, A.G.; Ruiz, N. The bacterial lipid II flippase MurJ functions by an alternating-access mechanism. J. Biol. Chem. 2019, 294, 981–990.
- Rubino, F.A.; Kumar, S.; Ruiz, N.; Walker, S.; Kahne, D.E. Membrane Potential Is Required for MurJ Function. J. Am. Chem. Soc. 2018, 140, 4481–4484.
- Fay, A.; Dworkin, J. Bacillus subtilis homologs of MviN (MurJ), the putative Escherichia coli lipid II flippase, are not essential for growth. J. Bacteriol. 2009, 191, 6020–6028.
- Meeske, A.J.; Sham, L.T.; Kimsey, H.; Koo, B.M.; Gross, C.A.; Bernhardt, T.G.; Rudner, D.Z. MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2015, 112, 6437–6442.
- Meeske, A.J.; Riley, E.P.; Robins, W.P.; Uehara, T.; Mekalanos, J.J.; Kahne, D.; Walker, S.; Kruse, A.C.; Bernhardt, T.G.; Rudner, D.Z. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 2016, 537, 634–638.
- Mohammadi, T.; van Dam, V.; Sijbrandi, R.; Vernet, T.; Zapun, A.; Bouhss, A.; Diepeveen-de Bruin, M.; Nguyen-Distèche, M.; de Kruijff, B.; Breukink, E. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J. 2011, 30, 1425–1432.
- Liu, X.; Kumar, S.; Ruiz, N.; Walker, S.; Kahne, D.E. FtsW activity and lipid II synthesis are required for recruitment of MurJ to midcell during cell division in Escherichia coli. Mol. Microbiol. 2018, 109, 855–884.
- Manat, G.; El Ghachi, M.; Auger, R.; Baouche, K.; Olatunji, S.; Kerff, F.; Touzé, T.; Mengin-Lecreulx, D.; Bouhss, A. Membrane Topology and Biochemical Characterization of the Escherichia coli BacA Undecaprenyl-Pyrophosphate Phosphatase. PLoS ONE 2015, 10, e0142870.
- Zhao, H.; Sun, Y.; Peters, J.M.; Gross, C.A.; Garner, E.C.; Helmann, J.D. Depletion of Undecaprenyl Pyrophosphate Phosphatases Disrupts Cell Envelope Biogenesis in Bacillus subtilis. J. Bacteriol. 2016, 198, 2925–2935.
- Szwedziak, P.; Löwe, J. Do the divisome and elongasome share a common evolutionary past? Curr. Opin. Microbiol. 2013, 16, 745–751.
- Garner, E.C.; Bernard, R.; Wang, W.; Zhuang, X.; Rudner, D.Z.; Mitchison, T. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science 2011, 333, 222–225.
- Domínguez-Escobar, J.; Chastanet, A.; Crevenna, A.H.; Fromion, V.; Wedlich-Söldner, R.; Carballido-López, R. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science 2011, 333, 225–228.
- Carballido-López, R.; Formstone, A.; Li, Y.; Ehrlich, S.D.; Noirot, P.; Errington, J. Actin homolog MreBH governs cell morphogenesis by localization of the cell wall hydrolase LytE. Dev. Cell 2006, 11, 399–409.
- Patel, Y.; Zhao, H.; Helmann, J.D. A regulatory pathway that selectively up-regulates elongasome function in the absence of class A PBPs. Elife 2020, 9, e57902.
- Errington, J. Bacterial morphogenesis and the enigmatic MreB helix. Nat. Rev. Microbiol. 2015, 13, 241–248.
- de Boer, P.; Crossley, R.; Rothfield, L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature 1992, 359, 254–256.
- RayChaudhuri, D.; Park, J.T. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature 1992, 359, 251–254.
- Gamba, P.; Veening, J.W.; Saunders, N.J.; Hamoen, L.W.; Daniel, R.A. Two-step assembly dynamics of the Bacillus subtilis divisome. J. Bacteriol. 2009, 191, 4186–4194.
- Du, S.; Lutkenhaus, J. Assembly and activation of the Escherichia coli divisome. Mol. Microbiol. 2017, 105, 177–187.
- Navarro, P.P.; Vettiger, A.; Ananda, V.Y.; Llopis, P.M.; Allolio, C.; Bernhardt, T.G.; Chao, L.H. Cell wall synthesis and remodelling dynamics determine division site architecture and cell shape in Escherichia coli. Nat. Microbiol. 2022, 7, 1621–1634.
- den Blaauwen, T.; de Pedro, M.A.; Nguyen-Distèche, M.; Ayala, J.A. Morphogenesis of rod-shaped sacculi. FEMS Microbiol. Rev. 2008, 32, 321–344.
- Goffin, C.; Ghuysen, J.M. Multimodular penicillin-binding proteins: An enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 1998, 62, 1079–1093.
- Sassine, J.; Xu, M.; Sidiq, K.R.; Emmins, R.; Errington, J.; Daniel, R.A. Functional redundancy of division specific penicillin-binding proteins in Bacillus subtilis. Mol. Microbiol. 2017, 106, 304–318.
- Wei, Y.; Havasy, T.; McPherson, D.C.; Popham, D.L. Rod shape determination by the Bacillus subtilis class B penicillin-binding proteins encoded by pbpA and pbpH. J. Bacteriol. 2003, 185, 4717–4726.
- Daniel, R.A.; Harry, E.J.; Errington, J. Role of penicillin-binding protein PBP 2B in assembly and functioning of the division machinery of Bacillus subtilis. Mol. Microbiol. 2000, 35, 299–311.
- McPherson, D.C.; Driks, A.; Popham, D.L. Two class A high-molecular-weight penicillin-binding proteins of Bacillus subtilis play redundant roles in sporulation. J. Bacteriol. 2001, 183, 6046–6053.
- Claessen, D.; Emmins, R.; Hamoen, L.W.; Daniel, R.A.; Errington, J.; Edwards, D.H. Control of the cell elongation-division cycle by shuttling of PBP1 protein in Bacillus subtilis. Mol. Microbiol. 2008, 68, 1029–1046.
- Cleverley, R.M.; Rutter, Z.J.; Rismondo, J.; Corona, F.; Tsui, H.T.; Alatawi, F.A.; Daniel, R.A.; Halbedel, S.; Massidda, O.; Winkler, M.E.; et al. The cell cycle regulator GpsB functios as cytosolic adaptor for multiple cell wall enzymes. Nat. Commun. 2019, 10, 261.
- Paradis-Bleau, C.; Markovski, M.; Uehara, T.; Lupoli, T.J.; Walker, S.; Kahne, D.E.; Bernhardt, T.G. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell 2010, 143, 1110–1120.
- Typas, A.; Banzhaf, M.; van den Berg van Saparoea, B.; Verheul, J.; Biboy, J.; Nichols, R.J.; Zietek, M.; Beilharz, K.; Kannenberg, K.; von Rechenberg, M.; et al. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell 2010, 143, 1097–1109.
- Sardis, M.F.; Bohrhunter, J.L.; Greene, N.G.; Bernhardt, T.G. The LpoA activator is required to stimulate the peptidoglycan polymerase activity of its cognate cell wall synthase PBP1a. Proc. Natl. Acad. Sci. USA 2021, 118, e210889411.
- Kermani, A.A.; Biboy, J.; Vollmer, D.; Vollmer, W. Outer membrane-anchoring enables LpoB to regulate peptidoglycan synthesis rate. Cell Surf. 2022, 8, 100086.
- Delisle, J.; Cordier, B.; Audebert, S.; Pophillat, M.; Cluzel, C.; Espinosa, L.; Grangeasse, C.; Galinier, A.; Doan, T. Characterization of TseB: A new actor in cell wall elongation in Bacillus subtilis. Mol. Microbiol. 2021, 116, 1099–1112.
- Stamsås, G.A.; Myrbråten, I.S.; Straume, D.; Salehian, Z.; Veening, J.W.; Håvarstein, L.S.; Kjos, M. CozEa and CozEb play overlapping and essential roles in controlling cell division in Staphylococcus aureus. Mol. Microbiol. 2018, 109, 615–632.
- Fenton, A.K.; El Mortaji, L.; Lau, D.T.; Rudner, D.Z.; Bernhardt, T.G. CozE is a member of the MreCD complex that directs cell elongation in Streptococcus pneumoniae. Nat. Microbiol. 2016, 2, 16237.
- Fenton, A.K.; Manuse, S.; Flores-Kim, J.; Garcia, P.S.; Mercy, C.; Grangeasse, C.; Bernhardt, T.G.; Rudner, D.Z. Phosphorylation-dependent activation of the cell wall synthase PBP2a in Streptococcus pneumoniae by MacP. Proc. Natl. Acad. Sci. USA 2018, 115, 2812–2817.
- Winther, A.R.; Kjos, M.; Herigstad, M.L.; Håvarstein, L.S.; Straume, D. EloR interacts with the lytic transglycosylase MltG at midcell in Streptococcus pneumoniae R6. J. Bacteriol. 2021, 203, e00691-20.
- Stamsås, G.A.; Straume, D.; Ruud Winther, A.; Kjos, M.; Frantzen, C.A.; Håvarstein, L.S. Identification of EloR (Spr1851) as a regulator of cell elongation in Streptococcus pneumoniae. Mol. Microbiol. 2017, 105, 954–967.
- Hashimoto, M.; Ooiwa, S.; Sekiguchi, J. Synthetic lethality of the lytE cwlO genotype in Bacillus subtilis is caused by lack of D,L-endopeptidase activity at the lateral cell wall. J. Bacteriol. 2012, 194, 796–803.
- Dobihal, G.S.; Flores-Kim, J.; Roney, I.J.; Wang, X.; Rudner, D.Z. The WalR-WalK Signaling Pathway Modulates the Activities of both CwlO and LytE through Control of the Peptidoglycan Deacetylase PdaC in Bacillus subtilis. J. Bacteriol. 2022, 204, e0053321.
- Brunet, Y.R.; Wang, X.; Rudner, D.Z. SweC and SweD are essential co-factors of the FtsEX-CwlO cell wall hydrolase complex in Bacillus subtilis. PLoS Genet. 2019, 15, e1008296.
- Du, S.; Pichoff, S.; Lutkenhaus, J. FtsEX acts on FtsA to regulate divisome assembly and activity. Proc. Natl. Acad. Sci. USA 2016, 113, E5052–E5061.
- Du, S.; Henke, W.; Pichoff, S.; Lutkenhaus, J. How FtsEX localizes to the Z ring and interacts with FtsA to regulate cell division. Mol. Microbiol. 2019, 112, 881–895.
- Straume, D.; Piechowiak, K.W.; Kjos, M.; Håvarstein, L.S. Class A PBPs: It is time to rethink traditional paradigms. Mol. Microbiol. 2021, 116, 41–52.
- McPherson, D.C.; Popham, D.L. Peptidoglycan synthesis in the absence of class A penicillin-binding proteins in Bacillus subtilis. J. Bacteriol. 2003, 185, 1423–1431.
- Emami, K.; Guyet, A.; Kawai, Y.; Devi, J.; Wu, L.J.; Allenby, N.; Daniel, R.A.; Errington, J. RodA as the missing glycosyltransferase in Bacillus subtilis and antibiotic discovery for the peptidoglycan polymerase pathway. Nat. Microbiol. 2017, 2, 16253.
- Pazos, M.; Vollmer, W. Regulation and function of class A Penicillin-binding proteins. Curr. Opin. Microbiol. 2021, 60, 80–87.
- Welsh, M.A.; Schaefer, K.; Taguchi, A.; Kahne, D.; Walker, S. Direction of Chain Growth and Substrate Preferences of Shape, Elongation, Division, and Sporulation-Family Peptidoglycan Glycosyltransferases. J. Am. Chem. Soc. 2019, 141, 12994–12997.
- Taguchi, A.; Welsh, M.A.; Marmont, L.S.; Lee, W.; Sjodt, M.; Kruse, A.C.; Kahne, D.; Bernhardt, T.G.; Walker, S. FtsW is a peptidoglycan polymerase that is functional only in complex with its cognate penicillin-binding protein. Nat. Microbiol. 2019, 4, 587–594.
- Boylan, R.J.; Mendelson, N.H. Initial characterization of a temperature-sensitive rod--mutant of Bacillus subtilis. J. Bacteriol. 1969, 100, 1316–1321.
- Sjodt, M.; Rohs, P.D.A.; Gilman, M.S.A.; Erlandson, S.C.; Zheng, S.; Green, A.G.; Brock, K.P.; Taguchi, A.; Kahne, D.; Walker, S.; et al. Structural coordination of polymerization and crosslinking by a SEDS-bPBP peptidoglycan synthase complex. Nat. Microbiol. 2020, 5, 813–820.
- Marmont, L.S.; Bernhardt, T.G. A conserved subcomplex within the bacterial cytokinetic ring activates cell wall synthesis by the FtsW-FtsI synthase. Proc. Natl. Acad. Sci. USA 2020, 117, 23879–23885.
- Yang, X.; McQuillen, R.; Lyu, Z.; Phillips-Mason, P.; De La Cruz, A.; McCausland, J.W.; Liang, H.; DeMeester, K.E.; Santiago, C.C.; Grimes, C.L.; et al. A two-track model for the spatiotemporal coordination of bacterial septal cell wall synthesis revealed by single-molecule imaging of FtsW. Nat. Microbiol. 2021, 6, 584–593.
- Vigouroux, A.; Cordier, B.; Aristov, A.; Alvarez, L.; Özbaykal, G.; Chaze, T.; Oldewurtel, E.R.; Matondo, M.; Cava, F.; Bikard, D.; et al. Class-A penicillin binding proteins do not contribute to cell shape but repair cell-wall defects. Elife 2020, 9, e51998.
- Dion, M.F.; Kapoor, M.; Sun, Y.; Wilson, S.; Ryan, J.; Vigouroux, A.; van Teeffelen, S.; Oldenbourg, R.; Garner, E.C. Bacillus subtilis cell diameter is determined by the opposing actions of two distinct cell wall synthetic systems. Nat. Microbiol. 2019, 4, 1294–1305.
- Brunet, Y.R.; Habib, C.; Brogan, A.P.; Artzi, L.; Rudner, D.Z. Intrinsically disordered protein regions are required for cell wall homeostasis in Bacillus subtilis. Genes Dev. 2022, 36, 970–984.
- Straume, D.; Piechowiak, K.W.; Olsen, S.; Stamsås, G.A.; Berg, K.H.; Kjos, M.; Heggenhougen, M.V.; Alcorlo, M.; Hermoso, J.A.; Håvarstein, L.S. Class A PBPs have a distinct and unique role in the construction of the pneumococcal cell wall. Proc. Natl. Acad. Sci. USA 2020, 117, 6129–6138.
More
Information
Subjects:
Microbiology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.6K
Revisions:
2 times
(View History)
Update Date:
08 May 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




