Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Iwona Ciereszko | -- | 3625 | 2023-05-05 08:00:13 | | | |
| 2 | Violetta Katarzyna Macioszek | + 157 word(s) | 3782 | 2023-05-05 08:29:59 | | | | |
| 3 | Rita Xu | Meta information modification | 3782 | 2023-05-05 08:35:46 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Macioszek, V.K.; Jęcz, T.; Ciereszko, I.; Kononowicz, A.K. Jasmonic Acid in Plant Response to Necrotrophic Fungi. Encyclopedia. Available online: https://encyclopedia.pub/entry/43826 (accessed on 07 February 2026).
Macioszek VK, Jęcz T, Ciereszko I, Kononowicz AK. Jasmonic Acid in Plant Response to Necrotrophic Fungi. Encyclopedia. Available at: https://encyclopedia.pub/entry/43826. Accessed February 07, 2026.
Macioszek, Violetta Katarzyna, Tomasz Jęcz, Iwona Ciereszko, Andrzej Kiejstut Kononowicz. "Jasmonic Acid in Plant Response to Necrotrophic Fungi" Encyclopedia, https://encyclopedia.pub/entry/43826 (accessed February 07, 2026).
Macioszek, V.K., Jęcz, T., Ciereszko, I., & Kononowicz, A.K. (2023, May 05). Jasmonic Acid in Plant Response to Necrotrophic Fungi. In Encyclopedia. https://encyclopedia.pub/entry/43826
Macioszek, Violetta Katarzyna, et al. "Jasmonic Acid in Plant Response to Necrotrophic Fungi." Encyclopedia. Web. 05 May, 2023.
Copy Citation
Jasmonic acid (JA) and its derivatives, all named jasmonates, are the simplest phytohormones which regulate multifarious plant physiological processes including development, growth and defense responses to various abiotic and biotic stress factors. Moreover, jasmonate plays an important mediator’s role during plant interactions with necrotrophic oomycetes and fungi in the process of activation defense responses.
jasmonates
necrotrophic fungi
defense response
circadian clock
1. Introduction
Jasmonates (JAs) are one of the structurally simplest plant hormones. The term ‘jasmonates’ describes the group of oxylipin phytohormones, derivatives of jasmonic acid (JA), that come into existence in cytosol, such as methyl ester of JA (MeJA), cis-jasmone, jasmonic acid glucoside (JA-Glc), 12-hydroxyjasmonic acid (tuberonic acid, 12-OH-JA) or JA-isoleucine conjugate (JA-Ile), that regulate diverse developmental and physiological processes [1][2]. JA plays multifarious roles in plant physiological processes, i.e., growth and development [3], circadian rhythm of metabolism [4], senescence and cold acclimation [5], as well as the response to abiotic and biotic stresses [6][7]. The special function, however, of jasmonic acid is performing as a signal mediator in defense against herbivorous insects [8] and necrotrophic pathogens [9]. During plant defense response, JA not only induces the expression of pathogenesis-related (PR) genes [10] but also regulates the secondary metabolism promoting synthesis of flavonoids, glucosinolates, terpenoids and phytoalexins [11][12], as well as lignin deposition that enhances the mechanical structure of cell walls [13][14]. Jasmonates levels vary depending on plant species and environmental conditions; thus, their concentration in response to stress is an individual quality of a plant [15].
The hormonal character of jasmonates, although hitherto widely accepted, was ultimately confirmed by the discovery of the JA-specific receptor complex. CORONATINE INSENSITIVE1 (COI1) protein, first described by Xie et al. [16], was proven to bind directly to JA-Ile and serve as a jasmonate receptor [17]. The bioactive form of JA-isoleucine conjugate is (+)-7-iso-JA-L-Ile, whereas its (-)-JA-L-Ile epimer was shown to be an inactive, although more stable, form. The pH changes promote conversion of (+)-7-iso-JA-L-Ile to the inactive (-)-JA-L-Ile form, thus providing a simple mechanism that can rapidly and reversibly regulate hormone activity through epimerization [18]. The perception of JA-Ile conjugate is crucial for interaction of the COI1 and Jasmonate-ZIM (Zinc-finger Inflorescence Meristem) domain (JAZ) repressor protein. It was proven, that in Arabidopsis and tomato only that this form of jasmonate, unlike the other JA derivatives such as methyl jasmonate (MeJA) or JA precursor 12-oxo-phytodienoic acid (OPDA), promotes binding JAZ1 by COI1 [19][20]. Forming the COI1-JAZ1 complex does not involve any JA-Ile-induced enzymatic modifications, as JA-Ile promotes the direct physical interactions between these two proteins [21].
Contrary to biotrophic pathogens that feed on living host tissues, necrotrophic fungi obtain nutrients by killing plant cells and feeding on dying or dead host tissues. Necrotrophic fungal pathogens attack either a broad spectrum of host plant species or a narrow host range, or even, rather rarely, like many biotrophic fungi, a single host plant species [22][23]. Necrotrophic fungi cause substantial crop yield loss during all steps of crop agriculture from seed, through seedlings and young plant stages, to mature, ready to harvest, plants and also postharvest during storage. Moreover, they generate more devastating economic impact on food production world-wide than biotrophic fungi [24][25]. Extensively studied model necrotrophic fungi such as generalists Botrytis cinerea [26] and Sclerotinia sclerotiorum [27] or a specialist that infects plants of Brassicaceae family—Alternaria brassicicola [28]—induce JA pathway in resistant and to a lesser extent also in susceptible host plants. Infection of plant host cells by a necrotrophic fungus is accomplished mostly either by a repertoire of fungal cell wall degrading enzymes (CWDE) and plethora of toxins or by a more intricate mechanism containing secreted effector proteins and plant receptors, although this last possibility is only currently being broadly discussed and supported by genomic studies in regard to necrotrophic fungi [29]. Upon perception of necrotrophic fungi by host cellular receptors, signal transduction through secondary messengers (e.g., reactive oxygen species, ROS) triggers plant resistance responses leading, among other events, to JA biosynthesis and activation of a JA-dependent signaling cascade including a set of transcription factors (TFs) and following over-expression of defense-related JA marker genes such as, e.g., plant defensin (e.g., PDF1.2) and/or thionin (e.g., THI 2.1) [30][31][32]. The complexity of not yet fully explored JA-dependent defense responses of plants to necrotrophic fungi and the possibility of using their many features in contemporary agricultural technologies as an alternative to for example fungicides is one of the most interesting areas in modern plant science.
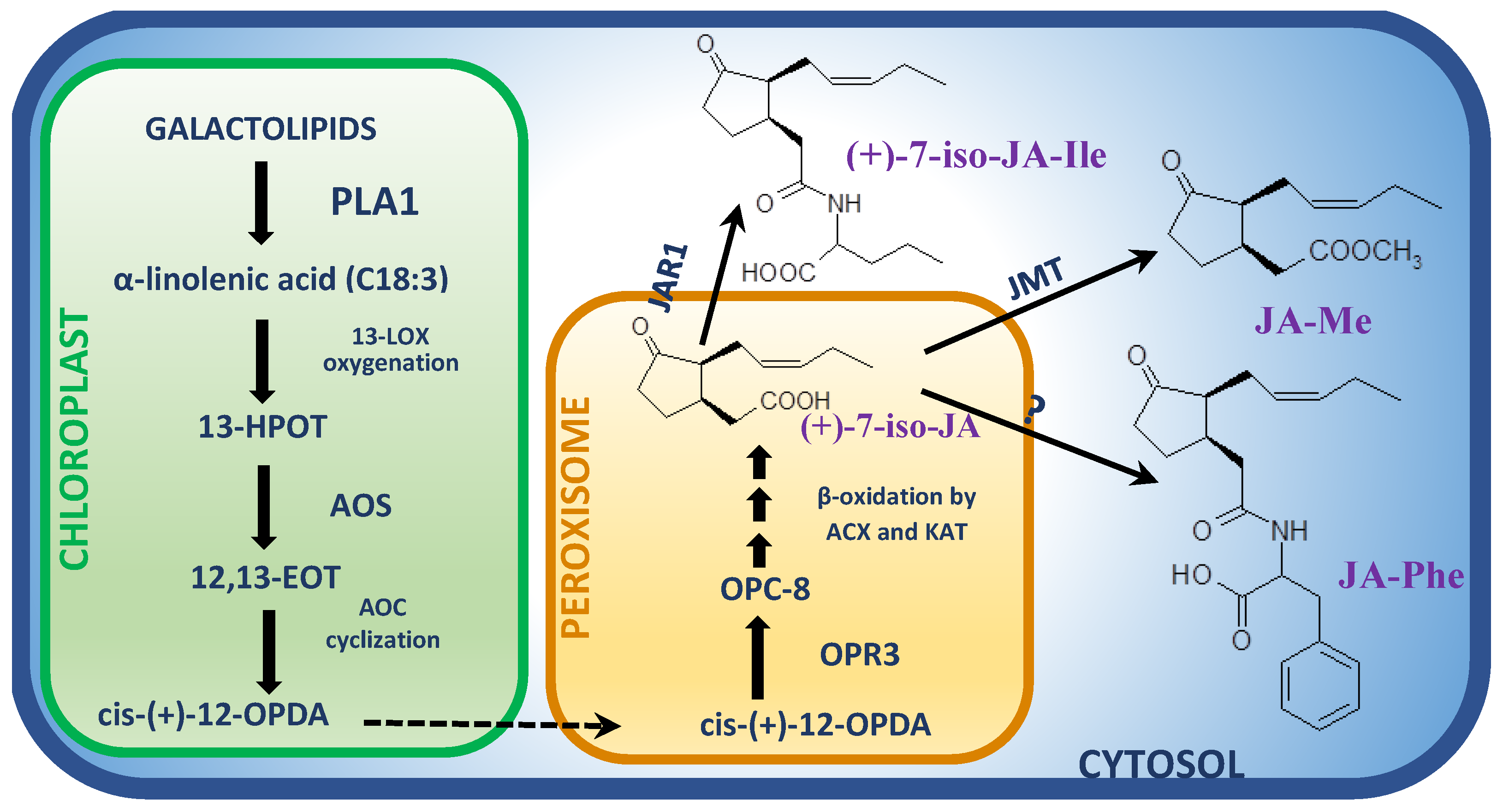
Figure 1. Biosynthesis pathway of jasmonates in Arabidopsis active during necrotrophic interactions. Detailed description in the text. Abbreviations: ACX, acyl-CoA oxidase; AOC, allene oxide cyclase; AOS, allene oxide synthase; 12,13-EOT, 12,13(S)-epoxy-9(Z),11,15(Z)-octadecatrienoic acid; (+)-7-iso-JA, (+)-7-iso-jasmonic acid; (+)-7-iso-JA-Ile, (+)-7-iso-jasmonoyl-L-isoleucine; JMT, JA carboxyl methyl transferase; JAR1, Jasmonate-Resistant synthase; JA-Me, methyl ester of JA; JA-Phe, jasmonoyl-phenylalanine; 13-HPOT, (13S)-hydroperoxy-octadecatrienoic acid; KAT, l-3-ketoacyl-CoA thiolase; 13-LOX, 13-lipoxygenase; OPC-8, 3-oxo-2-(2′-[Z]-pentenyl)cyclopentane-1-octanoic acid; cis-(+)-12-OPDA, cis-(+)-12-oxo-phytodienoic acid; OPR3, 12-oxo-phytodienoic acid reductase; PLA1, phospholipase A1.
The initial step in JA biosynthesis is oxygenation of α-linolenic acid in the C-13 position by lipoxygenase (LOX) (Figure 1) [33]. Tomato mutants impaired in TmLOXD (wound-induced 13-lipooxygenase) function were unable to produce JA. Moreover, the significantly increased accumulation of JA as well as enhanced resistance to B. cinerea in tomato plants overexpressing LOXD gene was observed [34]. The fatty acid hyperoxide resulting from α-LeA oxygenation is subsequently dehydrated by allene oxide synthase (AOS) to unstable allene oxide. In the presence of allene oxide cyclase (AOC), allene oxide is transformed into 12-oxo-phytodienoic acid (OPDA) enantiomer, 9S,13S/cis-(+)-OPDA, and it is the last step of JA biosynthesis that takes place in chloroplasts (Figure 1) [2]. The role of AOC in JA-dependent response to necrotrophic infection was confirmed in the Oryza sativa–Magnaporthe oryzae pathosystem, in which the rice mutants impaired in AOC production showed reduced production of JA and increased susceptibility to the pathogen [35]. In peroxisomes, cis (+)-OPDA is further converted into (+)-7-iso-JA by 12-oxo-phytodienoic acid reductase (OPR) and three β-oxidation steps involving acyl-CoA oxidase (ACX) and l-3-ketoacyl-CoA thiolase (KAT) (Figure 1) [1][2]. Tomato plants with a silenced OPR3 gene displayed a significant increase in susceptibility to B. cinerea accompanied by the dramatically decreased production of both OPDA and JA-Ile [36]. Consequently, in double opr7/opr8 maize (Zea mays) mutants, the reduced biosynthesis of JA as well as a diminished resistance to oomycete Pythium aristoporum, was observed [37].
In the next step of JA biosynthesis in cytosol, (+)-7-iso-JA may be subsequently conjugated with an amino acid by JAR1 (JASMONATE RESISTANT1) synthase, which is able to bind amino acids exclusively to jasmonic acid molecule (Figure 1) [38]. Different members of JAR family may synthetize rather rarely the JA conjugates with different amino acids such as valine (Val), leucine (Leu) and phenylalanine (Phe) [30]; however, the most biologically substantial conjugate JA-Ile is provided by JAR1 [39]. The Arabidopsis jar1 mutant showed increased susceptibility to both the S. sclerotiorum strain deprived of Sclerotinia sclerotiorum integrin-like (SSITL) protein suppressing host resistance as well as to the wild type B. cinerea isolate [40]. Accordingly, in rice plants challenged with Magnaporthe grisea infection, a gradual increase in expression of OsJAR1, but not the OsJAR2 gene, was observed from 48 to 72 hpi. Simultaneously, the elevated OsJAR1 expression was accompanied by induction of endogenous JA-Ile, but not JA-Phe levels, within the same time period [41]. In agreement with the above findings, the content of (+)-7-iso-JA-Ile was found to be significantly elevated in wheat Fhb1 plants inoculated with F. graminearum in comparison to mock-inoculated plants [42], providing yet more evidence for the JA-Ile as a crucial jasmonate in defense against necrotrophic fungi. Metabolite profiling studies of Arabidopsis plants infected with B. cinerea showed the maximum peak of JA-Ile accumulation at 3 days post-inoculation (dpi) [43]. The intensity and duration of JA responses are controlled to a large degree by the precise balance between biosynthesis and catabolism of JA-Ile. It was demonstrated that CYP94B3, CYP94C1 and CYP94B1 genes, the members of Cyt P450 family, play a key role in JA-Ile catabolic inactivation [44][45][46][47]. These genes encode JA-Ile 12-hydroxylase, which is an enzyme catalyzing the conversion of JA-Ile to biologically inactive hydroxylated forms. The disease symptoms in B. cinerea-infected Arabidopsis lines overexpressing CYP94B3 and CYP94C1 genes (B3-OE and C1-OE, respectively) were much stronger in comparison to wild type plants. Moreover, the expression levels of JA defense cascade marker genes, PDF1.2 and PR4, were strongly impaired in infected OE lines. These findings clearly indicate that CYP94B3 and CYP94C1 are integral components of the fungus-induced metabolic pathway controlling the abundance of JA-Ile [43]. In general, JA and its precursors contents increase in plant cultivar resistant to necrotrophic fungi more than in susceptible ones.
In the context of defense response against necrotrophic fungal infection, the concurrent/independent operation of another jasmonate forms alternative to JA-Ile conjugate should be considered. Analogous yet variant phenomenon revealed the significant accumulation of JA-Phe conjugate and its cyp94-oxidized forms in Arabidopsis plants infected with B. cinerea, suggesting that precisely controlled levels of JA-Phe may also be involved in responses to necrotrophic pathogens [48]. In maize, infection by Cochliobolus heterostrophus resulted in the local production of 9-lipoxygenase (LOX)-derived 10-oxo-11-phytoenoic acid (10-OPEA), 10-OPDA and a series of related 12- and 14-carbon cyclopente(a)nones, which apart from displaying direct phytoalexin activity, mediate defense gene expression [49]. Similarly, in tomato plants infected with B. cinerea, OPDA played a major role in defense response not only as a precursor of JA but also as an autonomous mediator [36].
The role of methyl ester of jasmonic acid (MeJA) as a mediator in defense against necrotrophs was also suggested [50]. Only a few studies have provided, however indirectly, further evidence supporting this theory. Fungal elicitor alamethicin isolated from Trichoderma viridae was revealed to cause significant induction of gene encoding JA carboxyl methyl transferase (JMT), a key enzyme catalyzing the conversion of JA to MeJA, in poplar (Populus trichocarpa) leaves within 2 h after treatment [51]. Consistently, the transcriptional activation of JMT was observed in Brassica juncea-Alternaria brassicicola pathosystem at 2 dpi [52]. However, it has to be emphasized that an exogenous application of MeJA to plants before or simultaneously with a necrotrophic fungus induce in many pathosystems a sufficient defense response to restrict a necrotroph development and limit lesions spreading [53][54][55][56][57].
2. JA Biosynthesis Genes Induced in Response to Necrotrophic Fungi Infection
The need for accumulation of JA levels effective for signal transduction in response to pathogen infection compel host plants into reprogramming the transcriptional activity of JA-biosynthesis genes. Accordingly, numerous transcriptomic surveys confirmed that genes encoding enzymes involved in JA biosynthesis are induced upon necrotrophic fungi infection, suggesting the direct and pathogen-responsive transcriptomic regulation of JA abundance in planta.
2.1. Phospholipase (PL) Genes
As mentioned above, the primary role of A1 family phospholipases is releasing α-linolenic acid for further JA biosynthesis; although possible, this seems to be uncertain in the case of response to necrotrophic fungi infection. However, deep transcriptome sequencing experiments revealed the significant up-regulation of PLA1 genes upon pathogen attack, the involvement of yet another PLs gene has to be considered in at least JA biosynthesis regulation. In Arabidopsis plants infected with B. cinerea, the induction of A1 as well as Dγ1 phospholipase genes (observed at 18 hpi), was preceded by the up-regulation of PLA2 gene (12 hpi), whereby the elevated levels of all these genes transcripts were detectable also at 24 hpi [58]. In earlier research on the same pathosystem, no significant up-regulation of A1 family phospholipase genes has been observed. However, induction of A2α, Dγ1 and Dδ1 phospholipases encoding genes at 18 hpi was revealed, whereas the phospholipase Dγ2 gene was shown to be down-regulated at that time point [59]. Moreover, elevated levels of A2, A2β, Dα1 and Dβ1 phospholipase gene transcripts in B. cinerea-infected Arabidopsis plants were detected (Table 1) [60]. Nevertheless, elevated transcript levels for phospholipase A1γ and Dβ1 in wild tomato (Solanum lycopersicoides) at 24 h after B. cinerea infection were revealed [61]. In lettuce (Lactuca sativa), in plants infected with B. cinerea, up-regulation of three phospholipase A1 and four phospholipase D (γ1, ζ1, ζ2 and one of unknown isoform) encoding genes were observed at 48 hpi (Table 1) [62]. The up-regulation of phospholipase A1, Dβ1 and Dα2 genes was detected in pooled samples of chrysanthemum (Chrysanthemum morifolium) leaves, collected at five time points between 0 and 72 h after inoculation with Alternaria tenuissima [63]. Comparison of transcriptomes of resistant (R) and susceptible (S) Brassica napus lines challenged with S. sclerotiorum infection revealed significant up-regulation of phospholipase A2α and Dζ2 genes in R lines at 48 h post-inoculation (hpi). No significant increase in expression level of PLA1 genes was observed in this case [64].
Table 1. Phospholipase (PL) genes encoding different isoforms of PLA and PLD active in various pathosystems.
In light of the above results, it has to be considered that trigger-up of jasmonate biosynthesis upon necrotrophic fungi infection is not exclusively regulated by the phospholipase A1 family genes and that the role of A2 and especially the D family of PLs genes may be underestimated here (Table 1). This conclusion is consistent with the findings of depleted production of JA and resistance level in PLDβ1 dysfunctional Arabidopsis mutants, suggesting a role of the Dβ1 phospholipase gene as a positive regulator of JA biosynthesis in response to B. cinerea [65].
2.2. Lipoxygenase (LOX) Genes
Plant lipoxygenases are often classified according to a positional specificity for the oxygenation of polyunsaturated fatty acids (PUFAs). Thus, plants produce two classes of lipoxygenases 13-LOX and 9-LOX inserting O2 to C-13 or C-9 position of hydrocarbon backbone of linolenic acid, respectively [66]. However, only 13-LOXs participate in JA biosynthesis. From six genes encoding lipoxygenases in A. thaliana, four genes encode LOX2, LOX3, LOX4 and LOX6 enzymes that show 13S-lipoxygenase activity, contain chloroplast signaling peptides, and were proven to function in JA biosynthesis in Arabidopsis [1][67][68]. Analysis of RNA sequencing-based transcriptomics revealed that Arabidopsis plants challenged with B. cinerea infection displayed the elevated expression of LOX2 and LOX4 genes at 18 hpi compared to control plants [58][59]. Quite confusingly, in a previous study, the down-regulation of LOX2 in Arabidopsis plants starting 20 h after inoculation with B. cinerea was observed; however, the LOX4 gene was shown to be up-regulated within that time [60]. The authors speculated that such differences in regulation of the genes belonging to the same pathway may reflect distinct roles of particular LOX genes in the biosynthesis of JA in response to different stimuli. However, LOX2 down-regulation was also observed in susceptible Brassica oleracea inoculated with A. brassicicola at a later stage of infection (48 hpi) [69]. In phenotypically resistant Brassica napus genotypes, when comparing susceptible plants, the LOX2 gene was found to be up-regulated at 24 h, whereas LOX3 and LOX4 genes were up-regulated at 48 h after inoculation with S. sclerotiorum [64][70][71]. Similarly, expression of LOX2 and LOX4 genes was induced in lettuce plants inoculated with B. cinerea at 48 hpi [62].
Surprisingly, no significant induction of 13S-lipoxygenase genes was observed neither in cucumber (Cucumis sativa) [72] nor in S. lycopersicoides plants [61] and S. lycopersicum fruits [73] infected with B. cinerea. However, tomato (S. lycopersicum) mutants with a dysfunctional 13S-lipoxygenase D (TomLOXD) gene displayed severely compromised resistance to B. cinerea. Consistently, the overexpression of TomLOXD resulted in elevated JA biosynthesis and enhanced resistance to this pathogen [34]. The above results suggest that in the case of LOX genes the regulation of their product abundance may be driven by the mechanism different than transcriptional control.
2.3. Allene Oxide Synthase (AOS) and Allene Oxide Cyclase (AOC) Genes
Allene oxide synthase (AOS) catalyzes the synthesis of LOX-produced 9-/13-HPOT (polyunsaturated fatty acids hydroperoxides) to the unstable epoxide, 12,13-EOT (12,13-epoxyoctadecatrienoic acid), which is further cyclized to 12-oxo-phytodienoic acid (OPDA) by allene oxide cyclase (AOC). Similar to LOXs, only 13-AOS functions in JA biosynthesis. Either 13-AOS and AOC genes encode a plastid-transit peptide, indicating that OPDA synthesis is localized in chloroplast [67]. In Arabidopsis, a single copy of AOS gene and four genes of AOC have been identified [74][75].
The induction of the AOS gene in both resistant (R) and susceptible (S) B. napus genotypes was revealed at 24 h after inoculation with S. sclerotiorum; however, the higher level of its expression was observed in R genotypes at that time point [64]. The up-regulation of AOS gene was also observed in Arabidopsis after inoculation with B. cinerea (18 hpi) [59], lettuce plants (48 hpi) [62], as well as in green and ripe tomato fruits (1 dpi) [73].
A significant up-regulation of AOC2 gene was observed in resistant B. napus genotypes 48 h after inoculation with S. sclerotiorum [71]. Nevertheless, in most recent studies, no significant differences in AOC2 expression level were found between R and S genotypes for this pathosystem [64]. The latter authors, however, observed the enhanced up-regulation of the AOC3 gene at 24 hpi and the AOC4 gene at 48 hpi in B. napus R genotypes when compared to S plants. The up-regulation of AOC2 and AOC3 genes was observed in Arabidopsis plants 18 h after inoculation with B. cinerea [58]. These results are unanimous with previous research on this pathosystem in which the induced expression of AOC2 and AOC3 was observed at 8 and 20 hpi, respectively [60]. However, in the latter experiment, the down-regulation of the AOC4 gene was observed after 20 hpi, similar to the LOX2 manner of expression yet inconsistent with the other members of this pathway. Confusingly, no significant changes were found in any of the AOC gene expressions in Arabidopsis plants tested at 18 h after inoculation with B. cinerea [59]. Similar to that, no regulation of AOC genes was detected in tomato (S. lycopersicum) fruits [73] and cucumber (C. sativa) plants [72] infected with this pathogen. In the latter case, the operation of a signaling pathway alternative to JA-mediated one may be speculated, as none of the genes involved in jasmonate biosynthesis displayed a regulation in infection-triggered manner.
2.4. Oxo-Phytodienoic Acid Reductase (OPR) Genes
The family of oxo-phytodienoic acid (OPDA) reductases (OPRs) comprises at least 3 members in tomato, 6 in Arabidopsis, 6 in pea, 8 in maize and 10 in rice [76]. As described above, the silencing of the OPR3 gene in tomato as well as disruption of OPR7 and OPR8 genes in maize resulted in decreased production of JA and diminished resistance to necrotrophic fungi [36][37], supporting the idea that jasmonic acid and not OPDA plays a crucial role in defense to this group of pathogens. The up-regulation of OPR1 and OPR3 genes in susceptible and resistant B. napus genotypes infected with S. sclerotiorum was observed, with no significant differences in expression levels between the two phenotypic groups [64].
Quite unexpectedly, no up-regulation of the OPR3 gene in Arabidopsis upon infection with B. cinerea was revealed. However, 24 h after the combined challenge with B. cinerea and herbivore pest Pieris rapae, the induction of this gene was observed, suggesting that in that case the mechanical wounding stimulus had a bigger effect on JA biosynthesis than of necrotrophic infection alone [58]. These findings are in accordance with earlier research [60] that also reported no time-course differences in OPR genes expression in Arabidopsis plants during B. cinerea infection. The explanation for such expression observed in the above-mentioned experiments seems unobtainable at the moment, especially as the up-regulation of the OPR3 gene was -yet revealed in another transcriptomic study in this pathosystem [59].
3. JA-Mediated Response to Necrotrophic Infection Regulated by Circadian Clock and Photoperiod
The circadian clock, an endogenous time-keeping mechanism, adjusts biological processes of a plant in response to environmental signals, so that they are turned on at optimum times throughout the day [77][78]. Plant defenses are also rhythmically regulated to be expressed with full strength at the time of maximal susceptibility to infection or to synchronize with the time of the day when a pathogen is most abundant [79]. Arabidopsis plants show differential susceptibility to B. cinerea depending on the time of inoculation during the day [80]. It is speculated that plants can anticipate the timing of pathogen infection by time-specific defense pathway activation and thus maximize the response against a particular pathogen [81]. Consequently, the susceptibility of Arabidopsis to B. cinerea decreases after inoculation at early daytime (dawn) compared with night. Moreover, the state of decreased susceptibility persists under permanent light conditions and is disrupted in mutants impaired in circadian clock (CC) function. Moreover, the enhanced susceptibility to this pathogen has been lost in the jaz6 mutant, suggesting the key role of JA signal transduction via JAZ6 in rhythm-dependent susceptibility of Arabidopsis to B. cinerea [80]. As yet, the only evidence for the direct molecular interaction between CC and JA-mediated defense components comes from the plant response to bacteria P. syringae pv. tomato. As it was revealed, the circadian clock component TIME FOR COFFEE (TIC) rhythmically regulates the JA signaling pathway in Arabidopsis by inhibiting MYC2 protein accumulation and controlling transcriptional repression of COI1 in an evening-phase-specific manner [81]. In case of temporal variation in susceptibility to necrotrophic fungi, the operation of more complex functional CC network has been suggested, since among the transcription factors that responded more rapidly to infection at subjective dawn than subjective night, the target genes of core clock regulators were shown to be notably abundant [80]. Moreover, duration of the light period seems to influence not only regulation of plant response to biotic stress factors but also the development of an attacking pathogen [82][83]. Mustard plants (B. juncea) grown under different regimes of light periods showed variation not only in leaf size but also in necrosis formation in response to A. brassicicola. The light period over 16 h restricted leaf development and necrosis spreading [84]. However, how this phenomenon may be connected to a plant JA-dependent resistant response to A. brassicicola must be further explored [84]. Consistently, long day photoperiod enhanced Arabidopsis resistance to B. cinerea activating JA-dependent defense responses, e.g., expression of MYC2 gene [85]. Nevertheless, the JA-dependent influence of circadian clock and photoperiod on defense response to necrotrophic fungi requires further extensive investigations.
4. Conclusions
Negative impact of climatic changes and a growing human population requires harnessing new efficient technologies in agriculture to increase yield of crops and decrease to minimum the loss of yield and incomes due to the disadvantageous influence, among other factors, of pathogenic fungi [86]. One of the new approaches to create modern agricultural technologies, which fit into ecological trends leading mostly in Europe and North America, is the use of natural plant defense mechanisms against pathogens. Skilled use and/or manipulation of JA biosynthesis and JA-dependent signaling pathways can be a good basis for development of novel ‘green’ compounds that not only stimulate growth of plants but also increase the defense capacity of the whole plant with a long-lasting effect against attacks of various necrotrophic pathogens.
In recent years, many research groups all over the world have worked on JA biosynthesis and signaling in various crop species. However, further investigations should also focus exclusively on the JA-dependent signal transduction pathway and JA-responsive genes activation in plants resistant and susceptible to necrotrophic fungi under not only laboratory conditions but also in the field.
References
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 2017, 68, 1303–1321.
- Zhai, Q.; Yan, C.; Li, L.; Xie, D.; Li, C. Jasmonates. In Hormone Metabolism and Signaling in Plants; Li, J., Li, C., Smith, S.M., Eds.; Elsevier Ltd.: Amsterdam, The Netherlands, 2017; pp. 243–272.
- Huang, H.; Liu, B.; Liu, L.; Song, S. Jasmonate action in plant growth and development. J. Exp. Bot. 2017, 68, 1349–1359.
- Thines, B.; Parlan, E.V.; Fulton, E.C. Circadian Network Interactions with Jasmonate Signaling and Defense. Plants 2019, 8, 252.
- Hu, Y.; Jiang, Y.; Han, X.; Wang, H.; Pan, J.; Yu, D. Jasmonate regulates leaf senescence and tolerance to cold stress: Crosstalk with other phytohormones. J. Exp. Bot. 2017, 68, 1361–1369.
- Wang, Y.; Mostafa, S.; Zeng, W.; Jin, B. Function and Mechanism of Jasmonic Acid in Plant Responses to Abiotic and Biotic Stresses. Int. J. Mol. Sci. 2021, 22, 8568.
- Raza, A.; Charagh, S.; Zahid, Z.; Mubarik, M.S.; Javed, R.; Siddiqui, M.H.; Hasanuzzaman, M. Jasmonic acid: A key frontier in conferring abiotic stress tolerance in plants. Plant Cell Rep. 2021, 40, 1513–1541.
- Wang, J.; Wu, D.; Wang, Y.; Xie, D. Jasmonate action in plant defense against insects. J. Exp. Bot. 2019, 70, 3391–3400.
- Zhang, L.; Zhang, F.; Melotto, M.; Yao, J.; He, S.Y. Jasmonate signaling and manipulation by pathogens and insects. J. Exp. Bot. 2017, 68, 1371–1385.
- Ali, S.; Ganai, B.A.; Kamili, A.N.; Bhat, A.A.; Mir, Z.A.; Bhat, J.A.; Tyagi, A.; Islam, S.T.; Mushtaq, M.; Yadav, P.; et al. Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol. Res. 2018, 212–213, 29–37.
- Sohn, S.-I.; Pandian, S.; Rakkammal, K.; Largia, M.J.V.; Thamilarasan, S.K.; Balaji, S.; Zoclanclounon, Y.A.B.; Shilpha, J.; Ramesh, M. Jasmonates in plant growth and development and elicitation of secondary metabolites: An updated overview. Front. Plant Sci. 2022, 13, 942789.
- Wasternack, C.; Strnad, M. Jasmonates are signals in the biosynthesis of secondary metabolites—Pathways, transcription factors and applied aspects—A brief review. New Biotechnol. 2019, 48, 1–11.
- Onohata, T.; Gomi, K. Overexpression of jasmonate-responsive OsbHLH034 in rice results in the induction of bacterial blight resistance via an increase in lignin biosynthesis. Plant Cell Rep. 2020, 39, 1175–1184.
- Hu, Q.; Min, L.; Yang, X.; Jin, S.; Zhang, L.; Li, Y.; Ma, Y.; Qi, X.; Li, D.; Liu, H.; et al. Laccase GhLac1 Modulates Broad-Spectrum Biotic Stress Tolerance via Manipulating Phenylpropanoid Pathway and Jasmonic Acid Synthesis. Plant Physiol. 2018, 176, 1808–1823.
- Tamogami, S.; Agrawal, G.K.; Rakwal, R. Targeted Quantitative Analysis of Jasmonic Acid (JA) and Its Amino Acid Conjugates in Plant Using HPLC-Electrospray Ionization-Tandem Mass Spectrometry (ESI-LC-MS/MS). In Sample Preparation in Biological Mass Spectrometry; Ivanov, A., Lazarev, A., Eds.; Springer: Dordrecht, The Netherlands, 2011; pp. 869–875.
- Xie, D.; Feys, B.; James, S.; Nieto-Rostro, M.; Turner, J.G. COI1: An Arabidopsis Gene Required for Jasmonate-Regulated Defense and Fertility. Science 1998, 280, 1091–1094.
- Yan, J.; Zhang, C.; Gu, M.; Bai, Z.; Zhang, W.; Qi, T.; Cheng, Z.; Peng, W.; Luo, H.; Nan, F.; et al. The Arabidopsis CORONATINE INSENSITIVE1 Protein Is a Jasmonate Receptor. Plant Cell 2009, 21, 2220–2236.
- Fonseca, S.; Chini, A.; Hamberg, M.; Adie, B.; Porzel, A.; Kramell, R.; Miersch, O.; Wasternack, C.; Solano, R. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 2009, 5, 344–350.
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 2007, 448, 661–665.
- Melotto, M.; Mecey, C.; Niu, Y.; Chung, H.S.; Katsir, L.; Yao, J.; Zeng, W.; Thines, B.; Staswick, P.; Browse, J.; et al. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 2008, 55, 979–988.
- Katsir, L.; Schilmiller, A.L.; Staswick, P.E.; He, S.Y.; Howe, G.A. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Nat. Acad. Sci. USA 2008, 105, 7100–7105.
- Newman, T.E.; Derbyshire, M.C. The Evolutionary and Molecular Features of Broad Host-Range Necrotrophy in Plant Pathogenic Fungi. Front. Plant Sci. 2020, 11, 591733.
- Liao, C.-J.; Hailemariam, S.; Sharon, A.; Mengiste, T. Pathogenic strategies and immune mechanisms to necrotrophs: Differences and similarities to biotrophs and hemibiotrophs. Curr. Opin. Plant Biol. 2022, 69, 102291.
- Fisher, M.C.; Henk, D.A.; Briggs, C.J.; Brownstein, J.S.; Madoff, L.C.; Mccraw, S.L.; Gurr, S.J. Emerging fungal threats to animal, plant and ecosystem health. Nature 2012, 484, 186–194.
- Fones, H.N.; Bebber, D.P.; Chaloner, T.M.; Kay, W.T.; Steinberg, G.; Gurr, S.J. Threats to global food security from emerging fungal and oomycete crop pathogens. Nat. Food 2020, 1, 332–342.
- Bi, K.; Liang, Y.; Mengiste, T.; Sharon, A. Killing softly: A roadmap of Botrytis cinerea pathogenicity. Trends Plant Sci. 2023, 28, 2.
- Derbyshire, M.C.; Newman, T.E.; Khentry, Y.; Taiwo, A.O. The evolutionary and molecular features of the broad-host-range plant pathogen Sclerotinia sclerotiorum. Mol. Plant Pathol. 2022, 23, 1075–1090.
- Macioszek, V.K.; Lawrence, C.B.; Kononowicz, A.K. Infection cycle of Alternaria brassicicola on Brassica oleracea leaves under growth room conditions. Plant Pathol. 2018, 67, 1088–1096.
- Shao, D.; Smith, D.L.; Kabbage, M.; Roth, M.G. Effectors of Plant Necrotrophic Fungi. Front. Plant Sci. 2021, 12, 687713.
- Ghorbel, M.; Brini, F.; Sharma, A.; Landi, M. Role of jasmonic acid in plants: The molecular point of view. Plant Cell Rep. 2021, 40, 1471–1494.
- Sewelam, N.; El-Shetehy, M.; Mauch, F.; Maurino, V.G. Combined Abiotic Stresses Repress Defense and Cell Wall Metabolic Genes and Render Plants More Susceptible to Pathogen Infection. Plants 2021, 10, 1946.
- Nguyen, T.H.; Goossens, A.; Lacchini, E. Jasmonate: A hormone of primary importance for plant metabolism. Curr. Opin. Plant Biol. 2022, 67, 102197.
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058.
- Yan, L.; Zhai, Q.; Wei, J.; Li, S.; Wang, B.; Huang, T.; Du, M.; Sun, J.; Kang, L.; Li, C.-B.; et al. Role of Tomato Lipoxygenase D in Wound-Induced Jasmonate Biosynthesis and Plant Immunity to Insect Herbivores. PLoS Genet. 2013, 9, e1003964.
- Riemann, M.; Haga, K.; Shimizu, T.; Okada, K.; Ando, S.; Mochizuki, S.; Nishizawa, Y.; Yamanouchi, U.; Nick, P.; Yano, M.; et al. Identification of rice Allene Oxide Cyclasemutants and the function of jasmonate for defence against Magnaporthe oryzae. Plant J. 2013, 74, 226–238.
- Scalschi, L.; Sanmartín, M.; Camañes, G.; Troncho, P.; Sánchez-Serrano, J.J.; García-Agustín, P.; Vicedo, B. Silencing of OPR3 in tomato reveals the role of OPDA in callose deposition during the activation of defense responses against Botrytis cinerea. Plant J. 2015, 81, 304–315.
- Yan, Y.; Christensen, S.; Isakeit, T.; Engelberth, J.; Meeley, R.; Hayward, A.; Emery, R.J.N.; Kolomiets, M.V. Disruption of OPR7 and OPR8 Reveals the Versatile Functions of Jasmonic Acid in Maize Development and Defense. Plant Cell 2012, 24, 1420–1436.
- Staswick, P.E.; Tiryaki, I. The Oxylipin Signal Jasmonic Acid Is Activated by an Enzyme That Conjugates It to Isoleucine in Arabidopsis. Plant Cell 2004, 16, 2117–2127.
- Guranowski, A.; Miersch, O.; Staswick, P.E.; Suza, W.; Wasternack, C. Substrate specificity and products of side-reactions catalyzed by jasmonate:amino acid synthetase (JAR1). FEBS Lett. 2007, 581, 815–820.
- Zhu, W.; Wei, W.; Fu, Y.; Cheng, J.; Xie, J.; Li, G.; Yi, X.; Kang, Z.; Dickman, M.B.; Jiang, D. A Secretory Protein of Necrotrophic Fungus Sclerotinia sclerotiorum That Suppresses Host Resistance. PLoS ONE 2013, 8, e53901.
- Wakuta, S.; Suzuki, E.; Saburi, W.; Matsuura, H.; Nabeta, K.; Imai, R.; Matsui, H. OsJAR1 and OsJAR2 are jasmonyl-l-isoleucine synthases involved in wound- and pathogen-induced jasmonic acid signalling. Biochem. Biophys. Res. Commun. 2011, 409, 634–639.
- Gunnaiah, R.; Kushalappa, A.C.; Duggavathi, R.; Fox, S.; Somers, D.J. Integrated Metabolo-Proteomic Approach to Decipher the Mechanisms by Which Wheat QTL (Fhb1) Contributes to Resistance against Fusarium graminearum. PLoS ONE 2012, 7, e40695.
- Aubert, Y.; Widemann, E.; Miesch, L.; Pinot, F.; Heitz, T. CYP94-mediated jasmonoyl-isoleucine hormone oxidation shapes jasmonate profiles and attenuates defence responses to Botrytis cinerea infection. J. Exp. Bot. 2015, 66, 3879–3892.
- Koo, A.J.K.; Cooke, T.F.; Howe, G.A. Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-L-isoleucine. Proc. Nat. Acad. Sci. USA 2011, 108, 9298–9303.
- Koo, A.J.; Thireault, C.; Zemelis, S.; Poudel, A.N.; Zhang, T.; Kitaoka, N.; Brandizzi, F.; Matsuura, H.; Howe, G.A. Endoplasmic Reticulum-associated Inactivation of the Hormone Jasmonoyl-l-Isoleucine by Multiple Members of the Cytochrome P450 94 Family in Arabidopsis. J. Biol. Chem. 2014, 289, 29728–29738.
- Kitaoka, N.; Matsubara, T.; Sato, M.; Takahashi, K.; Wakuta, S.; Kawaide, H.; Matsui, H.; Nabeta, K.; Matsuura, H. Arabidopsis CYP94B3 Encodes Jasmonyl-L-Isoleucine 12-Hydroxylase, a Key Enzyme in the Oxidative Catabolism of Jasmonate. Plant Cell Physiol. 2011, 52, 1757–1765.
- Heitz, T.; Widemann, E.; Lugan, R.; Miesch, L.; Ullmann, P.; Desaubry, L.; Holder, E.; Grausem, B.; Kandel, S.; Miesch, M.; et al. Cytochromes P450 CYP94C1 and CYP94B3 Catalyze Two Successive Oxidation Steps of Plant Hormone Jasmonoyl-isoleucine for Catabolic Turnover. J. Biol. Chem. 2012, 287, 6296–6306.
- Widemann, E.; Grausem, B.; Renault, H.; Pineau, E.; Heinrich, C.; Lugan, R.; Ullmann, P.; Miesch, L.; Aubert, Y.; Miesch, M.; et al. Sequential oxidation of Jasmonoyl-Phenylalanine and Jasmonoyl-Isoleucine by multiple cytochrome P450 of the CYP94 family through newly identified aldehyde intermediates. Phytochemistry 2015, 117, 388–399.
- Christensen, S.A.; Huffaker, A.; Kaplan, F.; Sims, J.; Ziemann, S.; Doehlemann, G.; Jif, L.; Schmitz, R.J.; Kolomietsh, M.V.; Alborn, H.T.; et al. Maize death acids, 9-lipoxygenase–derived cyclopente(a)nones, display activity as cytotoxic phytoalexins and transcriptional mediators. Proc. Natl. Acad. Sci. USA 2015, 112, 11407–11412.
- Seo, H.S.; Song, J.T.; Cheong, J.-J.; Lee, Y.-H.; Lee, Y.-W.; Hwang, I.; Lee, J.S.; Choi, Y.D. Jasmonic acid carboxyl methyltransferase: A key enzyme for jasmonate-regulated plant responses. Proc. Natl. Acad. Sci. USA 2001, 98, 4788–4793.
- Zhao, N.; Yao, J.; Chaiprasongsuk, M.; Li, G.; Guan, J.; Tschaplinski, T.J.; Guo, H.; Chen, F. Molecular and biochemical characterization of the jasmonic acid methyltransferase gene from black cottonwood (Populus trichocarpa). Phytochemistry 2013, 94, 74–81.
- Meur, G.; Shukla, P.; Dutta-Gupta, A.; Kirti, P.B. Characterization of Brassica juncea–Alternaria brassicicola interaction and jasmonic acid carboxyl methyl transferase expression. Plant Gene 2015, 3, 1–10.
- Kępczyńska, E.; Król, P. The phytohormone methyl jasmonates as an activator of induced resistance against the necrotroph Alternaria porri f. sp. solani in tomato plants. J. Plant Interact. 2012, 7, 307–315.
- Yu, X.; Zhang, W.; Zhang, Y.; Zhang, X.; Lang, D.; Zhang, X. The roles of methyl jasmonate to stress in plants. Funct. Plant Biol. 2019, 46, 197–212.
- Wang, H.; Kou, X.; Wu, C.; Fan, G.; Li, T. Methyl jasmonate induces the resistance of postharvest blueberry to gray mold caused by Botrytis cinerea. J. Sci. Food Agric. 2020, 100, 4272–4281.
- Li, S.; Xiao, L.; Chen, M.; Cao, Q.; Luo, Z.; Kang, N.; Jia, M.; Chen, J.; Xiang, M. The involvement of the phenylpropanoid and jasmonate pathways in methyl jasmonate-induced soft rot resistance in kiwifruit (Actinidia chinensis). Front. Plant Sci. 2022, 13, 1097733.
- Dixit, S.; Grover, A.; Pushkar, S.; Singh, S.B. BA-induced SA accumulation causes higher susceptibility in B. juncea as compared to tolerant genotypes against A. brassicae. bioRxiv 2022. preprint.
- Coolen, S.; Proietti, S.; Hickman, R.; Olivas, N.H.D.; Huang, P.-P.; Van Verk, M.C.; Van Pelt, J.A.; Wittenberg, A.H.J.; De Vos, M.; Prins, M.; et al. Transcriptome dynamics of Arabidopsis during sequential biotic and abiotic stresses. Plant J. 2016, 86, 249–267.
- Sham, A.; Al-Azzawi, A.; Al-Ameri, S.; Al-Mahmoud, B.; Awwad, F.; Al-Rawashdeh, A.; Iratni, R.; AbuQamar, S. Transcriptome Analysis Reveals Genes Commonly Induced by Botrytis cinerea Infection, Cold, Drought and Oxidative Stresses in Arabidopsis. PLoS ONE 2014, 9, e113718.
- Windram, O.; Madhou, P.; McHattie, S.; Hill, C.; Hickman, R.; Cooke, E.; Jenkins, D.J.; Penfold, C.A.; Baxter, L.; Breeze, E.; et al. Arabidopsis Defense against Botrytis cinerea: Chronology and Regulation Deciphered by High-Resolution Temporal Transcriptomic Analysis. Plant Cell 2012, 24, 3530–3557.
- Smith, J.E.; Mengesha, B.; Tang, H.; Mengiste, T.; Bluhm, B.H. Resistance to Botrytis cinerea in Solanum lycopersicoides involves widespread transcriptional reprogramming. BMC Genom. 2014, 15, 334.
- De Cremer, K.; Mathys, J.; Vos, C.; Froenicke, L.; Michelmore, R.W.; Cammue, B.P.A.; De Coninck, B. RNAseq-based transcriptome analysis of Lactuca sativa infected by the fungal necrotroph Botrytis cinerea. Plant Cell Environ. 2013, 36, 1992–2007.
- Li, H.; Chen, S.; Song, A.; Wang, H.; Fang, W.; Guan, Z.; Jiang, J.; Chen, F. RNA-Seq derived identification of differential transcription in the chrysanthemum leaf following inoculation with Alternaria tenuissima. BMC Genom. 2014, 15, 9.
- Wu, J.; Zhao, Q.; Yang, Q.; Liu, H.; Li, Q.; Yi, X.; Cheng, Y.; Guo, L.; Fan, C.; Zhou, Y. Comparative transcriptomic analysis uncovers the complex genetic network for resistance to Sclerotinia sclerotiorum in Brassica napus. Sci. Rep. 2016, 6, 19007.
- Zhao, J.; Devaiah, S.P.; Wang, C.; Li, M.; Welti, R.; Wang, X. Arabidopsis phospholipase Dβ1 modulates defense responses to bacterial and fungal pathogens. New Phytol. 2013, 199, 228–240.
- Singh, P.; Yamshi, A.; Miszczuk, E.; Bajguz, A.; Hayat, S. Specific Roles of Lipoxygenases in Development and Responses to Stress in Plants. Plants 2022, 11, 979.
- Yan, Y.; Borrego, E.; Kolomiets, M.V. Jasmonate Biosynthesis, Perception and Function in Plant Development and Stress Responses. In Lipid Metabolism; Baez, R.V., Ed.; IntechOpen: London, UK, 2013; pp. 393–442.
- Viswanath, K.K.; Varakumar, P.; Pamuru, R.R.; Basha, S.J.; Mehta, S.; Rao, A.D. Plant Lipoxygenases and Their Role in Plant Physiology. J. Plant Biol. 2020, 63, 83–95.
- Macioszek, V.K.; Gapińska, M.; Zmienko, A.; Sobczak, M.; Skoczowski, A.; Oliwa, J.; Kononowicz, A.K. Complexity of Brassica oleracea–Alternaria brassicicola Susceptible Interaction Reveals Downregulation of Photosynthesis at Ultrastructural, Transcriptional, and Physiological Levels. Cells 2020, 9, 2329.
- Wang, Z.; Tan, X.; Zhang, Z.; Gu, S.; Li, G.; Shi, H. Defense to Sclerotinia sclerotiorum in oilseed rape is associated with the sequential activations of salicylic acid signaling and jasmonic acid signaling. Plant Sci. 2012, 184, 75–82.
- Wei, L.; Jian, H.; Lu, K.; Filardo, F.; Yin, N.; Liu, L.; Qu, C.; Li, W.; Du, H.; Li, J. Genome-wide association analysis and differential expression analysis of resistance to Sclerotinia stem rot in Brassica napus. Plant Biotechnol. J. 2015, 14, 1368–1380.
- Kong, W.; Chen, N.; Liu, T.; Zhu, J.; Wang, J.; He, X.; Jin, Y. Large-Scale Transcriptome Analysis of Cucumber and Botrytis cinerea during Infection. PLoS ONE 2015, 10, e0142221.
- Blanco-Ulate, B.; Vincenti, E.; Powell, A.L.T.; Cantu, D. Tomato transcriptome and mutant analyses suggest a role for plant stress hormones in the interaction between fruit and Botrytis cinerea. Front. Plant Sci. 2013, 4, 142.
- Stenzel, I.; Hause, B.; Miersch, O.; Kurz, T.; Maucher, H.; Weichert, H.; Ziegler, J.; Feussner, I.; Wasternack, C. Jasmonate biosynthesis and the allene oxide cyclase family of Arabidopsis thaliana. Plant Mol. Biol. 2003, 51, 895–911.
- Farmer, E.E.; Goossens, A. Jasmonates: What ALLENE OXIDE SYNTHASE does for plants. J. Exp. Bot. 2019, 70, 3373–3378.
- Schaller, A.; Stintzi, A. Enzymes in jasmonate biosynthesis—Structure, function, regulation. Phytochemistry 2009, 70, 1532–1538.
- Sharma, M.; Bhatt, D. The circadian clock and defence signalling in plants. Mol. Plant Pathol. 2015, 16, 210–218.
- Venkat, A.; Munee, S. Role of Circadian Rhythms in Major Plant Metabolic and Signaling Pathways. Front. Plant Sci. 2022, 13, 836244.
- Hevia, M.A.; Canessa, P.; Müller-Esparza, H.; Larrondo, L.F. A circadian oscillator in the fungus Botrytis cinerea regulates virulence when infecting Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2015, 112, 8744–8749.
- Ingle, R.A.; Stoker, C.; Stone, W.; Adams, N.; Smith, R.; Grant, M.; Carré, I.; Roden, L.C.; Denby, K.J. Jasmonate signalling drives time-of-day differences in susceptibility of Arabidopsis to the fungal pathogen Botrytis cinerea. Plant J. 2015, 84, 937–948.
- Shin, J.; Heidrich, K.; Sanchez-Villarreal, A.; Parker, J.E.; Davis, S.J. TIME FOR COFFEE Represses Accumulation of the MYC2 Transcription Factor to Provide Time-of-Day Regulation of Jasmonate Signaling in Arabidopsis. Plant Cell 2012, 24, 2470–2482.
- Roeber, V.M.; Schmülling, T.; Cortleven, A. The Photoperiod: Handling and Causing Stress in Plants. Front Plant Sci. 2022, 12, 781988.
- Shimizu, S.; Yamauchi, Y.; Ishikawa, A. Photoperiod Following Inoculation of Arabidopsis with Pyricularia oryzae (syn. Magnaporthe oryzae) Influences on the Plant–Pathogen Interaction. Int. J. Mol. Sci. 2021, 22, 5004.
- Macioszek, V.K.; Sobczak, M.; Skoczowski, A.; Oliwa, J.; Michlewska, S.; Gapińska, M.; Ciereszko, I.; Kononowicz, A.K. The Effect of Photoperiod on Necrosis Development, Photosynthetic Efficiency and ‘Green Islands’ Formation in Brassica juncea Infected with Alternaria brassicicola. Int. J. Mol. Sci. 2021, 22, 8435.
- Cagnola, J.I.; Cerdán, P.D.; Pacín, M.; Andrade, A.; Rodriguez, V.; Zurbriggen, M.D.; Legris, M.; Buchovsky, S.; Carrillo, N.; Chory, J.; et al. Long-Day Photoperiod Enhances Jasmonic Acid-Related Plant Defense. Plant Physiol. 2018, 178, 163–173.
- Bailey-Serres, J.; Parker, J.E.; Ainsworth, E.A.; Oldroyd, G.E.D.; Schroeder, J.I. Genetic strategies for improving crop yields. Nature 2019, 575, 109–118.
More
Information
Subjects:
Plant Sciences
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.3K
Revisions:
3 times
(View History)
Update Date:
05 May 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




