Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Su, X.; Qu, Y.; Mu, D. METTL3 in Neurophysiological Events. Encyclopedia. Available online: https://encyclopedia.pub/entry/43548 (accessed on 07 February 2026).
Su X, Qu Y, Mu D. METTL3 in Neurophysiological Events. Encyclopedia. Available at: https://encyclopedia.pub/entry/43548. Accessed February 07, 2026.
Su, Xiaojuan, Yi Qu, Dezhi Mu. "METTL3 in Neurophysiological Events" Encyclopedia, https://encyclopedia.pub/entry/43548 (accessed February 07, 2026).
Su, X., Qu, Y., & Mu, D. (2023, April 27). METTL3 in Neurophysiological Events. In Encyclopedia. https://encyclopedia.pub/entry/43548
Su, Xiaojuan, et al. "METTL3 in Neurophysiological Events." Encyclopedia. Web. 27 April, 2023.
Copy Citation
Methyltransferase-like 3 (METTL3) is a typical component of N6-methyladenosine writers that exhibits methyltransferase activity and deposits methyl groups on RNA. Accumulating studies have demonstrated the involvement of METTL3 in the regulation of neuro-physiological and pathological events.
METTL3
neurobiological events
neurological disorders
regulatory network
1. Introduction
N6-methyladenosine (m6A) is an epigenetic modification of RNAs [1], which involves adding (via writers) or removing (via erasers) a methyl group at the N6-position of adenosine [2]. Writers include methylases such as METTL3, METTL14, and WT1-associated protein (WTAP), and erasers include demethylases such as fat mass and obesity-associated protein (FTO) and AlkB homolog 5(ALKBH5) [3]. The modification is recognized by m6A reader proteins (such as YTHDF domain-containing proteins) [3] (Figure 1). Therefore, the m6A modification is a carefully controlled and regulated process. Specifically, the normal performance of m6A modification is critical for the regulation of physiological processes [4]; however, dysregulation of this modification contributes to multiple pathological processes [4]. In summary, it is easy to recognize the significance of m6A components in the regulation of physiological and pathological processes in the body.
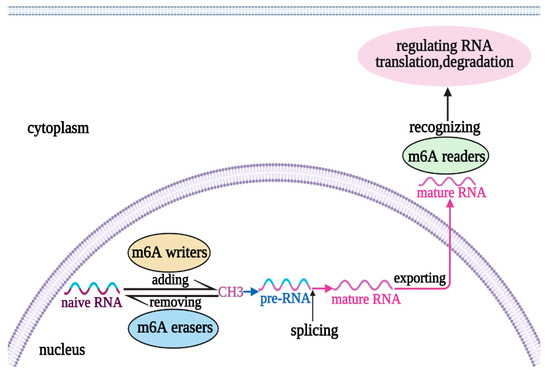
Figure 1. Mechanism of RNA m6A Modification. The process of m6A modification on RNA consists of adding and removing a methyl group by writers and erasers to form pre-RNA and then splicing to form the mature RNA, respectively. Finally, the mature RNA was nuclear-exported, and recognized by readers to regulate RNA translation and degradation. m6A, N6-methyladenosine.CH3, methyl group.
METTL3 is an important component of m6A writers, with typical structures and functions, that mainly acts as a methylase in m6A regulation [3]. Previous studies have indicated that METTL3 participates as a dynamic epigenetic regulator involved in the regulation of neurophysiological and -pathological events [5]. Physiological events include nerve cell biological functions, neurogenesis, learning and memory, and neurodevelopment. Pathological events include brain tumors (glioblastoma, GBM), neurodegenerative diseases (Alzheimer’s disease, AD), brain injuries (e.g., transient focal ischemia and traumatic brain injury (TBI)), and other brain disorders (e.g., cerebral arteriovenous malformations, typical neuropathic pain, and postoperative cognitive dysfunction).
2. Structure and Function of METTL3
METTL3, a 70 kDa protein also named MT-A70, was first identified and isolated from mammalian cell nuclear extracts in 1997 [6]. It belongs to the conserved family of methyltransferases, possessing a methyltransferase domain [MTD] that catalyzes methyl transfer to adenosine [6]. The N-terminus of METTL3 contains two Cys-Cys-Cys-His (CCCH)-type zinc finger (ZnF) motifs common in RNA-binding proteins [7]. The helical structure at the N-terminus of METTL3, also known as the leader helix, is necessary for METTL3 and WTAP interaction. The binding site of METTL3 is located within the first 150 amino acids of WTAP, and the main interaction of METTL3 with METTL14 is mediated by the MTD [7]. Typically, METTL3 and METTL14 first form a heterodimeric enzyme complex (METTL3/METTL14) in the nucleus; the METTL3/METTL14 complex is restricted to nuclear speckles and then interacts with WTAP to form the m6A methyltransferase complex (METTL3/METTL14/WTAP) that is also called the m6A writer [8][9].
METTL3/METTL14 usually functions by adding the methyl group, while WTAP potentially regulates the recruitment of this complex to mRNA targets [10]. METTL3 is the key component of the m6A writer that has critical functions in the activity of the METTL3/METTL14/WTAP complex, including catalysis, binding, and translational regulation [10]. One study indicated that the RNA-binding capacity of METTL3 is highly reduced in the absence of WTAP and that it primarily acts as a methylase to exert its core catalytic functions during m6A modification, while METTL14 serves as the RNA-binding platform [11]. Therefore, the catalytic activity of the complex is solely conferred by METTL3, which favorably modifies substrate RNAs containing the GGACU consensus sequence. Another study indicated that METTL3 could recognize 3′-UTR m6A sites on target mRNAs in addition to methyltransferase activity and then promote protein translation of the transcript by facilitating translation loop formation via interaction with the eukaryotic translation initiation factor 3 (eIF3h) subunit [11]. Collectively, these findings suggest that the typical structure of METTL3 determines its specific functions. Moreover, these functions have been implicated in the physiological and pathological processes of the nervous system.
3. METTL3 in Neurophysiological Events
METTL3 influences the behavior of RNA in nerve cells, which in turn regulates cell biological functions. Based on the roles of METTL3 in nerve cells, it further regulates many neurobiological events such as neurogenesis, learning and memory, and neurodevelopment.
3.1. METTL3 with Neurogenesis
Neurogenesis is the formation of new neurons and glial cells from neural stem cells [12]. Neural stem cells (NSCs) can differentiate into neural progenitor cells and glial progenitor cells; the latter produce glial cells such as astrocytes, oligodendrocytes, and microglia. The regulated process of neurogenesis produces an incredible diversity of neurons to carry out the complex functions of the brain [12]. Therefore, neurogenesis is accompanied by the regulation of nerve cell biological functions. Currently, several studies have indicated the multifaceted roles and mechanisms of METTL3 in the regulation of nerve cell biological processes [13]. For example, Choi et al. [14] demonstrated the role of METTL3 in cell reprogramming, reporting that METTL3 knockdown decreased the efficiency of direct lineage reprogramming, whereas METTL3 overexpression increased the efficiency of the induced neuronal cell (iN) generation [14]. The transcription factor B-cell translocation gene 2(Btg2) is a functional target of METTL3 for efficient iN generation [14]. Together, these findings indicate the importance of METTL3 modification in remodeling cell fate transition into iNs. In addition, METTL3 is also involved in the regulation of nerve cell proliferation. Chen et al. reported that the depletion of METTL3 significantly reduced m6A levels in adult NSCs, inhibiting their proliferation [15]. Meanwhile, another study in glioma stem-like cells (GSCs) also indicated that METTL3 expression is downregulated [16]. Further investigation identified SOX2 as its target in maintaining GSC stability. Moreover, silencing METTL3 promoted the proliferation of GSCs, which was achieved by the alternative splicing of isoform switches and modulating the nonsense-mediated mRNA decay of splicing factors in GBM [16][17]. Barring the above cell biological functions, cell differentiation plays a critical role during a cell’s life and has also been reported to be regulated by METTL3. For example, Yoon et al. found that METTL3 knockdown led to the prolongation of the cell cycle and maintenance of radial glial cells, which hindered cell differentiation [18]. Besides, Visvanathan et al. reported that METTL3 expression is downregulated in GSCs during differentiation by targeting SOX2 [17]. Collectively, these studies demonstrated that the downregulated expression of METTL3 actively participated in inhibiting the proliferation, reprogramming, and differentiation of nerve cells via different pathways, hinting that METTL3 might regulate neurogenesis as well.
Accumulating evidence suggests the involvement of METTL3 in neurogenesis regulation. One study indicated that the ablation of RNA-binding motif protein 15 (RBM15, a subunit of the m6A methyltransferase complex) expression in cultured neuronal cells and the developing cortex augmented chromatin remodeling factor Brg1/Brm-associated factor 155 (BAF155) expression, which is one of the integral subunits of the SWI/SNF-like complex that uses ATP-derived energy to regulate nucleosome occupancy and chromatin architecture [19]. Conversely, RBM15 overexpression decreased BAF155 expression. Mechanistically, transcriptional profiling found that BAF155 mRNA degradation by RBM15 depends on METTL3 activity, which disrupts the ability of BAF155 to produce the apical radial glial progenitors that are a hallmark of the genesis of basal radial glial progenitor cells [19]. This detailed the key roles of METTL3 in regulating glial progenitor genesis, which is a significant step in neurogenesis. Another study found that the depletion of METTL3 inhibited neuronal development, disrupted the differentiation of adult NSCs more toward glial lineage, and affected the morphological maturation of newborn neurons in the adult brain [15]. Further mechanistic investigation revealed that METTL3 knockdown decreased both histone methyltransferase Ezh2 protein expression and consequent H3K27me3 levels [15]. Furthermore, Ezh2 overexpression could rescue the neurogenesis and neuronal development defects induced by METTL3 depletion. These findings indicate that METTL3-mediated m6A modification plays an important role in regulating neurogenesis and neuronal development by targeting Ezh2 [15]. In addition, in an adult hippocampal neurogenesis study, it was shown that the knock-down of METTL3 in NSCs dramatically reduced proliferation and neuronal genesis, while enhancing glial differentiation [20]. Mechanically, METTL3 enhances the stability and translation efficiency of Lrp2 mRNA by relying on the reader protein Ythdc2, which in turn promotes neurogenesis [20]. Consistently, depletion of METTL3 in mice also showed reduced hippocampal neurogenesis, decreased spatial memory, and depression-like behavior [20]. Overexpression of either METTL3 or Lrp2 in the hippocampus of depressed mice may rescue these behavioral deficits [20]. Collectively, these findings demonstrate that METTL3 modulates adult hippocampal neurogenesis by targeting the Ythdc2-Lrp2 axis (Figure 2).
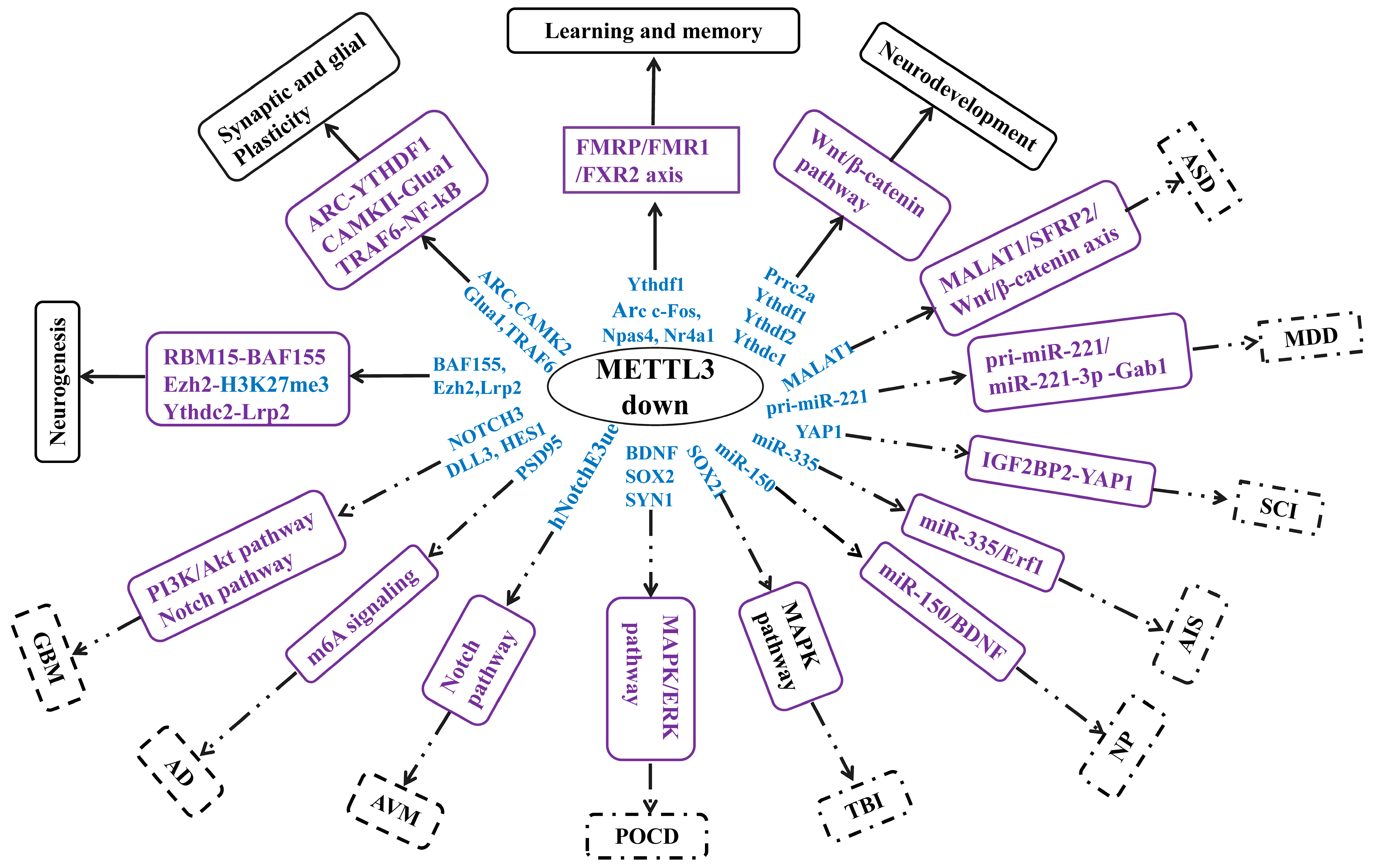
Figure 2. Illustration of the regulatory network of METTL3 in the nervous system. The targets and signaling pathways of METTL3 in neurophysiological and neuropathological events are summarized. Researchers found that low expression of METTL3 exhibits different effects, which exert beneficial roles to promote neurological events such as promoting neurogenesis, while possibly exacerbating neuropathological events, such as worsening GBM. Blue wording, targets. Purple wording, signaling pathways. Solid box, neurophysiological events. Dotted box, neuropathological events. METTL3, methyltransferase-like3; METTL3 downregulation facilitated glioma (GBM) occurrence by altering the PI3K/Akt pathway, and promoted glioma development via activating the Notch pathway that targeted NOTCH3, DLL3, and HES1; AD, METTL3 downregulation worsens Aβ-induced synaptic damage in Alzheimer’s disease (AD) via m6A signaling that is mainly targeted to upregulation of PSD95; downregulation of METTL3 significantly affected angiogenesis of human endothelial cells in arteriovenous malformation (AVM) by reducing the level of hNotch E3ue and activating the Notch signaling pathway; METTL3 downregulation participates in sevoflurane-induced postoperative cognitive dysfunction (POCD) via suppressing the MAPK/ERK pathway that targeted BDNF, SOX2, and SYN1; METTL3 downregulation triggered traumatic brain injury (TBI) development via the MAPK signaling pathway that downregulated SOX21 expression; NP, METTL3 downregulation triggers neuropathic pain (NP) occurrence via the miR-150/BDNF pathway; METTL3 downregulation triggers acute ischemic stroke (AIS) occurrence via the miR-335/Erf1 axis. NOTCH3, Notch homolog 3; DLL3, Delta-like 3; PSD95, postsynaptic density protein 95; hNotch E3ue, heterodimeric Notch E3 ubiquitin ligase; BDNF, neurotrophic factor; SYN1, synapsin 1; IEGs, immediate early genes; Ezh2, enhancer of Zeste homolog 2.
3.2. METTL3 with Synaptic Plasticity and Glial Plasticity
Consistently, recent studies have also hinted at the role and mechanisms of METTL3 in regulating synaptic and glial plasticity. For example, the study by Mareen Engel et al. group reported that METTL3 was upregulated in the amygdala (AMY) after acute stress [21], which rescued the synaptic plasticity impairment as time goes on via upregulating the m6A methylation of its target gene Ntrk1, Creb1, and Nr3c1 in the hippocampus, suggesting that METTL3 upregulation in AMY is closely related to maintaining synaptic plasticity [21]. In addition, a study on the freezing and anoxia of the wood frog brain indicated that METTL3 was significantly upregulated, coupled with a significant increase in stress granule (SG) markers TIAR and TIA-1, indicating the beneficial role of METTL3 in balancing synaptic plasticity to confront the low-oxygen stress [22]. Moreover, loss of Mettl3 decreased the levels of nuclear Ythdc1, which in turn leads to stress resilience. Overall, these data suggest that METTL3-mediated m6A modification on YTHDC1 in Drosophila dampens the brain’s biological response to stress [23]. Activity-regulated cytoskeleton-associated protein (ARC) plays an important role in the synaptic plasticity of memory consolidation [24]. The study from analyzing the brain of AD patients and Aβ-induced cell models indicated that METTL3 was downregulated to inhibit ARC expression via YTHDF1-dependent m6A modification, which then impaired the synaptic plasticity to influence memory capacity, while overexpression of METTL3 rescued ARC expression after Aβ treatment [24]. These findings demonstrate the role of low levels of METTL3 in triggering or exacerbating synaptic plasticity via the ARC-YTHDF1 axis. Moreover, the study from the Ricardo Castro-Hernándeza group reported that neurons with METTL3 knockdown significantly impaired the synaptic translation of calcium/calmodulin-dependent protein kinase 2 (CAMK2) and AMPA-selective glutamate receptor 1 (Glua1) mRNAs [25]. Consistently, the knockdown of METTL3 was also associated with decreased neuronal activity [25]. Collectively, these findings provide evidence for the idea that METTL3-mediated down-regulation of the m6A-modification mechanism contributes to synaptic function impairment via impairing the synaptic proteins CAMKII and Glua1 synthesis.
Consistent with the regulation of synaptic plasticity, there have been several studies hinting at the role and mechanism of METTL3 in regulating glial plasticity. For example, Wen et al. reported that METTL3 expression was increased in LPS-induced microglial inflammation, which subsequently upregulated TRAF6 m6A modification to activate the NF-κB pathway [26]. Consistently, inhibiting NF-κB attenuated METTL3-mediated microglial activation [26]. Collectively, these findings suggested that the upregulated level of METTL3 triggered the pro-inflammation activity of microglia by activating the TRAF6-NF-κB pathway. Furthermore, in a retina-specific METTL3 conditional knockout mouse model [27], Xin et al. reported that the deletion of METTL3 promoted the degradation of retinal progenitor cell (RPC) transcripts and subsequently promoted the transition of RPCs into glial cells, indicating that a low level of METTL3 triggers glial cell formation, which is usually related to neurological disease pathogenesis [27].
3.3. METTL3 with Neurodevelopment
Emerging studies have indicated that METTL3 is temporally and spatially regulated during neurodevelopment and aging [28]. METTL3 mRNA expression shows a distinct tissue-specific methylation profile that is associated with tissue-specific developmental processes. For example, in cortical-specific conditional FTO and METTL3 double knockout mice, Du et al. showed that the resultant severe brain defects were caused by METTL3 and not FTO deletion [29]. Moreover, METTL3 deletion elevated translation of Prrc2a, Ythdf1, Ythdf2, and Ythdc1 [29]. In summary, this uncovered a profound role of METTL3 in regulating the translation of major mRNAs that control proper cortical development by influencing “reader” proteins. In addition, depletion of m6A by METTL3 knockout also leads to a prolonged cell cycle and maintenance of radial glial cells, which hinders cortical neurogenesis, suggesting the role of METTL3 in maintaining normal neurogenesis [18] (Figure 2).
The hippocampus and AMY are two regions that respond to several behaviors, including memory. The study showed that METTL3 depletion alters not only the steady-state transcriptome in adult hippocampal neurons, but also the transcriptomic response to fear conditioning stress, including regulation of several genes involved in neuronal circuit function and pointing out a function of m6A/m in regulating neuronal circuits [21]. Moreover, it has also been revealed that METTL3 knockout significantly hypermethylated genes in the AMY, which regulate behavioral and hormonal stress responses, fear, and anxiety [21]. Collectively, these data suggest that METTL3 is critical for maintaining the normal development of the hippocampus and AMY.
Furthermore, METTL3 is actively involved in the regulation of the developing mouse cerebellum. Wang et al. inactivated METTL3 specifically in the developing mouse brain and reported severe developmental defects in the cerebellum [30]. Further analysis indicated that METTL3-mediated m6A participates in cerebellar development by controlling the mRNA stability of genes related to cerebellar development or apoptosis and by regulating the alternative splicing of pre-mRNAs of synapse-associated genes [30]. Another study showed the spatiotemporal-specific expression of METTL3 in the mouse cerebellum, and ectopic expression of METTL3 mediated by lentiviral transduction led to disorganized structures of both Purkinje and glial cells [31] (Figure 2).
Apart from its roles in cortical and cerebellar development, METTL3 can regulate neural tube development (NTD). Zhang et al. reported that METTL3 was enriched in HT-22 cells, and its depletion reduced cell proliferation and increased apoptosis by suppressing the Wnt/β-catenin signaling pathway [32]. The research revealed the important role of METTL3 in regulating NTD. Another study indicated that the knockdown of METTL3 caused anteriorization of neurulas and tailbud embryos along with the loss of the neural crest and neuronal cells in the Xenopus [33]. The mechanistic analysis determined that canonical Wnt signaling was inhibited in METTL3 morphants, which might explain the neural patterning defects of the morphants [33]. This addressed the multiple roles of METTL3 during Xenopus neurulation in anteroposterior neural patterning, neural crest specification, and neuronal cell differentiation, which further supports the hypothesis that METTL3 plays significant roles during NTD (Figure 2).
Taken together, these findings suggest that the down-regulated expression of METTL3 plays a negative role in spatiotemporal fashions in both early and late brain development.
3.4. METTL3 with Learning and Memory
Learning and memory are dominant functions of the brain. Hippocampal neurogenesis is critical in forming new memories [34]. A study has shown that dysregulated neurogenesis in the hippocampus severely impairs learning and memory tasks that are dependent on the hippocampus [35].
As METTL3 broadly participates in the process of neurogenesis, the neurogenesis-dependent functions of learning and memory are also greatly altered by METTL3. Shi et al. reported that mice with hippocampus-specific knockdown of METTL3 exhibited learning and memory defects, as well as impaired hippocampal synaptic transmission and long-term potentiation, whereas overexpression of METTL3 rescued the neurobehavioral abilities and synaptic defects in mice by targeting YTH N6-methyladenosine RNA binding protein 1(YTHDF1) [36]. This emphasized the significant roles of METTL3 in maintaining normal hippocampal function, suggesting that the abundant METTL3 in the wild-type mouse hippocampus is positively correlated with learning efficacy, and overexpression of METTL3 significantly enhances long-term memory consolidation [36]. Another study reported that METTL3 depletion in the mouse hippocampus could reduce memory consolidation ability by translational regulation of immediate early genes, including Arc, c-Fos, Egr1, Npas4, and Nr4a1, while unimpaired learning outcomes could be achieved if the function of METTL3 was restored [37]. These findings uncovered a direct role of METTL3 modification in regulating long-term memory formation. Apart from the role of METTL3 in memory formation, it was also found to be associated with the regulation of memory types, such as fear memory [21]. METTL3 deletion in adult neurons altered the FMRP/FMR1 and FXR2 transcriptome response to fear and synaptic plasticity, and finally increased fear memory [21]. Moreover, the study reported that neither deletion of METTL3 nor FTO in mice showed altered anxiety-like behavior or locomotion, but researchers observed significant changes in spontaneous digging behavior [21] (Figure 2).
References
- Niu, Y.; Zhao, X.; Wu, Y.-S.; Li, M.-M.; Wang, X.-J.; Yang, Y.-G. N6-methyl-adenosine (m6A) in RNA: An Old Modification with A Novel Epigenetic Function. Genom. Proteom. Bioinform. 2013, 11, 8–17.
- Yang, Y.; Hsu, P.J.; Chen, Y.-S.; Yang, Y.G. Dynamic transcriptomic m(6)A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018, 28, 616–624.
- Balacco, D.L.; Soller, M. The m(6)A Writer: Rise of a Machine for Growing Tasks. Biochemistry 2019, 58, 363–378.
- Yang, C.; Hu, Y.; Zhou, B.; Bao, Y.; Li, Z.; Gong, C.; Yang, H.; Wang, S.; Xiao, Y. The role of m6A modification in physiology and disease. Cell Death Dis. 2020, 11, 1–16.
- Mathoux, J.; Henshall, D.C.; Brennan, G.P. Regulatory Mechanisms of the RNA Modification m6A and Significance in Brain Function in Health and Disease. Front. Cell. Neurosci. 2021, 15, 671932.
- Luo, Q.; Mo, J.; Chen, H.; Hu, Z.; Wang, B.; Wu, J.; Liang, Z.; Xie, W.; Du, K.; Peng, M.; et al. Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4. Nat. Commun. 2022, 13, 5636.
- Wang, X.; Feng, J.; Xue, Y.; Guan, Z.; Zhang, D.; Liu, Z.; Gong, Z.; Huang, J.; Tang, C.; Zou, T.; et al. Structural basis of N6-adenosine methylation by the METTL3–METTL14 complex. Nature 2016, 534, 575–578.
- Scholler, E.; Weichmann, F.; Treiber, T.; Ringle, S.; Treiber, N.; Flatley, A.; Feederle, R.; Bruckmann, A.; Meister, G. Interactions, localization, and phosphorylation of the m(6)A generating METTL3-METTL14-WTAP complex. RNA 2018, 24, 499–512.
- Huang, Q.; Mo, J.; Liao, Z.; Chen, X.; Zhang, B. The RNA m(6)A writer WTAP in diseases: Structure, roles, and mechanisms. Cell Death Dis. 2022, 13, 852.
- Wang, P.; Doxtader, K.A.; Nam, Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol. Cell 2016, 63, 306–317.
- Choe, J.; Lin, S.; Zhang, W.; Liu, Q.; Wang, L.; Ramirez-Moya, J.; Du, P.; Kim, W.; Tang, S.; Sliz, P.; et al. mRNA circularization by METTL3–eIF3h enhances translation and promotes oncogenesis. Nature 2018, 561, 556–560.
- Marques, B.L.; Maciel, G.F.; Brito, M.R.; Dias, L.D.; Scalzo, S.; Santos, A.K.; Kihara, A.H.; Santiago, H.d.C.; Parreira, R.C.; Birbrair, A.; et al. Regulatory mechanisms of stem cell differentiation: Biotechnological applications for neurogenesis. Semin. Cell Dev. Biol. 2023, 144, 11–19.
- Moroz-Omori, E.V.; Huang, D.; Bedi, R.K.; Cheriyamkunnel, S.J.; Bochenkova, E.; Dolbois, A.; Rzeczkowski, M.D.; Li, Y.; Wiedmer, L.; Caflisch, A. METTL3 Inhibitors for Epitranscriptomic Modulation of Cellular Processes. Chemmedchem 2021, 16, 3035–3043.
- Choi, H.; Baek, S.; Cho, B.; Kim, S.; Kim, J.; Chang, Y.; Shin, J.; Kim, J. Epitranscriptomic N6-Methyladenosine Modification Is Required for Direct Lineage Reprogramming into Neurons. ACS Chem. Biol. 2020, 15, 2087–2097.
- Chen, J.; Zhang, Y.-C.; Huang, C.; Shen, H.; Sun, B.; Cheng, X.; Zhang, Y.-J.; Yang, Y.-G.; Shu, Q.; Yang, Y.; et al. m6A Regulates Neurogenesis and Neuronal Development by Modulating Histone Methyltransferase Ezh2. Genom. Proteom. Bioinform. 2019, 17, 154–168.
- Visvanathan, A.; Patil, V.; Arora, A.; Hegde, A.S.; Arivazhagan, A.; Santosh, V.; Somasundaram, K. Essential role of METTL3-mediated m6A modification in glioma stem-like cells maintenance and radioresistance. Oncogene 2018, 37, 522–533.
- Li, F.; Yi, Y.; Miao, Y.; Long, W.; Long, T.; Chen, S.; Cheng, W.; Zou, C.; Zheng, Y.; Wu, X.; et al. N6-Methyladenosine Modulates Nonsense-Mediated mRNA Decay in Human Glioblastoma. Cancer Res. 2019, 79, 5785–5798.
- Yoon, K.-J.; Ringeling, F.R.; Vissers, C.; Jacob, F.; Pokrass, M.; Jimenez-Cyrus, D.; Su, Y.; Kim, N.-S.; Zhu, Y.; Zheng, L.; et al. Temporal Control of Mammalian Cortical Neurogenesis by m6A Methylation. Cell 2017, 171, 877–889.e17.
- Xie, Y.; Castro-Hernández, R.; Sokpor, G.; Pham, L.; Narayanan, R.; Rosenbusch, J.; Staiger, J.F.; Tuoc, T. RBM15 Modulates the Function of Chromatin Remodeling Factor BAF155 Through RNA Methylation in Developing Cortex. Mol. Neurobiol. 2019, 56, 7305–7320.
- Xu, B.; Li, Q.; Wu, Y.; Wang, H.; Xu, J.; Liu, H.; Xuan, A. Mettl3-mediated m(6) A modification of Lrp2 facilitates neurogenesis through Ythdc2 and elicits antidepressant-like effects. FASEB J. 2022, 36, e22392.
- Engel, M.; Eggert, C.; Kaplick, P.M.; Eder, M.; Röh, S.; Tietze, L.; Namendorf, C.; Arloth, J.; Weber, P.; Rex-Haffner, M.; et al. The Role of m6A/m-RNA Methylation in Stress Response Regulation. Neuron 2018, 99, 389–403.e9.
- Wade, S.; Hadj-Moussa, H.; Storey, K.B. mRNA m 6 A methylation in wood frog brain is maintained during freezing and anoxia. J. Exp. Zool. Part A Ecol. Integr. Physiol. 2023, 339, 325–334.
- Perlegos, A.E.; Shields, E.J.; Shen, H.; Liu, K.F.; Bonini, N.M. Mettl3-dependent m6A modification attenuates the brain stress response in Drosophila. Nat. Commun. 2022, 13, 5387.
- Xu, C.; Huang, H.; Zhang, M.; Zhang, P.; Li, Z.; Liu, X.; Fang, M. Methyltransferase-Like 3 Rescues the Amyloid-beta protein-Induced Reduction of Activity-Regulated Cytoskeleton Associated Protein Expression via YTHDF1-Dependent N6-Methyladenosine Modification. Front. Aging Neurosci. 2022, 14, 890134.
- Castro-Hernández, R.; Berulava, T.; Metelova, M.; Epple, R.; Centeno, T.P.; Richter, J.; Kaurani, L.; Pradhan, R.; Sakib, M.S.; Burkhardt, S.; et al. Conserved reduction of m 6 A RNA modifications during aging and neurodegeneration is linked to changes in synaptic transcripts. Proc. Natl. Acad. Sci. USA 2023, 120, e2204933120.
- Wen, L.; Sun, W.; Xia, D.; Wang, Y.; Li, J.; Yang, S. The m6A methyltransferase METTL3 promotes LPS-induced microglia inflammation through TRAF6/NF-kappaB pathway. Neuroreport 2022, 33, 243–251.
- Xin, Y.; He, Q.; Liang, H.; Zhang, K.; Guo, J.; Zhong, Q.; Chen, D.; Li, J.; Liu, Y.; Chen, S. m(6)A epitranscriptomic modification regulates neural progenitor-to-glial cell transition in the retina. Elife 2022, 11, e79994.
- Shafik, A.M.; Zhang, F.; Guo, Z.; Dai, Q.; Pajdzik, K.; Li, Y.; Kang, Y.; Yao, B.; Wu, H.; He, C.; et al. N6-methyladenosine dynamics in neurodevelopment and aging, and its potential role in Alzheimer’s disease. Genome Biol. 2021, 22, 17.
- Du, K.; Zhang, Z.; Zeng, Z.; Tang, J.; Lee, T.; Sun, T. Distinct roles of Fto and Mettl3 in controlling development of the cerebral cortex through transcriptional and translational regulations. Cell Death Dis. 2021, 12, 700.
- Wang, C.-X.; Cui, G.-S.; Liu, X.; Xu, K.; Wang, M.; Zhang, X.-X.; Jiang, L.-Y.; Li, A.; Yang, Y.; Lai, W.-Y.; et al. METTL3-mediated m6A modification is required for cerebellar development. PLoS Biol. 2018, 16, e2004880.
- Ma, C.; Chang, M.; Lv, H.; Zhang, Z.-W.; Zhang, W.; He, X.; Wu, G.; Zhao, S.; Zhang, Y.; Wang, D.; et al. RNA m6A methylation participates in regulation of postnatal development of the mouse cerebellum. Genome Biol. 2018, 19, 68.
- Zhang, L.; Cao, R.; Li, D.; Sun, Y.; Zhang, J.; Wang, X.; Khan, A.; Liu, Z.; Niu, B.; Xu, J. Ethionine-mediated reduction of S-adenosylmethionine is responsible for the neural tube defects in the developing mouse embryo-mediated m6A modification and is involved in neural tube defects via modulating Wnt/beta-catenin signaling pathway. Epigenetics Chromatin 2021, 14, 52.
- Kim, H.; Jang, S. RNA m6A Methyltransferase Mettl3 Regulates Spatial Neural Patterning in Xenopus laevis. Mol. Cell. Biol. 2021, 41, e0010421.
- Farioli-Vecchioli, S.; Ricci, V.; Middei, S. Adult Hippocampal Neurogenesis in Alzheimer’s Disease: An Overview of Human and Animal Studies with Implications for Therapeutic Perspectives Aimed at Memory Recovery. Neural Plast. 2022, 2022, 9959044.
- Toda, T.; Parylak, S.L.; Linker, S.B.; Gage, F.H. The role of adult hippocampal neurogenesis in brain health and disease. Mol. Psychiatry 2018, 24, 67–87.
- Shi, H.; Zhang, X.; Weng, Y.-L.; Lu, Z.; Liu, Y.; Lu, Z.; Li, J.; Hao, P.; Zhang, Y.; Zhang, F.; et al. m6A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature 2018, 563, 249–253.
- Zhang, Z.; Wang, M.; Xie, D.; Huang, Z.; Zhang, L.; Yang, Y.; Ma, D.; Li, W.; Zhou, Q.; Yang, Y.-G.; et al. METTL3-mediated N6-methyladenosine mRNA modification enhances long-term memory consolidation. Cell Res. 2018, 28, 1050–1061.
More
Information
Subjects:
Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
621
Revisions:
2 times
(View History)
Update Date:
27 Apr 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




