Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | VIjay Kumar | -- | 3362 | 2023-04-21 05:43:05 | | | |
| 2 | Jessie Wu | + 415 word(s) | 3777 | 2023-04-21 06:35:45 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Naik, B.; Kumar, V.; Rizwanuddin, S.; Chauhan, M.; Choudhary, M.; Gupta, A.K.; Kumar, P.; Kumar, V.; Saris, P.E.J.; Rather, M.A.; et al. Genomics Improve Abiotic Stress Tolerance in Tomato Plant. Encyclopedia. Available online: https://encyclopedia.pub/entry/43311 (accessed on 08 February 2026).
Naik B, Kumar V, Rizwanuddin S, Chauhan M, Choudhary M, Gupta AK, et al. Genomics Improve Abiotic Stress Tolerance in Tomato Plant. Encyclopedia. Available at: https://encyclopedia.pub/entry/43311. Accessed February 08, 2026.
Naik, Bindu, Vijay Kumar, Sheikh Rizwanuddin, Mansi Chauhan, Megha Choudhary, Arun Kumar Gupta, Pankaj Kumar, Vivek Kumar, Per Erik Joakim Saris, Muzamil Ahmad Rather, et al. "Genomics Improve Abiotic Stress Tolerance in Tomato Plant" Encyclopedia, https://encyclopedia.pub/entry/43311 (accessed February 08, 2026).
Naik, B., Kumar, V., Rizwanuddin, S., Chauhan, M., Choudhary, M., Gupta, A.K., Kumar, P., Kumar, V., Saris, P.E.J., Rather, M.A., Bhuyan, S., Neog, P.R., Mishra, S., & Rustagi, S. (2023, April 21). Genomics Improve Abiotic Stress Tolerance in Tomato Plant. In Encyclopedia. https://encyclopedia.pub/entry/43311
Naik, Bindu, et al. "Genomics Improve Abiotic Stress Tolerance in Tomato Plant." Encyclopedia. Web. 21 April, 2023.
Copy Citation
Solanum lycopersicum L. (Solanaceae), generally known as tomato, is one of the most significant fruits that are nutritionally classified as a vegetable. It contains carotenoids (lycopene and carotene), phenolic compounds (flavonoids), vitamins (ascorbic acid, -tocopherol, vitamin A), glycoalkaloids (tomatine), and phytosterols (-sitosterol, campesterol, and stigmasterol).
abiotic stress
tomato
drought
1. Major Abiotic Stress in Tomato
Abiotic stress is a broad term that encompasses a variety of conditions such as temperature variations, osmotic pressure, drought, salinity, water logging, bruising, ozone, toxic compounds, excessive light, and UV radiation [1]. These are serious issues and cause a cascade of morphological, physiological, molecular, and biochemical changes, resulting in cellular damage and inhibition of general metabolism. Such environmental stressors include various factors that behave as ecological barriers that tremor cellular stability. They possess a negative impact on plant development and agricultural output [2]. Owing to the complexity of stress and its origin, finding particular abiotic stress is unusual; therefore, in responses to various stressors, a large number of stress-responsive components and pathways are prevalent (Figure 1).
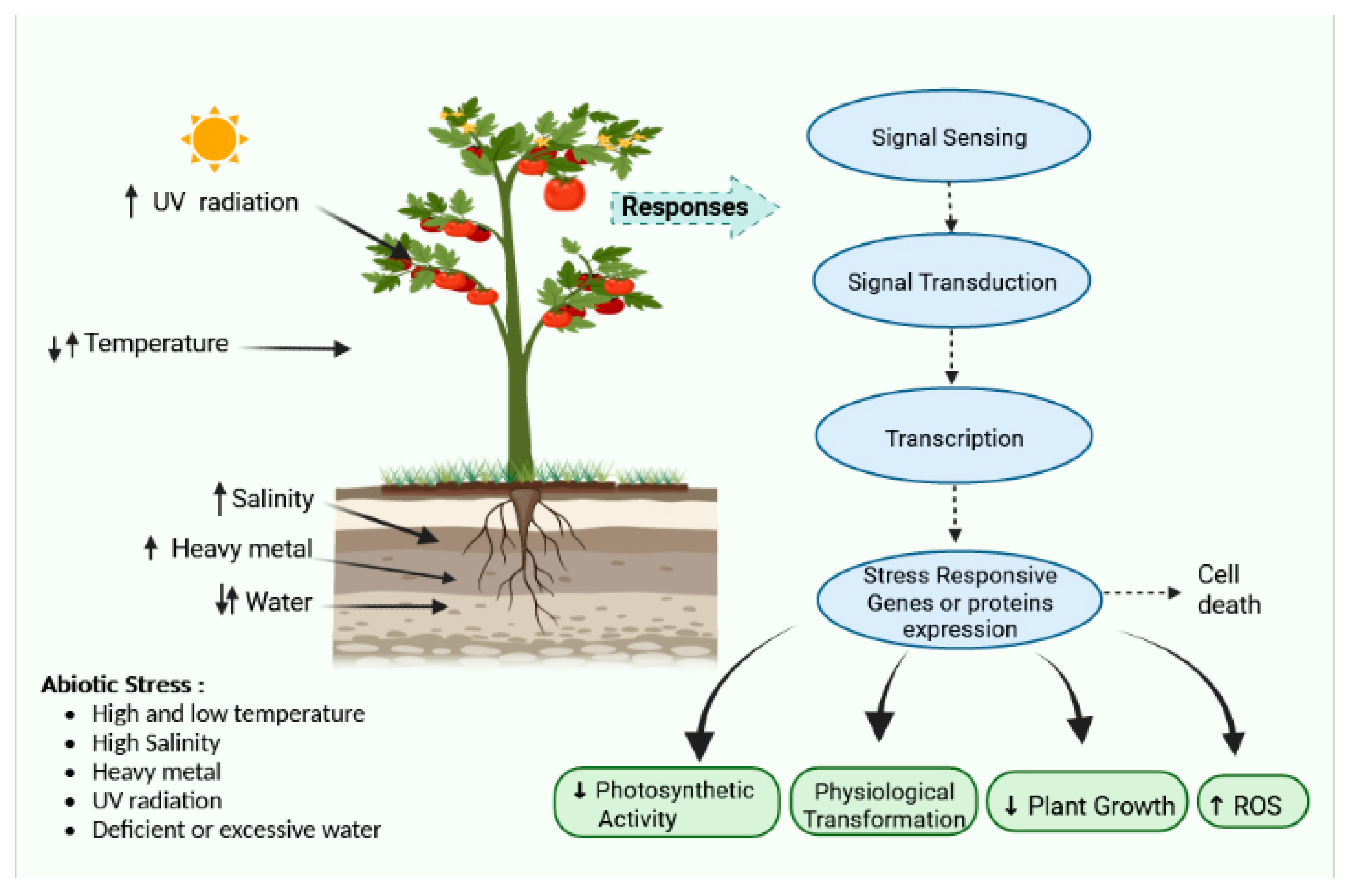
Figure 1. Effect of various stresses and their responses and pathways.
Temperature is an important abiotic factor that regulates cellular homeostasis. Heat increases water evaporation from the soil together with a greater leaf transpiration rate, resulting in water stress under field circumstances; it also increases the kinetics of biomolecules, which may result in protein misfolding [3], whereas cold reduces biomolecule and enzyme kinetics, resulting in lower cell membrane fluidity. Snow develops ice crystals in the soil, freezing the water residing within.
Water availability is the most significant abiotic element that has influenced and continues to influence plant evolution, and by 2050, more than half of all arable lands may be severely affected [4]. Drought reduces soil water balance, hampering osmotic adjustment, which is characterized by a reduction in the osmolarity of cytosol caused by the accumulation of various hydrophilic proteins (LEA proteins) and osmolytes. Drought causes stomatal closure in leaves, resulting in an imbalance between electron transport mechanisms and carbon absorption, leading to increased thermal energy dissipation and photoinhibition [5]. Anaerobiosis caused by flooding stress causes fermentation in plant roots. The cytoplasm of cells becomes more acidic during fermentation due to an increased concentration of organic acids, which inhibits the functioning of various enzymes. Induced salinity results in the buildup of numerous osmolytes and ions such as Na+, which may become toxic when accumulated in large amounts.
Heavy metal soil pollution is a global issue that is continuously becoming worse due to human activity, geochemical rock weathering, and other environmental factors including volcanic activity, acid rain, etc. Metal-contaminated soil is constantly being exposed to plants. The harmful effects include the adhesion of heavy metal ions to the sulfhydryl groups of proteins, deactivation of enzymes, removal of vital cations from specific binding sites, and generation of ROS, which causes oxidative damage to lipids, proteins, and nucleic acids [6]. In addition to limiting agricultural yields, heavy metal absorption poses a serious risk to both flora and fauna [7].
2. Physiological Transformation Due to Abiotic Stress in Tomato
There are certain phases of stress in plants, such as alarming, acclimation, resistance, exhaustion, and recovery. The differing proteome response exhibited in every phase is summarized in Figure 2. Despite this, other physiological, cytosolic, and molecular responses can also be observed, which are elaborated on in upcoming sections.
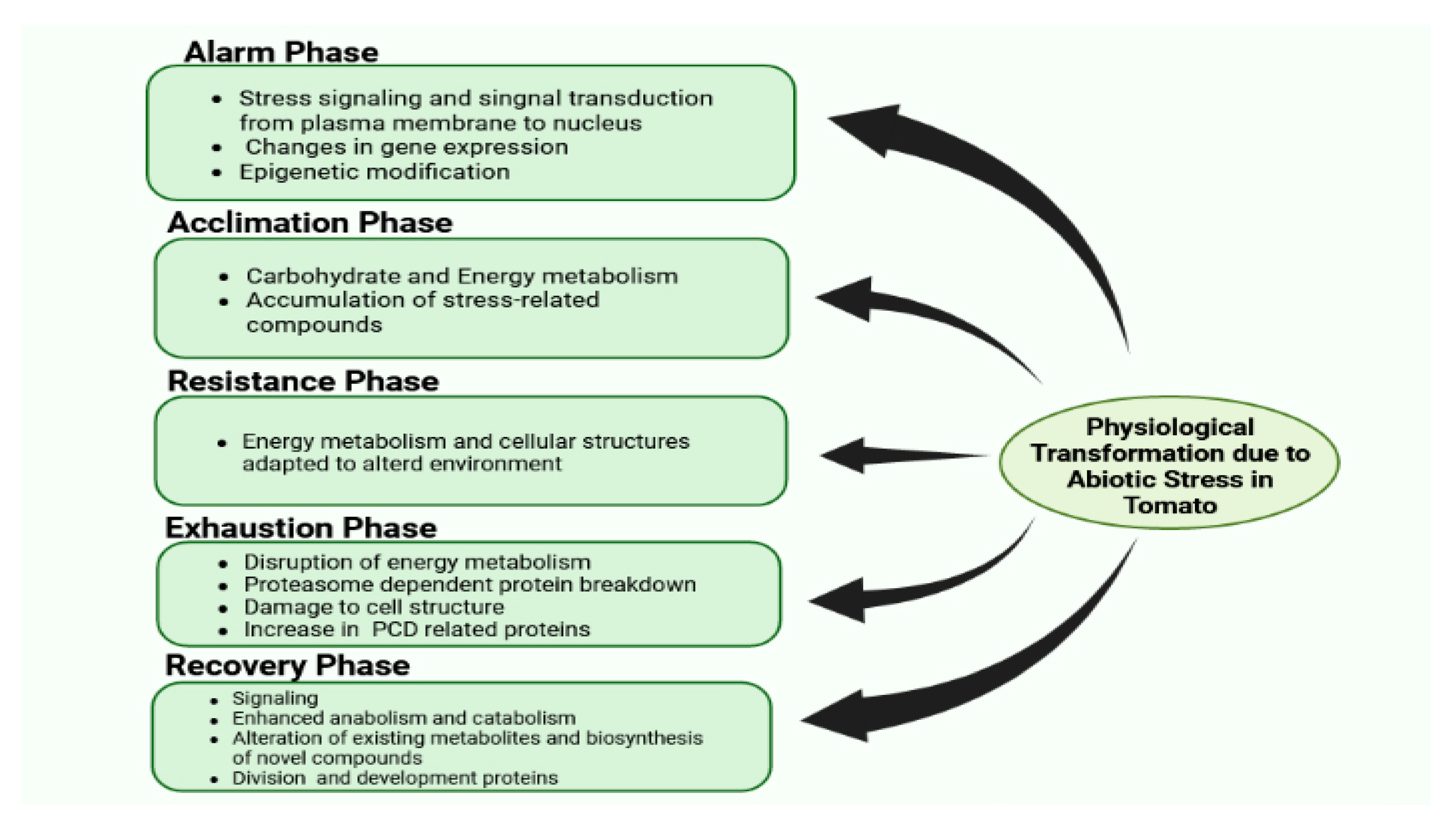
Figure 2. Physiological transformation due to abiotic stress in tomato.
According to various reports, proteins are involved in stress signaling (NDPK, involved in G-protein signaling and phosphate transfers), ROS scavenging (thioredoxin, glyoxalase I), detoxification, and chaperones (DnaK and HSP20); moreover, proteins are responsible for cell wall biosynthesis such as enzymes of the phenylpropanoid biosynthesis pathway [8]. Additionally, several cytoplasmic proteins, such as pentose phosphate pathway proteins and glucose metabolism, were found in the cell membrane. It is suggested that they might have a role in the manufacture of sugars as a component of osmotic alteration during dehydration stress or in ROS scavenging owing to NADPH generation.
ROS (singlet oxygen, superoxide, hydroxyl radicals, and hydrogen peroxide) are inherent byproducts released continuously during metabolic pathways; their formation and degradation are often reported to increase under environmental stresses [9], although ROS are also crucial communicating molecules [10] for numerous processes taking place in specific cell organelles. In drought-exposed sugar beets, cellular drying is also related to increased ROS production, leading to the activation of various ROS scavenging enzymes, including multiple thioredoxin (Trx) isoforms [11]. Accumulation of ROS causes oxidative stress, which destroys plant biomolecules and cellular components affecting the growth potential of plants [12].
Due to the closing of stomata, CO2 fixation is reduced, which causes a decrease in NADP+ regeneration via the Calvin cycle, and when this gets combined with altered photosystem activity as well as photosynthetic transport potential, it results in greater electron permeability to O2 [13][14]. Similar proteins such as GRP-like proteins-2 and APX (ascorbate peroxidase), as well as GPX (glutathione peroxidase), were discovered in the cell wall and leaves of tomatoes as abiotic stress-associated proteins. They are suggested to function as an integrating factor for the accumulation of cell wall elements and defense against oxidative stress respectively [15].
Ferritin- and osmotin-like proteins, which make up 13% of all known proteins, are crucial for detoxification. While ferritin stores iron in a soluble, nontoxic form that is readily accessible, ferrous iron is absorbed in the ferrous form and stored as ferric hydroxides when oxidized. According to reports, plants accumulate osmotin or proteins that resemble osmotin in response to a variety of stressors [16]. Small heat shock proteins (HSP) found in mitochondria make up 7% of all known proteins. The interruption of regular protein synthesis and the persistent production of HSPs are two characteristics of the heat shock response, which entails the temporal alteration of metabolic activity in the cell. The ability of plant mitochondria to withstand heat is greatly influenced by MT-sHSPs [15]. The upregulation of these proteins’ expression shows that they are crucial for avoiding the aggregation of denatured proteins and promoting protein refolding in response to stress [17].
Li et al. (2019) [18] developed slnpr1 mutants utilizing SlNPR1 from the variety of tomato ‘Ailsa Craig’ via the CRISPR/Cas9 system. The observation indicated various effects such as reduced drought tolerance, greater electrolytic leakage, increased stomatal aperture, hydrogen peroxide (H2O2) levels, higher malondialdehyde (MDA), and reduced antioxidant enzyme activity. The framework of study techniques and characterization in each section for the studies of genomics, proteomics, and metabolomics for tomato is given in Figure 3.
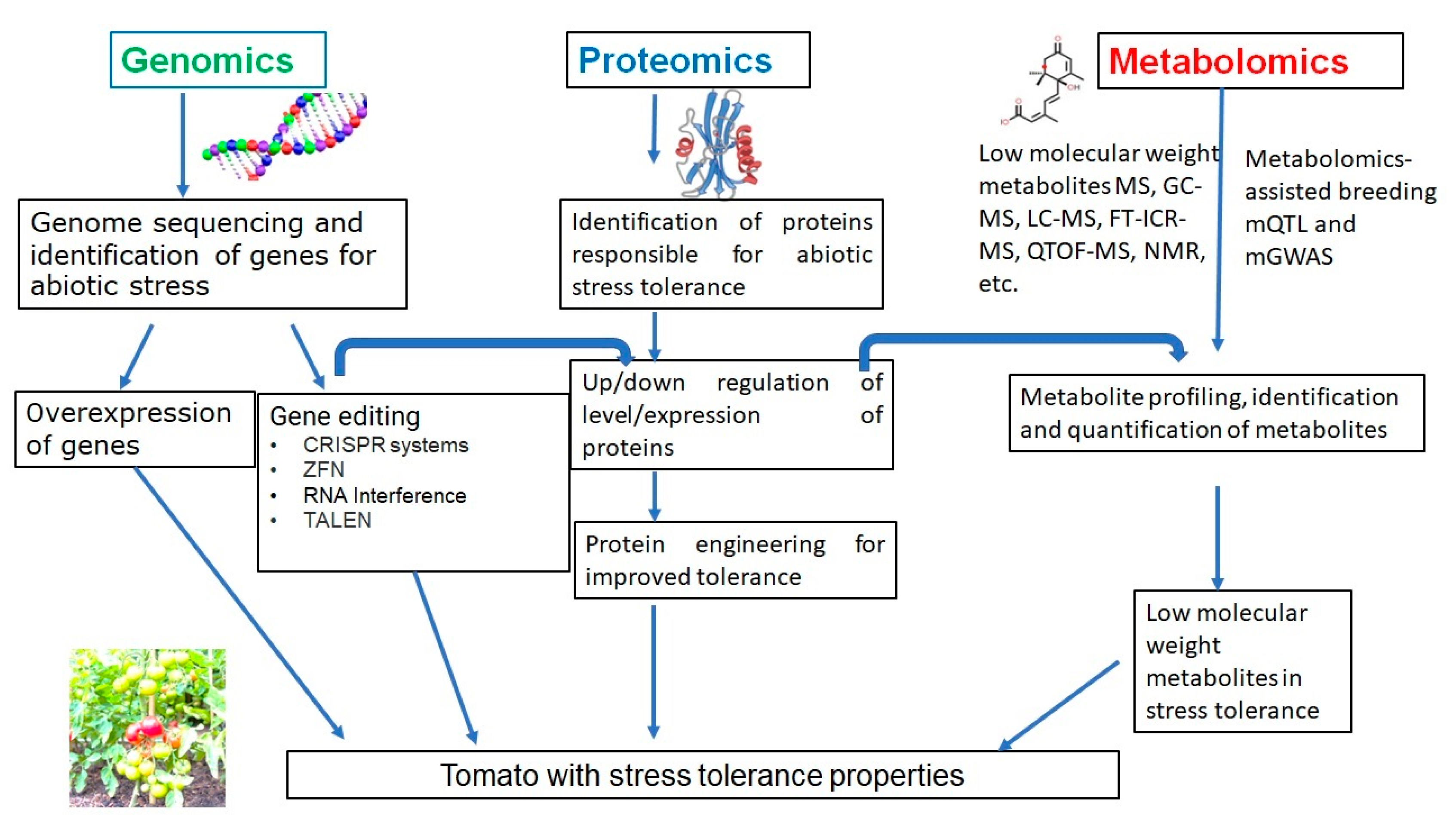
Figure 3. The framework of study techniques for the studies of genomics, proteomics, and metabolomics for tomato.
3. Genomics of Abiotic Stress
Solanaceous crop genomics is in an exciting stage of development after the sequencing of the potato and tomato genomes was recently completed. Tomato, a native of South America, has become one of the most widely utilized vegetable crops after spreading over the globe. It exhibits intriguing developmental traits, such as compound leaves, succulent fruits, short generation time, and easy diploid genetics. Together, these factors make the tomato a superb species for both basic and applied plant study [19]. Before the tomato genome sequencing research began, several genetic and genomic materials were available. To build a genetic map of tomatoes, scientists used morphological and isozyme markers [20]. They then determined which 12 linkage groups corresponded to the cytologically accessible chromosomes, which resulted in the building of an RFLP (restriction fragment length polymorphism) linkage map [21]. Therefore, breeders were able to locate quantitative trait loci (QTLs) as a result of the complete molecular linkage map, which helped them comprehend the genetic basis of many quantitative traits (Tomato Genome Consortium, 2012). On the website of the Solanaceae Genomics Network, there is a plethora of information about the genetic and genomic resources for tomatoes.
Recent developments in the genomics field have augmented the effective development of novel varieties. The ability to characterize genetic variation in the germplasm pool for almost any crop species has considerably improved, owing to molecular markers [22]. They are based on the polymorphism observed in any DNA sample, and molecular markers have numerous benefits compared to biochemical or morphological markers. Included in these benefits are simple assays, repeatability, ease of use, high availability, stability regardless of external or environmental conditions, and representation across entire genomes [23]. Numerous applications in genetics, molecular biology, genomics, and plant breeding—particularly tomatoes—have made extensive use of DNA markers. Numerous omics branches dealing primarily with the molecular elements of cellular biology have been developed recently because of technological advancements [24]. The genomics approach offers a comprehensive perspective for the structural–functional investigation of genes and the discovery of genetic variants, which can be employed to overcome abiotic stress in tomatoes.
3.1. Genome Sequencing of Tomato
Diverse abiotic stresses in open-field cultivation negatively influence tomato crop production, resulting in poor yield and fruit quality, thus fetching a lower price and not fulfilling the market requirement. To overcome the issues of abiotic stress (especially temperature and flood) in open fields, farmers tend to cultivate tomato crops in greenhouses [25]. In addition to being more expensive, greenhouse farming also results in a rapid buildup of NO3, PO4, and salt in the soil, which eventually causes the degradation of soil and polluting of the surface and groundwater. Therefore, increasing tomato cultivars’ ability to withstand abiotic stress is economically more advantageous and sustainable [26]. Abiotic stress conditions brought on by harsh water and temperature regimes, an imbalance in the nutritional content of the soil substrate, and elemental toxicity, along with high salinity, are the main issues limiting tomato development. In agricultural settings where many stressors frequently coexist, the abiotic pressures grow increasingly complex. It is vital to create resilient, high-yielding cultivars with improved tolerance to a series of abiotic stresses to meet the world’s food needs [27].
There have been various attempts to address certain stress aspects under controlled conditions, but this approach is not always workable because the plant response varies in the field when numerous factors and stresses are present at once [21][28]. Over the past ten years, conventional breeding has significantly improved a wide range of traits, including biotic and abiotic stress tolerance, yield components, and quality-related variables. However, conventional kinds are susceptible to numerous stresses at diverse locations. Given the genetic complexity and environmental interconnections, using more intensive and multidisciplinary methodologies provides a superior approach to improving stress tolerance in current crops [24]. The genomics of solanaceous crops is in a motivating stage of development after the sequencing of the potato and tomato genomes was recently completed.
3.2. Identification and Functional Validation of Genes Associated with Abiotic Stress Tolerance
One of the various strategies available for improving contemporary tomato plants is genetic engineering. Functional genomics has recently undergone technological advancements that have made it possible to identify the many gene families and processes that affect how plants respond to abiotic challenges, and consequently increase yield [19]. Genetic modification (GM) technology, in contrast to conventional selective breeding, enables faster and more effective acquisition of tomato plants resistant to abiotic challenges, increasing food production [19].
Yang et al. (2015) [29] conveyed that both the SlSnRK22.1 gene intricated in the regulation of abscisic acid signaling and the SlSnRK2.2 gene involved in osmotic stress signal transduction overexpressed and increased sensitivity to osmotic stress. Yu et al. (2016) [30] studied the low-temperature stress in tomatoes and observed that the SlMPK7 gene regulated ROS homeostasis through the initiation of cellular antioxidant systems and also modulated the transcription of stress-associated genes, resulting in improved tolerance to chilling. Liu et al. [31] studied the overexpression of ShDHN/Shabrochaites genes and observed the accumulation of protein with chaperone-like and detergent properties, resulting in enhanced tolerance to cold, drought, and salinity. Li et al. [32] stated that SpWRKY1 is a transcriptional factor, transcriptional regulation stress-related gene, and its overexpression resulted in enhanced abiotic tolerance. Shah et al. [33] reported that the AtDREB1A gene overexpressed under the Lip9 promoter and resulted in the expression of a transcriptional factor, transcriptional regulation stress-related gene, resulting in plant cold resistance. According to Meng et al. [34], overexpression of the LeAN2 gene led to the upregulation of several structural genes in the anthocyanin biosynthesis pathway, leading to anthocyanin accumulation, which improved heat resistance. The AtGRX gene is responsible for maintaining cellular redox homeostasis and is responsible for conferring chilling tolerance to tomato plants [35]. Similarly, Hu et al. [36] reported about the MdSOS2L1 gene, which is involved in signal transduction proteins and influences ion-driving transport mechanisms, and overexpression of the gene resulted in improved salt tolerance. Gong et al. [37] studied about SlSAM1 gene in tomatoes, which catalyzes the ATP and L-methionine conversion into S-adenosylmethionine, which is involved in polyamines and ethylene biosynthesis, resulting in plant alkali tolerance. Using small interfering RNA technology, Metwali et al. [38] improved tomato fruit quality under heat stress by silencing the vis-1 gene.
Researchers from all over the world have used a variety of techniques to produce tomato crops with different levels of abiotic stress tolerance [39]. GRAS transcription factors (TFs) have been shown by Habib et al. (2021) [40] to play an array of roles in biological processes. Abiotic stressors such as drought and salt stress were used to characterize how a tomato SlGRAS10 gene functions. Dwarf plants possessing shorter internode lengths, smaller leaves, and increased flavonoid accumulation were created when SlGRAS10 was downregulated via RNA interference (RNAi). These plants also showed heat tolerance. Abscisic acid (ABA), which is connected to members of the abscisic acid-responsive element binding factor (ABF) and abscisic acid-responsive element binding protein (AREB) subfamily of basic leucine zipper (bZIP) transcription factors, has been reported to play a significant role in how plants respond to abiotic stresses. The cultivated tomato contains the SlAREB1 and SlAREB2 components. Salinity and drought tolerance were promoted in the tissues of the leaves and roots by the overexpression of SlAREB1 and SlAREB2 [41]. The authors discovered genes that were upregulated in SlAREB1-overexpressing lines that encode proteins related to transcription regulation, oxidative stress, lipid transfer proteins (LTPs), and late embryogenesis abundant protein, showing that these genes might be participating in abiotic stress and could be reactive to pathogenic microbes. As per the structure of the gene, the composition of the motif, and phylogenetic investigation, Wang et al. [20] recognized 66 potential G2-like genes in tomatoes and categorized them into five groups (I to V). All 12 chromosomes had an unequal distribution of the G2-like genes. There were four tandemly duplicated SlGlk genes and nine pairs of tandemly duplicated gene segments. The SlGlks promoter regions contain a variety of CREs that are connected to hormones and stress, according to the cisregulatory elements (CREs) analysis. SlGlks were expressed in response to various abiotic stressors according to RNA-seq.
Gene expression, namely SlDWARF, SlCPD, and BIN2, was considerably elevated during drought stress in SLB3-silenced seedlings; however, TCH4-related gene expression was downregulated, according to Wang et al. [42]. These findings established that silencing the SLB3 gene decreased tomato plants’ ability to withstand drought and had an effect on the BR signaling transduction, which may be likely to blame for the difference in tomato plants’ ability to withstand drought. For biotic and abiotic stress responses. Bvindi et al. [43] examined the functions of tomato histone H3 lysine methyltransferases Set Domain Group 33 (SDG33) and SDG34. The H3K36 and H3K4 methylations, as well as the gene expression involved in a variety of processes and reactions to biotic and abiotic stimuli, were altered in the SDG33 and SDG34 mutants. The double mutant demonstrated better tolerance, consistent with separate and additional functions, whereas single mutants were still resistant to drought. Mutants enhanced recovery and survival when the drought ended and maintained a higher water status during it. Notably, decreased trimethylation in H3K4 and H3K36 and the production of negative regulators in stressed plants aid in the mutants’ ability to withstand stress. The tomato PDI gene family was thoroughly analyzed for the first time at the genome-wide level by Wai et al. [44], who discovered nineteen PDI genes in tomatoes, which were distributed unequally across eight tomato chromosomes out of twelve, with segmental duplications found for three paralogous gene pairs. The majority of the PDI genes showed variable expression across various fruit organs and developmental phases, according to expression profiling research. Additionally, the majority of the PDI genes were strongly activated by high temperature, salinity, and abscisic acid treatments, but only a small number of the genes were stimulated by freezing and stresses, including nutrition and water deficits. Dominate expression of the PDI gene family, SlPDI1-1, SlPDI1-3, SlPDI1-4, SlPDI2-1, SlPDI4-1, and SlPDI5-1 in response to ABA application and abiotic stresses suggested that they have a role in managing tomato abiotic stresses.
3.3. Genomic Approaches to Combat Abiotic Stress
Through a variety of mechanisms, including transcription, translation, regulation of calcium, phytohormone, sugar, and lipid signaling, as well as primary and secondary metabolism, plants respond to high temperatures and maintain life [24]. Numerous abiotic stimuli, such as heat stress during bud development, which results in stigma (style) exertion in tomato flowers, have a significant impact on the position and maturity of the male (anther cone) and female (style) organs (Krishna et al., 2019) [29]. Genetic diversity exists in heat stress resistance. Under heat stress, plants display a variety of physiological reactions, including leaf abscission and senescence, growth retardation of the shoots and roots, and fruit destruction. As a result, plant productivity is significantly reduced [45][46]. Plant development and agricultural productivity are harmed by unfavorable environmental variables such as salt stress, drought, and severe temperatures. GRAS genes are members of the family of plant-specific transcription factors (TFs), which are known to play a variety of functions in plant growth and development. According to several studies, the GRAS protein family is essential for plant growth and development in response to abiotic stressors [47]. The tomato SlGRAS10 gene’s functional characterization under abiotic conditions such as salt stress and drought was shown by Habib et al. [40]. Dwarf plants with shorter internodes and smaller leaves were created when SlGRAS10 was downregulated via RNA interference (RNAi). Additionally, compared to wild-type plants, SlGRAS10-RNAi plants were more resilient to abiotic stimuli such as salt, dehydration, and abscisic acid. By increasing the tomato plant’s osmotic potential, flavonoid biosynthesis, and ROS scavenging system, the researchers demonstrated the significant role of SlGRAS10 as a stress-tolerant transcription factor in a particular form of abiotic stress tolerance. A schematic illustration of the usage of genomic approaches for making abiotic stress-tolerant tomato plants is given in Figure 4.
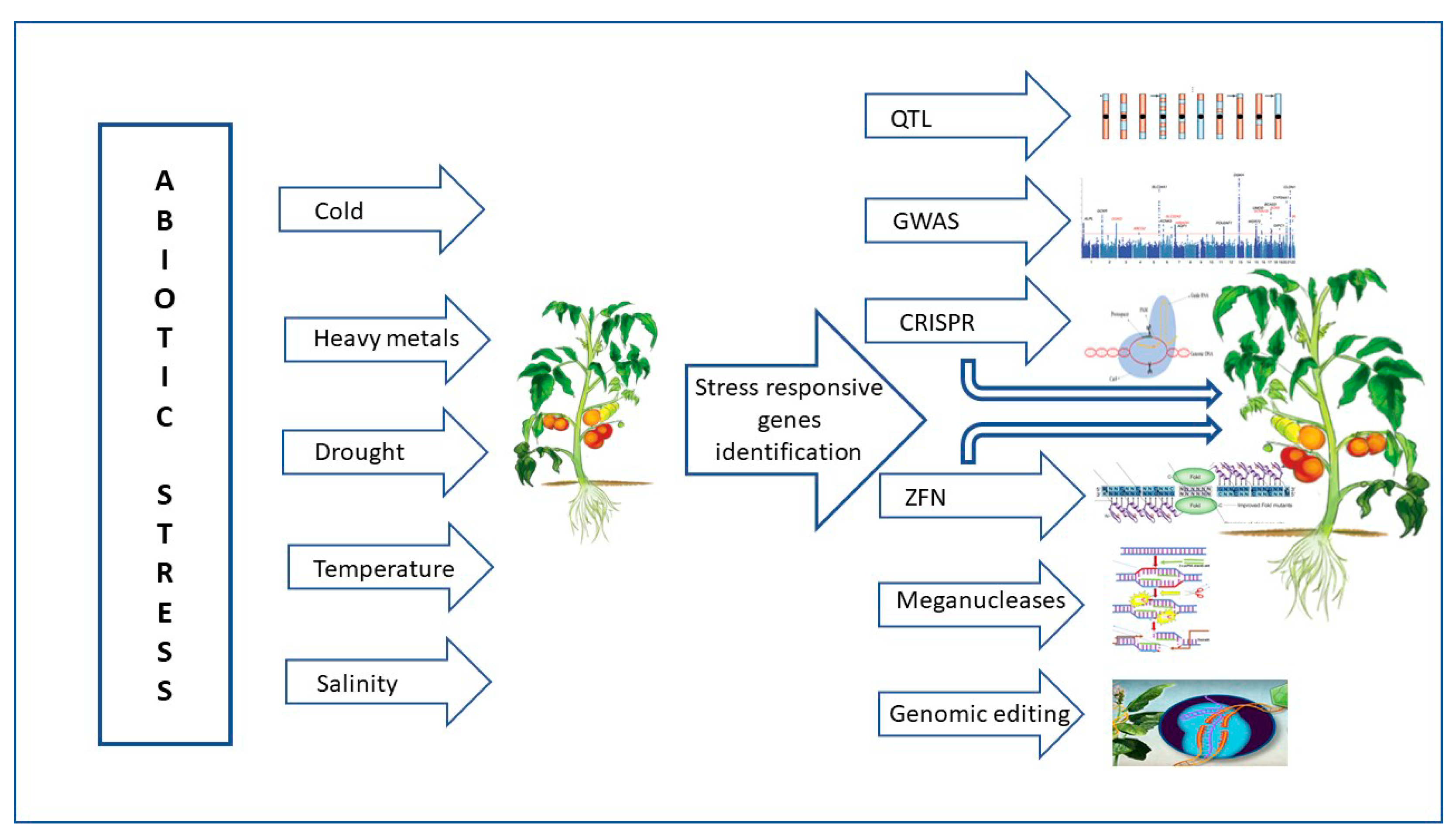
Figure 4. Schematic illustration of the usage of genomic approaches for making abiotic stress-tolerant tomato plants. The abiotic-stressed tomato plant shows restricted growth, while the plant with gene editing exhibits vigorous growth under abiotic stresses. QTL—quantitative trait loci; GWAS—genome-wide association studies; CRISPR—clustered regularly interspaced short palindromic repeats; ZFN—zinc-finger nucleases.
Gisbert et al. [48] demonstrated that tomato plants with the HAL1 gene, which comes from the yeast Saccharomyces cerevisiae, had increased tolerance to salinity. The ability of transgenic tomato lines to retain K+ under salt stress was shown to be higher than that of control plants when intracellular cation ratios (K+ to Na+) were considered. Therefore, by lowering long-term shoot Na+ retention, overexpression of the yeast gene HAL5 in tomatoes increases their ability to withstand salt stress. Regardless of the level of salt stress, this was the result of decreased Na+ transfer from roots to shoots. According to Kumari et al. [49], tomato plants overexpressing AtNHX1 had higher salt resistance. The researchers suggested that AtNHX1 was in charge of promoting active K+ absorption at the tonoplast and K+ dispersion inside cells.
The activities of the majority of the bZIP family members in tomatoes were investigated by Zhu and coworkers, and basic region/leucine zipper (bZIP) transcription factors function as essential regulators in ABA-mediated stress response in plants [25]. SlbZIP1 may have potential uses in the creation of salt- and drought-tolerant tomato cultivars, according to the researchers, who hypothesized that SlbZIP1 performs a critical function in salt and drought stress tolerance through altering an ABA-mediated pathway.
The expression of SlMYB102 was shown to be higher in ripe fruits and roots of tomato plants than in vegetative organs by Wang et al. [50]. Authors discovered additional regulatory components that are photo-responsive, abiotic stress-responsive, and hormone-responsive in the SlMYB102 promoter region. They emphasized that SlMYB102 may be implicated in the pathways for proline synthesis and C-repeat binding transcription factor (CBF), which boost tomato plant cold resistance. The genomics to combat abiotic stress in tomatoes is given in Table 1.
Table 1. Genomics to combat abiotic stress tolerance in tomato.
| S. No. | Gene/Origin | Function | Expression/Regulation | Results | References |
|---|---|---|---|---|---|
| 1 | SDG34 | Response to stress | Expression of negative stress response regulators and transcriptional repressors | Improvement of stress and pathogen tolerance | [43] |
| 2 | SlGRAS10 | Increasing osmotic potential, flavonoid production, and the ROS scavenging mechanism to increase abiotic stress tolerance | Downregulation | [40] | |
| 3 | BEL1-like genes | Numerous biological processes in plants are regulated by transcription factors, which are members of the superfamily of three-amino-loop-extension (TALE) proteins | Displayed various tissue-specific expression patterns and reacted to heat, cold, and drought stress | Plant growth and abiotic stress response | [51] |
| 4 | SlAIM1 | Salt and oxidative stress tolerance | Salt and oxidative stress tolerance is increased by SlAIM1 overexpression, but these two abiotic stimuli are made more sensitive by SlAIM1 silencing | Resistance to abiotic stress | [44][52] |
| 5 | TFs, s Cycling Dof Factor AtCDF3, AtDREB1a, NAC transcription factor JUNGBRUNNEN1 (AtJUB1) and AP2/ERF-like transcription factor CcHRD |
Increases abiotic stress tolerance of tomatoes, including cold, salt, and drought stress | Overexpression | Stress tolerance | [44][53] |
| 6 | SlMBP8, SlHB2, SlAGO4A | Tolerance to salt, drought stress | Overexpression | Tolerance to salt, drought stress | [54] |
| 7 | INVINH1 | Tolerance to cold stress | Tolerance to cold stress | [55] | |
| 8 | SlMBP8 | more tolerance to drought and salt stress | Gene silencing | More tolerance to drought and salt stress | [54] |
| 9 | SI PL | Resistance to pathogenic Botrytis cinerea and prolonged shelf life | Resistance to pathogenic Botrytis cinerea | [56] | |
| 10 | SlbZIP1 | Salt and drought stress tolerance | Expression | Salt and drought stress tolerance | [57] |
| 11 | SlMAPK3, SlMPK7 i | Resistance to chilling, cadmium, and drought stresses | Overexpression | Resistance to chilling, cadmium, and drought stresses | [58] |
| 12 | PpSnRK1α) | Accelerated metabolism of reactive oxygen species via upregulating antioxidase gene expression and antioxidant enzyme activity | Overexpression | Salt resistance | [59] |
| 13 | SlBZR1D | Salt tolerance and upregulated the expression of multiple stress-related genes | Overexpression and upregulation | Salt tolerance | [25] |
| 14 | SlNL33 | Ascorbate accumulation | suppressed expression | Stress tolerance | [60] |
| 15 | SlHY5 | Cold tolerance | Overexpression | [61] | |
| 16 | MdSWEET17 | Drought stress response and the regulation of fructose. | Expressed in tomatoes | Drought stress | [62] |
| 17 | SiDHN | Saussurea involucrata dehydrin gene overexpression | Overexpression | Cold and drought tolerance | [63] |
| 16 | SlHSP17.7 | Controlling Calcium Signaling and Phosphatidylglycerol Metabolism | Overexpression | Cold tolerance | [64] |
| 17 | SlABIG1 | Salt stress negative regulator gene | Knockout | Salt tolerance | [65] |
| 18 | Solyc03g020030 | Proteinase inhibitor-II | Gene silencing | Thermotolerance | [66] |
| 19 | SlDEAD23 and SlDEAD35 | Abiotic and biotic stress responses | Overexpression | Enhanced tolerance to salt and cold | [67] |
| 20 | SlGRAS10 | Improved the expression of superoxide dismutase, peroxidase, and catalase to lessen the impact of reactive oxygen species | Downregulation by RNA interference | Abiotic stress tolerance | [40] |
| 21 | SlLBD40 | A negative regulator of drought tolerance, it was implicated in JA signaling. | CRISPR/Cas9 targeted mutagenesis (knockout) | Drought tolerance | [68] |
| 22 | SlMAPK3i | Removing ROS buildup and increasing the expression of genes associated with ethylene signaling | Over-expression | Salt stress tolerance |
[69] |
References
- Rehem, B.C.; Bertolde, F.Z.; de Almeida, A.A.F. Regulation of Gene Expression in Response to Abiotic Stress in Plants; InTech: London, UK, 2012; pp. 13–38.
- Kosová, K.; Vítámvás, P.; Prášil, I.T.; Renaut, J. Plant proteome changes under abiotic stress—Contribution of proteomics studies to understanding plant stress response. J. Proteom. 2011, 74, 1301–1322.
- Kosová, K.; Vítámvás, P.; Urban, M.O.; Klíma, M.; Roy, A.; Prášil, I.T. Biological Networks Underlying Abiotic Stress Tolerance in Temperate Crops—A Proteomic Perspective. Int. J. Mol. Sci. 2015, 16, 20913–20942.
- Mustafa, G.; Akhtar, M.S.; Abdullah, R. Global Concern for Salinity on Various Agro-Ecosystems. In Salt Stress, Microbes, and Plant Interactions: Causes and Solution; Springer: Singapore, 2019; pp. 1–19.
- Kosová, K.; Vítámvás, P.; Urban, M.O.; Prášil, I.T.; Renaut, J. Plant Abiotic Stress Proteomics: The Major Factors Determining Alterations in Cellular Proteome. Front. Plant Sci. 2018, 9, 122.
- Hossain, Z.; Komatsu, S. Contribution of proteomic studies towards understanding plant heavy metal stress response. Front. Plant Sci. 2012, 3, 310.
- Ahsan, N.; Renaut, J.; Komatsu, S. Recent developments in the application of proteomics to the analysis of plant responses to heavy metals. Proteomics 2009, 9, 2602–2621.
- Pandey, A.; Rajamani, U.; Verma, J.; Subba, P.; Chakraborty, N.; Datta, A.; Chakraborty, S.; Chakraborty, N. Identification of Extracellular Matrix Proteins of Rice (Oryza sativa L.) Involved in Dehydration-Responsive Network: A Proteomic Approach. J. Proteome Res. 2010, 9, 3443–3464.
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467.
- Møller, I.M.; Jensen, P.E.; Hansson, A. Oxidative Modifications to Cellular Components in Plants. Annu. Rev. Plant Biol. 2007, 58, 459–481.
- Hajheidari, M.; Eivazi, A.; Buchanan, B.B.; Wong, J.H.; Majidi, I.; Salekdeh, G.H. Proteomics Uncovers a Role for Redox in Drought Tolerance in Wheat. J. Proteome Res. 2007, 6, 1451–1460.
- Suzuki, N.; Koussevitzky, S.; Mittler, R.; Miller, G. ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 2012, 35, 259–270.
- Ahammed, G.J.; Zhou, Y.H.; Xia, X.J.; Mao, W.H.; Shi, K.; Yu, J.Q. Brassinosteroid regulates secondary metabolism in tomato towards enhanced tolerance to phenanthrene. Biol. Plant. 2013, 57, 154–158.
- El-Esawi, M.A.; Elkelish, A.; Soliman, M.; Elansary, H.O.; Zaid, A.; Wani, S.H. Serratia marcescens BM1 Enhances Cadmium Stress Tolerance and Phytoremediation Potential of Soybean Through Modulation of Osmolytes, Leaf Gas Exchange, Antioxidant Machinery, and Stress-Responsive Genes Expression. Antioxidants 2020, 9, 43.
- Mousavi, A.; Hotta, Y. Glycine-Rich Proteins: A Class of Novel Proteins. Appl. Biochem. Biotechnol. 2005, 120, 169–174.
- Rai, G.; Parveen, A.; Jamwal, G.; Basu, U.; Kumar, R.; Rai, P.; Sharma, J.; Alalawy, A.; Al-Duais, M.; Hossain, M.; et al. Leaf Proteome Response to Drought Stress and Antioxidant Potential in Tomato (Solanum lycopersicum L.). Atmosphere 2021, 12, 1021.
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252.
- Li, R.; Liu, C.; Zhao, R.; Wang, L.; Chen, L.; Yu, W.; Zhang, S.; Sheng, J.; Shen, L. CRISPR/Cas9-Mediated SlNPR1 mutagenesis reduces tomato plant drought tolerance. BMC Plant Biol. 2019, 19, 38.
- Gerszberg, A.; Hnatuszko-Konka, K. Tomato tolerance to abiotic stress: A review of most often engineered target sequences. Plant Growth Regul. 2017, 83, 175–198.
- Wang, Z.-Y.; Zhao, S.; Liu, J.-F.; Zhao, H.-Y.; Sun, X.-Y.; Wu, T.-R.; Pei, T.; Wang, Y.; Liu, Q.-F.; Yang, H.-H.; et al. Genome-wide identification of Tomato Golden 2-Like transcription factors and abiotic stress related members screening. BMC Plant Biol. 2022, 22, 82.
- Ranjan, A.; Ichihashi, Y.; Sinha, N.R. The tomato genome: Implications for plant breeding, genomics and evolution. Genome Biol. 2012, 13, 167.
- Dheer, P.; Rautela, I.; Sharma, V.; Dhiman, M.; Sharma, A.; Sharma, N.; Sharma, M.D. Evolution in crop improvement approaches and future prospects of molecular markers to CRISPR/Cas9 system. Gene 2020, 753, 144795.
- Cambiaso, V.; Pratta, G.R.; da Costa, J.H.P.; Zorzoli, R.; Francis, D.M.; Rodríguez, G.R. Whole genome re-sequencing analysis of two tomato genotypes for polymorphism insight in cloned genes and a genetic map construction. Sci. Hortic. 2019, 247, 58–66.
- Chaudhary, J.; Khatri, P.; Singla, P.; Kumawat, S.; Kumari, A.; R, V.; Vikram, A.; Jindal, S.K.; Kardile, H.; Kumar, R.; et al. Advances in Omics Approaches for Abiotic Stress Tolerance in Tomato. Biology 2019, 8, 90.
- Zhu, M.; Meng, X.; Cai, J.; Li, G.; Dong, T.; Li, Z. Basic leucine zipper transcription factor SlbZIP1 mediates salt and drought stress tolerance in tomato. BMC Plant Biol. 2018, 18, 83.
- Szymanski, J.; Levin, Y.; Savidor, A.; Breitel, D.; Chappell-Maor, L.; Heinig, U.; Töpfer, N.; Aharoni, A. Label-free deep shotgun proteomics reveals protein dynamics during tomato fruit tissues development. Plant J. 2017, 90, 396–417.
- Kausar, R.; Wang, X.; Komatsu, S. Crop Proteomics under Abiotic Stress: From Data to Insights. Plants 2022, 11, 2877.
- Krishna, R.; Karkute, S.G.; Ansari, W.A.; Jaiswal, D.K.; Verma, J.P.; Singh, M. Transgenic tomatoes for abiotic stress tolerance: Status and way ahead. 3 Biotech 2019, 9, 143.
- Yang, Y.; Tang, N.; Xian, Z.; Li, Z. Two SnRK2 protein kinases genes play a negative regulatory role in the osmotic stress response in tomato. Plant Cell Tissue Organ Cult. 2015, 122, 421–434.
- Yu, L.; Yan, J.; Yang, Y.; He, L.; Zhu, W. Enhanced Tolerance to Chilling Stress in Tomato by Overexpression of a Mitogen-Activated Protein Kinase, SlMPK7. Plant Mol. Biol. Rep. 2016, 34, 76–88.
- Liu, H.; Yu, C.; Li, H.; Ouyang, B.; Wang, T.; Zhang, J.; Wang, X.; Ye, Z. Overexpression of ShDHN, a dehydrin gene from Solanum habrochaites enhances tolerance to multiple abiotic stresses in tomato. Plant Sci. 2015, 231, 198–211.
- Li, J.-B.; Luan, Y.-S.; Liu, Z. SpWRKY1 mediates resistance to Phytophthora infestans and tolerance to salt and drought stress by modulating reactive oxygen species homeostasis and expression of defense-related genes in tomato. Plant Cell Tissue Organ Cult. 2015, 123, 67–81.
- Shah, S.H.; Ali, S.; Jan, S.A.; Din, J.U.; Ali, G.M. Piercing and incubation method of in planta transformation producing stable transgenic plants by overexpressing DREB1A gene in tomato (Solanum lycopersicum Mill.). Plant Cell Tissue Organ Cult. 2015, 120, 1139–1157.
- Meng, X.; Wang, J.-R.; Wang, G.-D.; Liang, X.-Q.; Li, X.-D.; Meng, Q.-W. An R2R3-MYB gene, LeAN2, positively regulated the thermo-tolerance in transgenic tomato. J. Plant Physiol. 2015, 175, 1–8.
- Hu, Y.; Wu, Q.; Sprague, S.A.; Park, J.; Oh, M.; Rajashekar, C.B.; Koiwa, H.; Nakata, P.A.; Cheng, N.; Hirschi, K.D.; et al. Tomato expressing Arabidopsis glutaredoxin gene AtGRXS17 confers tolerance to chilling stress via modulating cold responsive components. Hortic. Res. 2015, 2, 15051.
- Hu, D.-G.; Ma, Q.-J.; Sun, C.-H.; Sun, M.-H.; You, C.-X.; Hao, Y.-J. Overexpression of MdSOS2L1, a CIPK protein kinase, increases the antioxidant metabolites to enhance salt tolerance in apple and tomato. Physiol. Plant. 2016, 156, 201–214.
- Gong, B.; Li, X.; Vandenlangenberg, K.M.; Wen, D.; Sun, S.; Wei, M.; Li, Y.; Yang, F.; Shi, Q.; Wang, X. Overexpression of S-adenosyl-l-methionine synthetase increased tomato tolerance to alkali stress through polyamine metabolism. Plant Biotechnol. J. 2014, 12, 694–708.
- Metwali, E.M.R.; Soliman, H.I.A.; Fuller, M.; Almaghrabi, O.A. Improving fruit quality in tomato (Lycopersicum esculentum Mill) under heat stress by silencing the vis 1 gene using small interfering RNA technology. Plant Cell, Tissue Organ Cult. 2015, 121, 153–166.
- Naeem, M.; Shahzad, K.; Saqib, S.; Shahzad, A.; Nasrullah; Younas, M.; Afridi, M.I. The Solanum melongena COP1LIKE manipulates fruit ripening and flowering time in tomato (Solanum lycopersicum). Plant Growth Regul. 2022, 96, 369–382.
- Habib, S.; Lwin, Y.; Li, N. Down-Regulation of SlGRAS10 in Tomato Confers Abiotic Stress Tolerance. Genes 2021, 12, 623.
- Orellana, S.; Yañez, M.; Espinoza, A.; Verdugo, I.; González, E.; Ruiz-Lara, S.; Casaretto, J.A. The transcription factor SlAREB1 confers drought, salt stress tolerance and regulates biotic and abiotic stress-related genes in tomato. Plant Cell Environ. 2010, 33, 2191–2208.
- Wang, Z.-Y.; Bao, Y.-F.; Pei, T.; Wu, T.-R.; DU, X.; He, M.-X.; Wang, Y.; Liu, Q.-F.; Yang, H.-H.; Jiang, J.-B.; et al. Silencing the SLB3 transcription factor gene decreases drought stress tolerance in tomato. J. Integr. Agric. 2020, 19, 2699–2708.
- Bvindi, C.; Lee, S.; Tang, L.; Mickelbart, M.V.; Li, Y.; Mengiste, T. Improved pathogen and stress tolerance in tomato mutants of SET domain histone 3 lysine methyltransferases. New Phytol. 2022, 235, 1957–1976.
- Wai, A.H.; Naing, A.H.; Lee, D.-J.; Kil Kim, C.; Chung, M.-Y. Molecular genetic approaches for enhancing stress tolerance and fruit quality of tomato. Plant Biotechnol. Rep. 2020, 14, 515–537.
- Pan, C.; Yang, D.; Zhao, X.; Jiao, C.; Yan, Y.; Lamin-Samu, A.T.; Wang, Q.; Xu, X.; Fei, Z.; Lu, G. Tomato stigma exsertion induced by high temperature is associated with the jasmonate signalling pathway. Plant Cell Environ. 2019, 42, 1205–1221.
- Bineau, E.; Diouf, I.; Carretero, Y.; Duboscq, R.; Bitton, F.; Djari, A.; Zouine, M.; Causse, M. Genetic diversity of tomato response to heat stress at the QTL and transcriptome levels. Plant J. 2021, 107, 1213–1227.
- Sidhu, N.S.; Pruthi, G.; Singh, S.; Bishnoi, R.; Singla, D. Genome-wide identification and analysis of GRAS transcription factors in the bottle gourd genome. Sci. Rep. 2020, 10, 14338.
- Gisbert, C.; Rus, A.M.; Bolarín, M.C.; López-Coronado, J.M.; Arrillaga, I.; Montesinos, C.; Caro, M.; Serrano, R.; Moreno, V. The Yeast HAL1 Gene Improves Salt Tolerance of Transgenic Tomato. Plant Physiol. 2000, 123, 393–402.
- Kumari, P.H.; Kumar, S.A.; Sivan, P.; Katam, R.; Suravajhala, P.; Rao, K.S.; Varshney, R.K.; Kishor, P.B.K. Overexpression of a Plasma Membrane Bound Na+/H+ Antiporter-Like Protein (SbNHXLP) Confers Salt Tolerance and Improves Fruit Yield in Tomato by Maintaining Ion Homeostasis. Front. Plant Sci. 2016, 7, 2027.
- Wang, M.; Hao, J.; Chen, X.; Zhang, X. SlMYB102 expression enhances low-temperature stress resistance in tomato plants. PeerJ 2020, 8, e10059.
- He, Y.; Yang, T.; Yan, S.; Niu, S.; Zhang, Y. Identification and characterization of the BEL1-like genes reveal their potential roles in plant growth and abiotic stress response in tomato. Int. J. Biol. Macromol. 2022, 200, 193–205.
- AbuQamar, S.; Luo, H.; Laluk, K.; Mickelbart, M.V.; Mengiste, T. Crosstalk between biotic and abiotic stress responses in tomato is mediated by the AIM1 transcription factor. Plant J. 2009, 58, 347–360, Erratum in: Plant J. 2009, 60, 929.
- Thirumalaikumar, V.P.; Devkar, V.; Mehterov, N.; Ali, S.; Ozgur, R.; Turkan, I.; Mueller-Roeber, B.; Balazadeh, S. NAC transcription factor JUNGBRUNNEN1 enhances drought tolerance in tomato. Plant Biotechnol. J. 2018, 16, 354–366.
- Yin, W.; Hu, Z.; Cui, B.; Guo, X.; Hu, J.; Zhu, Z.; Chen, G. Suppression of the MADS-box gene SlMBP8 accelerates fruit ripening of tomato (Solanum lycopersicum). Plant Physiol. Biochem. 2017, 118, 235–244.
- Xu, X.-X.; Hu, Q.; Yang, W.-N.; Jin, Y. The roles of cell wall invertase inhibitor in regulating chilling tolerance in tomato. BMC Plant Biol. 2017, 17, 195.
- Yang, L.; Huang, W.; Xiong, F.; Xian, Z.; Su, D.; Ren, M.; Li, Z. Silencing of SlPL, which encodes a pectate lyase in tomato, confers enhanced fruit firmness, prolonged shelf-life and reduced susceptibility to grey mould. Plant Biotechnol. J. 2017, 15, 1544–1555.
- Chaudhary, P.; Sharma, A.; Singh, B.; Nagpal, A.K. Bioactivities of phytochemicals present in tomato. J. Food Sci. Technol. 2018, 55, 2833–2849.
- Muhammad, T.; Zhang, J.; Ma, Y.; Li, Y.; Zhang, F.; Zhang, Y.; Liang, Y. Overexpression of a Mitogen-Activated Protein Kinase SlMAPK3 Positively Regulates Tomato Tolerance to Cadmium and Drought Stress. Molecules 2019, 24, 556.
- Jia, C.; Zhao, S.; Bao, T.; Zhao, P.; Peng, K.; Guo, Q.; Gao, X.; Qin, J. Tomato BZR/BES transcription factor SlBZR1 positively regulates BR signaling and salt stress tolerance in tomato and Arabidopsis. Plant Sci. 2021, 302, 110719.
- Ye, J.; Liu, G.; Chen, W.; Zhang, F.; Li, H.; Ye, Z.; Zhang, Y. Knockdown of SlNL33 accumulates ascorbate, enhances disease and oxidative stress tolerance in tomato (Solanum lycopersicum). Plant Growth Regul. 2019, 89, 49–58.
- Han, N.; Fan, S.; Zhang, T.; Sun, H.; Zhu, Y.; Gong, H.; Guo, J. SlHY5 is a necessary regulator of the cold acclimation response in tomato. Plant Growth Regul. 2020, 91, 1–12.
- Lu, J.; Sun, M.-H.; Ma, Q.-J.; Kang, H.; Liu, Y.-J.; Hao, Y.-J.; You, C.-X. MdSWEET17, a sugar transporter in apple, enhances drought tolerance in tomato. J. Integr. Agric. 2019, 18, 2041–2051.
- Guo, X.; Zhang, L.; Wang, X.; Zhang, M.; Xi, Y.; Wang, A.; Zhu, J. Overexpression of Saussurea involucrata dehydrin gene SiDHN promotes cold and drought tolerance in transgenic tomato plants. PLoS ONE 2019, 14, e0225090.
- Zhang, N.; Zhao, H.; Shi, J.; Wu, Y.; Jiang, J. Functional characterization of class I SlHSP17.7 gene responsible for tomato cold-stress tolerance. Plant Sci. 2020, 298, 110568.
- Ding, F.; Qiang, X.; Jia, Z.; Li, L.; Hu, J.; Yin, M.; Xia, S.; Chen, B.; Qi, J.; Li, Q.; et al. Knockout of a novel salt responsive gene SlABIG1 enhance salinity tolerance in tomato. Environ. Exp. Bot. 2022, 200, 104903.
- Balyan, S.; Rao, S.; Jha, S.; Bansal, C.; Das, J.R.; Mathur, S. Characterization of novel regulators for heat stress tolerance in tomato from Indian sub-continent. Plant Biotechnol. J. 2020, 18, 2118–2132.
- Pandey, S.; Muthamilarasan, M.; Sharma, N.; Chaudhry, V.; Dulani, P.; Shweta, S.; Jha, S.; Mathur, S.; Prasad, M. Characterization of DEAD-box family of RNA helicases in tomato provides insights into their roles in biotic and abiotic stresses. Environ. Exp. Bot. 2019, 158, 107–116.
- Liu, L.; Zhang, J.; Xu, J.; Li, Y.; Guo, L.; Wang, Z.; Zhang, X.; Zhao, B.; Guo, Y.-D.; Zhang, N. CRISPR/Cas9 targeted mutagenesis of SlLBD40, a lateral organ boundaries domain transcription factor, enhances drought tolerance in tomato. Plant Sci. 2020, 301, 110683.
- Shu, P.; Li, Y.; Li, Z.; Sheng, J.; Shen, L. SlMAPK3 enhances tolerance to salt stress in tomato plants by scavenging ROS accumulation and up-regulating the expression of ethylene signaling related genes. Environ. Exp. Bot. 2022, 193, 104698.
More
Information
Subjects:
Agriculture, Dairy & Animal Science
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.9K
Revisions:
2 times
(View History)
Update Date:
21 Apr 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




