
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Daniel Elson | -- | 1490 | 2023-04-19 06:17:38 | | | |
| 2 | Beatrix Zheng | + 2 word(s) | 1492 | 2023-04-19 07:09:46 | | |
Video Upload Options
The aryl hydrocarbon receptor (AhR) is a ligand-activated transcription factor involved in regulating a wide range of biological responses. A diverse array of xenobiotics and endogenous small molecules bind to the receptor and drive unique phenotypic responses. Due in part to its role in mediating toxic responses to environmental pollutants, AhR activation has not been traditionally viewed as a viable therapeutic approach. Nonetheless, the expression and activation of AhR can inhibit the proliferation, migration, and survival of cancer cells, and many clinically approved drugs transcriptionally activate AhR. Identification of novel select modulators of AhR-regulated transcription that promote tumor suppression is an active area of investigation. The development of AhR-targeted anticancer agents requires a thorough understanding of the molecular mechanisms driving tumor suppression.
1. Role of AhR in Xenobiotic Metabolism
2. The AhR Signaling Pathway
3. Role of Co-Regulators in AhR Signaling
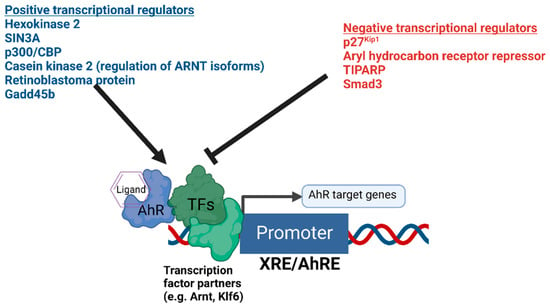
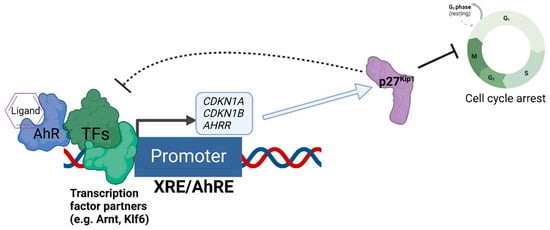
References
- McIntosh, B.E.; Hogenesch, J.B.; Bradfield, C.A. Mammalian Per-Arnt-Sim Proteins in Environmental Adaptation. Annu. Rev. Physiol. 2010, 72, 625–645.
- Denison, M.S.; Soshilov, A.A.; He, G.; DeGroot, D.E.; Bin Zhao, B. Exactly the Same but Different: Promiscuity and Diversity in the Molecular Mechanisms of Action of the Aryl Hydrocarbon (Dioxin) Receptor. Toxicol. Sci. 2011, 124, 1–22.
- Ciolino, H.P.; Macdonald, C.J.; Memon, O.S.; Bass, S.E.; Yeh, G.C. Sulindac regulates the aryl hydrocarbon receptor-mediated expression of Phase 1 metabolic enzymes in vivo and in vitro. Carcinogenesis 2005, 27, 1586–1592.
- Poland, A.P.; Glover, E.; Robinson, J.R.; Nebert, D.W. Genetic expression of aryl hydrocarbon hydroxylase activity. Induction of monooxygenase activities and cytochrome P1-450 formation by 2,3,7,8 tetrachlorodibenzo p dioxin in mice genetically “nonresponsive” to other aromatic hydrocarbons. J. Biol. Chem. 1974, 249, 5599–5606.
- National Academies of Sciences, Engineering, and Medicine. Veterans and Agent Orange: Update 2014; The National Academies Press: Washington, DC, USA, 2016.
- Antonsson, C.; Whitelaw, M.L.; McGuire, J.; Gustafsson, J.A.; Poellinger, L. Distinct roles of the molecular chaperone hsp90 in modulating dioxin receptor function via the basic helix-loop-helix and PAS domains. Mol. Cell. Biol. 1995, 15, 756–765.
- Tsuji, N.; Fukuda, K.; Nagata, Y.; Okada, H.; Haga, A.; Hatakeyama, S.; Yoshida, S.; Okamoto, T.; Hosaka, M.; Sekine, K.; et al. The activation mechanism of the aryl hydrocarbon receptor (AhR) by molecular chaperone HSP90. FEBS Open Bio 2014, 4, 796–803.
- Pappas, B.; Yang, Y.; Wang, Y.; Kim, K.; Chung, H.J.; Cheung, M.; Ngo, K.; Shinn, A.; Chan, W.K. p23 protects the human aryl hydrocarbon receptor from degradation via a heat shock protein 90-independent mechanism. Biochem. Pharmacol. 2018, 152, 34–44.
- Meyer, B.K.; Pray-Grant, M.G.; Heuvel, J.P.V.; Perdew, G.H. Hepatitis B Virus X-Associated Protein 2 Is a Subunit of the Unliganded Aryl Hydrocarbon Receptor Core Complex and Exhibits Transcriptional Enhancer Activity. Mol. Cell. Biol. 1998, 18, 978–988.
- Enan, E.; Matsumura, F. Identification of c-Src as the integral component of the cytosolic Ah receptor complex, transducing the signal of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) through the protein phosphorylation pathway. Biochem. Pharmacol. 1996, 52, 1599–1612.
- Petrulis, J.R.; Perdew, G.H. The role of chaperone proteins in the aryl hydrocarbon receptor core complex. Chem. Biol. Interact. 2002, 141, 25–40.
- Yao, E.F.; Denison, M.S. DNA sequence determinants for binding of transformed Ah receptor to a dioxin-responsive enhancer. Biochemistry 1992, 31, 5060–5067.
- Swanson, H.; Tullis, K.; Denison, M.S. Binding of transformed Ah receptor complex to a dioxin responsive transcriptional enhancer: Evidence for two distinct heteromeric DNA-binding forms. Biochemistry 1993, 32, 12841–12849.
- Tanos, R.; Patel, R.D.; Murray, I.A.; Smith, P.B.; Patterson, A.D.; Perdew, G.H. Aryl hydrocarbon receptor regulates the cholesterol biosynthetic pathway in a dioxin response element-independent manner. Hepatology 2012, 55, 1994–2004.
- Jackson, D.P.; Li, H.; Mitchell, K.A.; Joshi, A.D.; Elferink, C.J. Ah Receptor–Mediated Suppression of Liver Regeneration through NC-XRE–Driven p21Cip1Expression. Mol. Pharmacol. 2014, 85, 533–541.
- Apetoh, L.; Quintana, F.J.; Pot, C.; Joller, N.; Xiao, S.; Kumar, D.; Burns, E.J.; Sherr, D.H.; Weiner, H.L.; Kuchroo, V.K. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat. Immunol. 2010, 11, 854–861.
- Wilson, S.R.; Joshi, A.D.; Elferink, C.J. The Tumor Suppressor Kruppel-Like Factor 6 Is a Novel Aryl Hydrocarbon Receptor DNA Binding Partner. J. Pharmacol. Exp. Ther. 2013, 345, 419–429.
- Kim, D.W.; Gazourian, L.; Quadri, S.A.; Romieu-Mourez, R.; Sherr, D.H.; Sonenshein, G.E. The RelA NF-kappaB subunit and the aryl hydrocarbon receptor (AhR) cooperate to transactivate the c-myc promoter in mammary cells. Oncogene 2000, 19, 5498–5506.
- Vogel, C.F.A.; Matsumura, F. A new cross-talk between the aryl hydrocarbon receptor and RelB, a member of the NF-κB family. Biochem. Pharmacol. 2009, 77, 734–745.
- Reisz-Porszasz, S.; Probst, M.R.; Fukunaga, B.N.; Hankinson, O. Identification of functional domains of the aryl hydrocarbon receptor nuclear translocator protein (ARNT). Mol. Cell. Biol. 1994, 14, 6075–6086.
- Crews, S.T.; Fan, C.-M. Remembrance of things PAS: Regulation of development by bHLH–PAS proteins. Curr. Opin. Genet. Dev. 1999, 9, 580–587.
- Lindebro, M.; Poellinger, L.; Whitelaw, M. Protein-protein interaction via PAS domains: Role of the PAS domain in positive and negative regulation of the bHLH/PAS dioxin receptor-Arnt transcription factor complex. EMBO J. 1995, 14, 3528–3539.
- Pongratz, I.; Antonsson, C.; Whitelaw, M.L.; Poellinger, L. Role of the PAS Domain in Regulation of Dimerization and DNA Binding Specificity of the Dioxin Receptor. Mol. Cell. Biol. 1998, 18, 4079–4088.
- Andersson, P.; McGuire, J.; Rubio, C.; Gradin, K.; Whitelaw, M.L.; Pettersson, S.; Hanberg, A.; Poellinger, L. A constitutively active dioxin/aryl hydrocarbon receptor induces stomach tumors. Proc. Natl. Acad. Sci. USA 2002, 99, 9990–9995.
- Schulte, K.W.; Green, E.; Wilz, A.; Platten, M.; Daumke, O. Structural Basis for Aryl Hydrocarbon Receptor-Mediated Gene Activation. Structure 2017, 25, 1025–1033.e3.
- Hahn, M.E.; Karchner, S.I.; Merson, R.R. Diversity as opportunity: Insights from 600 million years of AHR evolution. Curr. Opin. Toxicol. 2017, 2, 58–71.
- Huang, X.; Powell-Coffman, J.A.; Jin, Y. The AHR-1 aryl hydrocarbon receptor and its co-factor the AHA-1 aryl hydrocarbon receptor nuclear translocator specify GABAergic neuron cell fate in C. elegans. Development 2004, 131, 819–828.
- Qin, H.; Powell-Coffman, J.A. The Caenorhabditis elegans aryl hydrocarbon receptor, AHR-1, regulates neuronal development. Dev. Biol. 2004, 270, 64–75.
- Sonnenfeld, M.; Ward, M.; Nystrom, G.; Mosher, J.; Stahl, S.; Crews, S. The Drosophila tango gene encodes a bHLH-PAS protein that is orthologous to mammalian Arnt and controls CNS midline and tracheal development. Development 1997, 124, 4571–4582.
- Di Giaimo, R.; Durovic, T.; Barquin, P.; Kociaj, A.; Lepko, T.; Aschenbroich, S.; Breunig, C.T.; Irmler, M.; Cernilogar, F.M.; Schotta, G.; et al. The Aryl Hydrocarbon Receptor Pathway Defines the Time Frame for Restorative Neurogenesis. Cell Rep. 2018, 25, 3241–3251.e5.
- Rothhammer, V.; Borucki, D.M.; Kenison, J.E.; Hewson, P.; Wang, Z.; Bakshi, R.; Sherr, D.H.; Quintana, F.J. Detection of aryl hydrocarbon receptor agonists in human samples. Sci. Rep. 2018, 8, 4970.
- Wheeler, J.L.H.; Martin, K.C.; Resseguie, E.; Lawrence, B.P. Differential Consequences of Two Distinct AhR Ligands on Innate and Adaptive Immune Responses to Influenza A Virus. Toxicol. Sci. 2014, 137, 324–334.
- Bohonowych, J.E.; Denison, M.S. Persistent Binding of Ligands to the Aryl Hydrocarbon Receptor. Toxicol. Sci. 2007, 98, 99–109.
- Ehrlich, A.K.; Pennington, J.M.; Bisson, W.H.; Kolluri, S.K.; Kerkvliet, N.I. TCDD, FICZ, and Other High Affinity AhR Ligands Dose-Dependently Determine the Fate of CD4+ T Cell Differentiation. Toxicol. Sci. 2018, 161, 310–320.
- Vogel, C.F.; Khan, E.M.; Leung, P.S.; Gershwin, M.E.; Chang, W.L.; Wu, D.; Haarmann-Stemmann, T.; Hoffmann, A.; Denison, M.S. Cross-talk between Aryl Hydrocarbon Receptor and the Inflammatory Response: A role for nuclear factor-κB. J. Biol. Chem. 2014, 289, 1866–1875.
- DiNatale, B.C.; Smith, K.; John, K.; Krishnegowda, G.; Amin, S.G.; Perdew, G.H. Ah Receptor Antagonism Represses Head and Neck Tumor Cell Aggressive Phenotype. Mol. Cancer Res. 2012, 10, 1369–1379.
- Fletcher, N.; Hanberg, A.; Håkansson, H. Hepatic Vitamin A Depletion Is a Sensitive Marker of 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) Exposure in Four Rodent Species. Toxicol. Sci. 2001, 62, 166–175.
- Korkalainen, M.; Tuomisto, J.; Pohjanvirta, R. The AH Receptor of the Most Dioxin-Sensitive Species, Guinea Pig, Is Highly Homologous to the Human AH Receptor. Biochem. Biophys. Res. Commun. 2001, 285, 1121–1129.
- Ramadoss, P.; Perdew, G.H. Use of 2-Azido-3-iodo-7,8-dibromodibenzo-p-dioxin as a Probe to Determine the Relative Ligand Affinity of Human versus Mouse Aryl Hydrocarbon Receptor in Cultured Cells. Mol. Pharmacol. 2004, 66, 129–136.
- Hubbard, T.D.; Murray, I.A.; Perdew, G.H. Indole and Tryptophan Metabolism: Endogenous and Dietary Routes to Ah Receptor Activation. Drug Metab. Dispos. 2015, 43, 1522–1535.
- Perkins, A.; Phillips, J.L.; Kerkvliet, N.I.; Tanguay, R.L.; Perdew, G.H.; Kolluri, S.K.; Bisson, W.H. A Structural Switch between Agonist and Antagonist Bound Conformations for a Ligand-Optimized Model of the Human Aryl Hydrocarbon Receptor Ligand Binding Domain. Biology 2014, 3, 645–669.
- Bisson, W.H.; Koch, D.C.; O’Donnell, E.F.; Khalil, S.M.; Kerkvliet, N.I.; Tanguay, R.L.; Abagyan, R.; Kolluri, S.K. Modeling of the Aryl Hydrocarbon Receptor (AhR) Ligand Binding Domain and Its Utility in Virtual Ligand Screening to Predict New AhR Ligands. J. Med. Chem. 2009, 52, 5635–5641.
- Wang, S.; Hankinson, O. Functional Involvement of the Brahma/SWI2-related Gene 1 Protein in Cytochrome P4501A1 Transcription Mediated by the Aryl Hydrocarbon Receptor Complex. J. Biol. Chem. 2002, 277, 11821–11827.
- Hankinson, O. Role of coactivators in transcriptional activation by the aryl hydrocarbon receptor. Arch. Biochem. Biophys. 2005, 433, 379–386.
- Watzky, M.; Huard, S.; Juricek, L.; Dairou, J.; Chauvet, C.; Coumoul, X.; Letessier, A.; Miotto, B. Hexokinase 2 is a transcriptional target and a positive modulator of AHR signalling. Nucleic Acids Res. 2022, 50, 5545–5564.
- Evans, B.R.; Karchner, S.I.; Allan, L.L.; Pollenz, R.S.; Tanguay, R.L.; Jenny, M.J.; Sherr, D.H.; Hahn, M.E. Repression of Aryl Hydrocarbon Receptor (AHR) Signaling by AHR Repressor: Role of DNA Binding and Competition for AHR Nuclear Translocator. Mol. Pharmacol. 2008, 73, 387–398.
- Ahmed, S.; Bott, D.; Gomez, A.; Tamblyn, L.; Rasheed, A.; Cho, T.; MacPherson, L.; Sugamori, K.S.; Yang, Y.; Grant, D.M.; et al. Loss of the Mono-ADP-ribosyltransferase, Tiparp, Increases Sensitivity to Dioxin-induced Steatohepatitis and Lethality. J. Biol. Chem. 2015, 290, 16824–16840.
- Grimaldi, G.; Rajendra, S.; Matthews, J. The aryl hydrocarbon receptor regulates the expression of TIPARP and its cis long non-coding RNA, TIPARP-AS1. Biochem. Biophys. Res. Commun. 2018, 495, 2356–2362.
- Hutin, D.; Tamblyn, L.; Gomez, A.; Grimaldi, G.; Soedling, H.; Cho, T.; Ahmed, S.; Lucas, C.; Kanduri, C.; Grant, D.M.; et al. Hepatocyte-Specific Deletion of TIPARP, a Negative Regulator of the Aryl Hydrocarbon Receptor, Is Sufficient to Increase Sensitivity to Dioxin-Induced Wasting Syndrome. Toxicol. Sci. 2018, 165, 347–360.
- Zhang, L.; Cao, J.; Dong, L.; Lin, H. TiPARP forms nuclear condensates to degrade HIF-1α and suppress tumorigenesis. Proc. Natl. Acad. Sci. USA 2020, 117, 13447–13456.
- Lu, P.; Cai, X.; Guo, Y.; Xu, M.; Tian, J.; Locker, J.; Xie, W. Constitutive Activation of the Human Aryl Hydrocarbon Receptor in Mice Promotes Hepatocarcinogenesis Independent of Its Coactivator Gadd45b. Toxicol. Sci. 2019, 167, 581–592.
- Weinert, B.T.; Narita, T.; Satpathy, S.; Srinivasan, B.; Hansen, B.K.; Schölz, C.; Hamilton, W.B.; Zucconi, B.E.; Wang, W.W.; Liu, W.R.; et al. Time-Resolved Analysis Reveals Rapid Dynamics and Broad Scope of the CBP/p300 Acetylome. Cell 2018, 174, 231–244.e12.
- Nakano, N.; Sakata, N.; Katsu, Y.; Nochise, D.; Sato, E.; Takahashi, Y.; Yamaguchi, S.; Haga, Y.; Ikeno, S.; Motizuki, M.; et al. Dissociation of the AhR/ARNT complex by TGF-β/Smad signaling represses CYP1A1 gene expression and inhibits benzepyrene-mediated cytotoxicity. J. Biol. Chem. 2020, 295, 9033–9051.
- Solaimani, P.; Damoiseaux, R.; Hankinson, O. Genome-Wide RNAi High-Throughput Screen Identifies Proteins Necessary for the AHR-Dependent Induction of CYP1A1 by 2,3,7,8-Tetrachlorodibenzo-p-dioxin. Toxicol. Sci. 2013, 136, 107–119.
- Solaimani, P.; Wang, F.; Hankinson, O. SIN3A, Generally Regarded as a Transcriptional Repressor, Is Required for Induction of Gene Transcription by the Aryl Hydrocarbon Receptor. J. Biol. Chem. 2014, 289, 33655–33662.
- Bourner, L.A.; Muro, I.; Cooper, A.M.; Choudhury, B.K.; Bailey, A.O.; Russell, W.K.; Khanipov, K.; Golovko, G.; Wright, C.W. AhR promotes phosphorylation of ARNT isoform 1 in human T cell malignancies as a switch for optimal AhR activity. Proc. Natl. Acad. Sci. USA 2022, 119, e2114336119.
- Elson, D.J.; Nguyen, B.D.; Wood, R.; Zhang, Y.; Puig-Sanvicens, V.; Kolluri, S.K. The cyclin-dependent kinase inhibitor p27 Kip1 interacts with the aryl hydrocarbon receptor and negatively regulates its transcriptional activity. FEBS Lett. 2022, 596, 2056–2071.
- Orlando, S.; Gallastegui, E.; Besson, A.; Abril, G.; Aligué, R.; Pujol, M.J.; Bachs, O. p27Kip1and p21Cip1collaborate in the regulation of transcription by recruiting cyclin–Cdk complexes on the promoters of target genes. Nucleic Acids Res. 2015, 43, 6860–6873.
- Pippa, R.; Espinosa, L.; Gundem, G.; García-Escudero, R.; Dominguez, A.; Orlando, S.; Gallastegui, E.; Saiz, C.; Besson, A.; Pujol, M.J.; et al. p27Kip1 represses transcription by direct interaction with p130/E2F4 at the promoters of target genes. Oncogene 2012, 31, 4207–4220.
- Puga, A.; Barnes, S.J.; Dalton, T.P.; Chang, C.-Y.; Knudsen, E.S.; Maier, M.A. Aromatic Hydrocarbon Receptor Interaction with the Retinoblastoma Protein Potentiates Repression of E2F-dependent Transcription and Cell Cycle Arrest. J. Biol. Chem. 2000, 275, 2943–2950.
- Elferink, C.J.; Ge, N.-L.; Levine, A. Maximal Aryl Hydrocarbon Receptor Activity Depends on an Interaction with the Retinoblastoma Protein. Mol. Pharmacol. 2001, 59, 664–673.
- Perearnau, A.; Orlando, S.; Islam, A.B.; Gallastegui, E.; Martínez, J.; Jordan, A.; Bigas, A.; Aligué, R.; Pujol, M.J.; Bachs, O. p27Kip1, PCAF and PAX5 cooperate in the transcriptional regulation of specific target genes. Nucleic Acids Res. 2017, 45, 5086–5099.
- Pérez-Luna, M.; Aguasca, M.; Perearnau, A.; Serratosa, J.; Martínez-Balbas, M.; Pujol, M.J.; Bachs, O. PCAF regulates the stability of the transcriptional regulator and cyclin-dependent kinase inhibitor p27Kip1. Nucleic Acids Res. 2012, 40, 6520–6533.




