
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Md. Motaher Hossain | -- | 1661 | 2023-04-12 16:42:16 | | | |
| 2 | Catherine Yang | Meta information modification | 1661 | 2023-04-13 02:44:51 | | | | |
| 3 | Catherine Yang | Meta information modification | 1661 | 2023-04-13 02:47:24 | | |
Video Upload Options
Sclerotinia sclerotiorum (Lib.) de Bary is a broad host-range fungus that infects an inclusive array of plant species and afflicts significant yield losses globally. S. sclerotiorum possesses an immense arsenal of disease weaponry to subsist and succeed under widespread environmental conditions. The massive pathogenic arsenal of S. sclerotiorum, including oxalic acid (OA), CWDEs, and small secretory proteins (effectors), has long been associated with virulence. A subtle interplay between these virulence factors serves various regulatory functions in host cells, allowing S. sclerotiorum to colonize the host, evade or inhibit the host defense system, and cause disease. Understanding and studying these mechanisms is crucial for detecting pathways of genetic interventions that could result in improved control of this disease.
1. Oxalic Acid, a Versatile Toxin and Broad-Spectrum Pathogenicity Factor
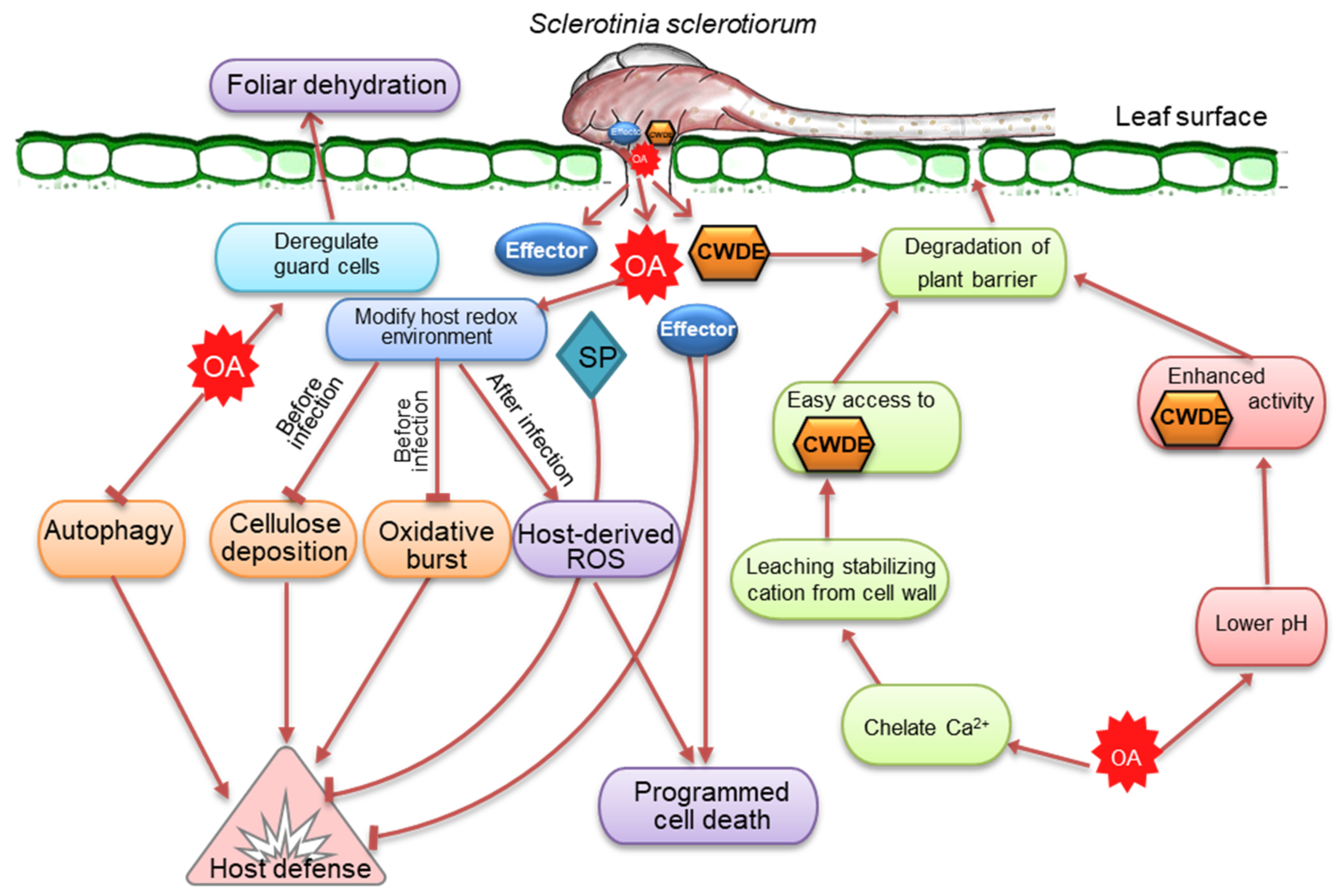
2. Arsenal of Cell Wall-Degrading Enzymes Reflects Broad Host Preference
3. Secretory Effectors, Critical Components in the Necrotrophic Infection Strategy
References
- Xu, T.; Li, J.; Yu, B.; Liu, L.; Zhang, X.; Liu, J.; Pan, H.; Zhang, Y. Transcription Factor SsSte12 was involved in mycelium growth and development in Sclerotinia sclerotiorum. Front. Microbiol. 2018, 9, 2476.
- Derbyshire, M.C.; Newman, T.E.; Khentry, Y.; Owolabi, T.A. The evolutionary and molecular features of the broad-host-range plant pathogen Sclerotinia sclerotiorum. Mol. Plant Pathol. 2022, 23, 1075–1090.
- Cessna, S.G.; Sears, V.E.; Low, P.S.; Dickman, M. Oxalic acid, a pathogenicity factor for Sclerotinia sclerotiorum, suppresses the oxidative burst of the host plant. Plant Cell. 2000, 12, 2191–2200.
- Liang, X.; Liberti, D.; Li, M.; Kim, Y.T.; Hutchens, A.; Wilson, R.; Rollins, J.A. Oxaloacetate acetylhydrolase gene mutants of Sclerotinia sclerotiorum do not accumulate oxalic acid but do produce limited lesions on host plants. Mol. Plant Pathol. 2015, 16, 559–571.
- Rollins, J.A.; Dickman, M.B. pH signaling in Sclerotinia sclerotiorum: Identification of a pacC/RIMI homolog. Appl. Environ. Microbiol. 2001, 67, 75–81.
- Williams, B.; Kabbage, M.; Kim, H.J.; Britt, R.; Dickman, M.B. Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog. 2011, 7, e1002107.
- Bateman, D.F.; Beer, S.V. Simultaneous production and synergistic action of oxalic acid and polygalacturonase during pathogenesis by Sclerotium rolfsii. Phytopathology 1965, 55, 204–211.
- Favaron, F.; Sella, L.; D’Ovidio, R. Relationships among endo-polygalacturonase, oxalate, pH, and plant polygalacturonase-inhibiting protein (PGIP) in the interaction between Sclerotinia sclerotiorum and soybean. Mol. Plant Microbe Interact. 2004, 17, 1402–1409.
- Guimaraes, R.L.; Stotz, H.U. Oxalate production by Sclerotinia sclerotiorum deregulates guard cells during infection. Plant Physiol. 2004, 136, 3703–3711.
- Kim, K.S.; Min, J.Y.; Dickman, M.B. Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Mol. Plant Microb. Interact. 2008, 21, 605–612.
- Heller, A.; Witt-Geiges, T. Oxalic acid has an additional, detoxifying function in Sclerotinia sclerotiorum pathogenesis. PLoS ONE 2013, 8, e72292.
- Fagundes-Nacarath, I.R.F.; Debona, D.; Oliveira, A.T.H.; Hawerroth, C.; Rodrigues, F.A. Biochemical responses of common bean to white mold potentiated by phosphites. Plant Physiol. Biochem. 2018, 132, 308–319.
- Dutton, M.V.; Evans, C.S. Oxalate production by fungi: Its role in pathogenicity and ecology in the soil environment. Can. J. Microbiol. 1996, 42, 881–895.
- Liang, X.F.; Rollins, J.A. Mechanisms of broad host range necrotrophic pathogenesis in Sclerotinia sclerotiorum. Phytopathology 2018, 108, 1128–1140.
- Liberti, D.; Rollins, J.A.; Dobinson, K.F. Peroxisomal carnitine acetyl transferase influences host colonization capacity in Sclerotinia sclerotiorum. Mol. Plant Microb. Interact. 2013, 26, 768–780.
- Rollins, J.; Cuomo, C.; Dickman, M.; Kohn, L. Genomics of Sclerotinia sclerotiorum. In Genomics of Plant-Associated Fungi and Oomycetes: Dicot Pathogens; Dean, R.A., Lichens-Park, A., Kole, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 1–17. ISBN 978-3-662-44056-8.
- Magro, P.; Marciano, P.; Lenna, P. Enzymatic oxalate decarboxylation in isolates of Sclerotinia sclerotiorum. FEMS Microbiol. Lett. 1988, 49, 49–52.
- Mäkelä, M.R.; Hildén, K.; Lundell, T.K. Oxalate decarboxylase: Biotechnological update and prevalence of the enzyme in filamentous fungi. Appl. Microbiol. Biotechnol. 2010, 87, 801–814.
- Held, M.A.; Jiang, N.; Basu, D.; Showalter, A.M.; Faik, A. Plant Cell Wall Polysaccharides: Structure and Biosynthesis. In Polysaccharides; Ramawat, K., Mérillon, J.M., Eds.; Springer: Cham, Switzerland, 2015; ISBN 978-3-319-03751-6.
- Blanco-Ulate, B.; Morales-Cruz, A.; Amrine, K.C.H.; Labavitch, J.M.; Powell, A.L.T.; Cantu, D. Genome-wide transcriptional profiling of Botrytis cinerea genes targeting plant cell walls during infections of different hosts. Front. Plant Sci. 2014, 5, 435.
- Amselem, J.; Cuomo, J.C.A.; van Kan, J.A.L.; Viaud, M.; Benito, E.P.; Couloux, A.; Coutinho, P.M.; de Vries, R.P.; Dyer, P.S.; Fillinger, S.; et al. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 2011, 7, e1002230.
- Kars, I.; Van Kan, J.A. Extracellular enzymes and metabolites involved in pathogenesis of Botrytis. In Botrytis: Biology, Pathology and Control; Elad, Y., Williamson, B., Tudzynski, P., Delen, N., Eds.; Springer: Dordrecht, The Netherlands, 2004; pp. 99–118. ISBN 978-1-4020-6586-6.
- Wagner, F.; Kusserow, H.; Schafer, W. Cloning and targeted disruption of two polygalacturonase genes in Penicillium olsonii. FEMS Microbiol. Lett. 2000, 186, 293–299.
- Garcia-Maceira, F.I.; Di Pietro, A.; Huertas-Gonzalez, M.D.; Ruiz-Roldan, M.C.; Roncero, M.I.G. Molecular characterization of an endopolygalacturonase from Fusarium oxysporum expressed during early stages of infection. Appl. Environ. Microbiol. 2001, 67, 2191–2196.
- Kars, I.; Krooshof, G.H.; Wagemakers, L.; Joosten, R.; Benen, J.A.; Van Kan, J.A. Necrotizing activity of five Botrytis cinerea endopolygalacturonases produced in Pichia pastoris. Plant J. 2005, 43, 213–225.
- Collins, T.; Gerday, C.; Feller, G. Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol. Rev. 2005, 29, 3–23.
- Yu, Y.; Xiao, J.; Du, J.; Yang, Y.; Bi, C.; Qing, L. Disruption of the gene encoding endo-β-1, 4-xylanase affects the growth and virulence of Sclerotinia sclerotiorum. Front. Microbiol. 2016, 7, 1787.
- Seifbarghi, S.; Borhan, M.H.; Wei, Y.; Coutu, C.; Robinson, S.J.; Hegedus, D.D. Changes in the Sclerotinia sclerotiorum transcriptome during Infection of Brassica napus. BMC Genom. 2017, 18, 266.
- Heard, S.; Brown, N.A.; Hammond-Kosack, K. An interspecies comparative analysis of the predicted secretomes of the necrotrophic plant pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS ONE 2015, 10, e0130534.
- Zuppini, A.; Navazio, L.; Sella, L.; Castiglioni, C.; Favaron, F.; Mariani, P. An endopolygalacturonase from Sclerotinia sclerotiorum induces calcium-mediated signaling and programmed cell death in soybean cells. Mol. Plant Microb. Interact. 2005, 18, 849–855.
- Wang, J.X.; Ma, H.X.; Chen, Y. Sensitivity of Sclerotinia sclerotiorum to boscalid in Jiangsu Province of China. Crop Prot. 2009, 28, 882–886.
- Dallal Bashi, Z.; Hegedus, D.D.; Buchwaldt, L.; Rimmer, S.R.; Borhan, M.H. Expression and regulation of Sclerotinia sclerotiorum necrosis and ethylene-inducing peptides (NEPs). Mol. Plant Pathol. 2010, 11, 43–53.
- Zhu, W.; Wei, W.; Fu, Y.; Cheng, J.; Xie, J.; Li, G.; Yi, X.; Kang, Z.; Dickman, M.B.; Jiang, D. A secretory protein of necrotrophic fungus Sclerotinia sclerotiorum that suppresses host resistance. PLoS ONE 2013, 8, e53901.
- Liang, X.; Rollins, J.A. An oxalate decarboxylase gene functions in the early infection processes of Sclerotinia sclerotiorum. Phytopathology 2013, 103, 81.
- Zhang, H.; Wu, Q.; Cao, S.; Zhao, T.; Chen, L.; Zhuang, P.; Zhou, X.; Gao, Z. A novel protein elicitor (SsCut) from Sclerotinia sclerotiorum induces multiple defense responses in plants. Plant Mol. Biol. 2014, 86, 495–511.
- Lyu, X.; Shen, C.; Fu, Y.; Xie, J.; Jiang, D.; Li, G.; Cheng, J. A small secreted virulence-related protein is essential for the necrotrophic interactions of Sclerotinia sclerotiorum with its host plants. PLoS Pathog. 2016, 12, e1005435.
- Lyu, X.; Shen, C.; Fu, Y.; Xie, J.; Jiang, D.; Li, G.; Cheng, J. The microbial opsin homolog Sop1 is involved in Sclerotinia sclerotiorum development and environmental stress response. Front. Microbiol. 2016, 6, 1504.
- Kabbage, M.; Williams, B.; Dickman, M.B. Cell death control: The interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum. PLoS Pathog. 2013, 9, e1003287.
- Xiao, X.; Xie, J.; Cheng, J.; Li, G.; Yi, X.; Jiang, D.; Fu, Y. Novel secretory protein Ss-Caf1 of the plant-pathogenic fungus Sclerotinia sclerotiorum is required for host penetration and normal sclerotial development. Mol. Plant Microb. Interact. 2014, 27, 40–55.
- Yu, Y.; Xiao, J.; Yang, Y.; Bi, C.; Qing, L.; Tan, W. Ss-Bi1 encodes a putative BAX inhibitor-1 protein that is required for full virulence of Sclerotinia sclerotiorum. Physiol. Mol. Plant Pathol. 2015, 90, 115–122.
- Xu, L.; Chen, W. Random T-DNA mutagenesis identifies a Cu/Zn superoxide dismutase gene as a virulence factor of Sclerotinia sclerotiorum. Mol. Plant Microb. Interact. 2013, 26, 431–441.
- Yarden, O.; Veluchamy, S.; Dickman, M.B.; Kabbage, M. Sclerotinia sclerotiorum catalase SCAT1 affects oxidative stress tolerance, regulates ergosterol levels and controls pathogenic development. Physiol. Mol. Plant Pathol. 2014, 85, 34–41.
- Pan, Y.; Xu, Y.; Li, X.; Yao, C.; Gao, Z. SsPemG1 encodes an elicitor-homologous protein and regulates pathogenicity in Sclerotinia sclerotiorum. Physiol. Mol. Plant Pathol. 2015, 92, 70–78.
- Yang, G.; Tang, L.; Gong, Y.; Xie, J.; Fu, Y.; Jiang, D.; Li, G.; Collinge, D.B.; Chen, W.; Cheng, J. A cerato-platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum. New Phytol. 2018, 217, 739–755.
- Seifbarghi, S.; Borhan, M.H.; Wei, Y.; Ma, L.; Coutu, C.; Bekkaoui, D.; Hegedus, D.D. Receptor-Like Kinases BAK1 and SOBIR1 are required for necrotizing activity of a novel group of Sclerotinia sclerotiorum necrosis-inducing effectors. Front. Plant Sci. 2020, 11, 1021.
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323.
- Wang, Y.; Wang, Y. Trick or treat: Microbial pathogens evolved apoplastic effectors modulating plant susceptibility to infection. Mol. Plant Microb. Interact. 2018, 31, 6–12.
- Guyon, K.; Balague, C.; Roby, D.; Raffaele, S. Secretome analysis reveals effector candidates associated with broad host range necrotrophy in the fungal plant pathogen Sclerotinia sclerotiorum. BMC Genom. 2014, 15, 336.
- Derbyshire, M.; Denton-Giles, M.; Hegedus, D.; Seifbarghy, S.; Rollins, J.; Van Kan, J.; Seidl, M.F.; Faino, L.; Mbengue, M.; Navaud, O.; et al. The complete genome sequence of the Phytopathogenic fungus Sclerotinia sclerotiorum reveals insights into the genome architecture of broad host range pathogens. Genome Biol. Evol. 2017, 9, 593–618.




