Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jakub Novosad | -- | 2148 | 2023-04-06 07:27:47 | | | |
| 2 | Conner Chen | Meta information modification | 2148 | 2023-04-10 08:12:11 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Novosad, J.; Krčmová, I.; Souček, O.; Drahošová, M.; Sedlák, V.; Kulířová, M.; Králíčková, P. Development, Phenotypes, and Functional Characteristics of Eosinophil Lineage. Encyclopedia. Available online: https://encyclopedia.pub/entry/42836 (accessed on 06 March 2026).
Novosad J, Krčmová I, Souček O, Drahošová M, Sedlák V, Kulířová M, et al. Development, Phenotypes, and Functional Characteristics of Eosinophil Lineage. Encyclopedia. Available at: https://encyclopedia.pub/entry/42836. Accessed March 06, 2026.
Novosad, Jakub, Irena Krčmová, Ondřej Souček, Marcela Drahošová, Vratislav Sedlák, Martina Kulířová, Pavlína Králíčková. "Development, Phenotypes, and Functional Characteristics of Eosinophil Lineage" Encyclopedia, https://encyclopedia.pub/entry/42836 (accessed March 06, 2026).
Novosad, J., Krčmová, I., Souček, O., Drahošová, M., Sedlák, V., Kulířová, M., & Králíčková, P. (2023, April 06). Development, Phenotypes, and Functional Characteristics of Eosinophil Lineage. In Encyclopedia. https://encyclopedia.pub/entry/42836
Novosad, Jakub, et al. "Development, Phenotypes, and Functional Characteristics of Eosinophil Lineage." Encyclopedia. Web. 06 April, 2023.
Copy Citation
The existence of eosinophils was documented histopathologically in the first half of the 19th century. However, the term “eosinophils” was first used by Paul Ehrlich in 1878. Since their discovery and description, their existence has been associated with asthma, allergies, and antihelminthic immunity. Eosinophils may also be responsible for various possible tissue pathologies in many eosinophil-associated diseases.
eosinophils
immunophenotype
asthma
1. Introduction
The existence of eosinophils was documented histopathologically in the first half of the 19th century in Gottlieb Gluge’s textbook of pathology from 1843. However, the term “eosinophils” was first used by Paul Ehrlich at the end of the 19th century (1878). At that time, their existence was associated with the pathogenesis of asthma. During the 20th century, the notion of the role of eosinophils in the human body was subject to considerable turbulence. While eosinophils were perceived as regulatory elements that dampen the proinflammatory activity of mediators released by mast cells during the 1970s and 1980s, in the later 1980s and 1990s, eosinophils began to be considered as terminal effector cells of immunopathological inflammation, causing tissue damage. The presence of eosinophils has traditionally been associated with defence against helminthic infections and with the pathogenesis of allergies. Since the beginning of the 21st century, theories about their physiological roles in the human body have been further revised. Furthermore, in 2010, J. J. Lee, the former president of the International Society for Eosinophils, proposed the concept of “LIAR” (Local Immunity And/or Remodelling/Repair) [1] in both health and disease. This impressive work has given rise to some controversies regarding the fundamental role of eosinophils in immune functions during the phylogeny, as follows: (1) Why have the host organisms evolved with a unique hematopoietic lineage as a defence mechanism against selected pathogens that are generally not life-threatening? (2) Why are the eosinophilic leukocytes absent from nearly all metazoans and present only among the five classes of vertebrates in the phylum Chordata (600 million years)? Furthermore, (3) if eosinophils were a prominent innate host defence against helminths across mammal species, why have pathogen-driven selective pressures not led to alternative or overlapping poietic pathways that could promote the expansion of these cells that would be independent of the eosinophils’ principal growth factor—interleukin-5 (IL-5)? [1].
Aside from the presumed antimicrobial functions, it has been suggested that eosinophils play roles in tissue morphogenesis, mucosal homeostasis, and metabolism, although this evidence has mainly come from preclinical models [2]. According to this theory, the regulatory and immunomodulating potential of eosinophils has been confirmed in the gastrointestinal tract (additionally in the small intestine, which represents the physiological reservoir for eosinophils in the body) [3], in adipose tissue [4], and in the lungs [5]. A common characteristic of tissues containing a high count of eosinophils is a high cellular turnover [1]. It has become apparent that mature eosinophils are not an immunologically homogeneous cell population but form more or less structurally and functionally different subtypes, which have separate development, localization, role and fate characteristics in tissues, especially during states of inflammation activation [5][6][7]. Therefore, a new classification scheme for eosinophil subsets has been proposed, which consists of eosinophil progenitors, steady-state eosinophils, and regulatory/resident eosinophils (rEos) and pro-inflammatory eosinophils (iEos) [5][8].
It has become evident that understanding such developmental phenotypes is inevitable. It can teach us about the origins and activities of functionally distinct eosinophilic cells during inflammation and explain the reasons for the successes and failures of biologics targeting this cell line.
2. Development, Phenotypes, and Functional Characteristics of the Eosinophil Lineage: Generation of Eosinophil Endotypes
In the current simplified paradigm, hematopoietic stem cells (HSC) from the bone marrow directly give rise to eosinophil/mast cell progenitors (EoMCP), from which eosinophil progenitors (EoP) develop and terminally differentiate into mature eosinophils. EoMCPs also differentiate into basophil progenitors (BaP) and mast cells. Aside from EoMCPs, megakaryocyte/erythroid progenitors (MEP), common lymphoid progenitors (CLP), and common myeloid progenitors (CMP) are generated from the HSCs. In the forthcoming steps, the CMP is a source of the subsequent progenitors, neutrophil and monocyte progenitors (NMP) and megakaryocyte/erythrocyte progenitors (MEP), which give rise to particular haematological elements [9]. These progenitors and mature cells reveal a distinct immunophenotype, which enables their detection using flow cytometry [10] (see Figure 1 and Table 1).
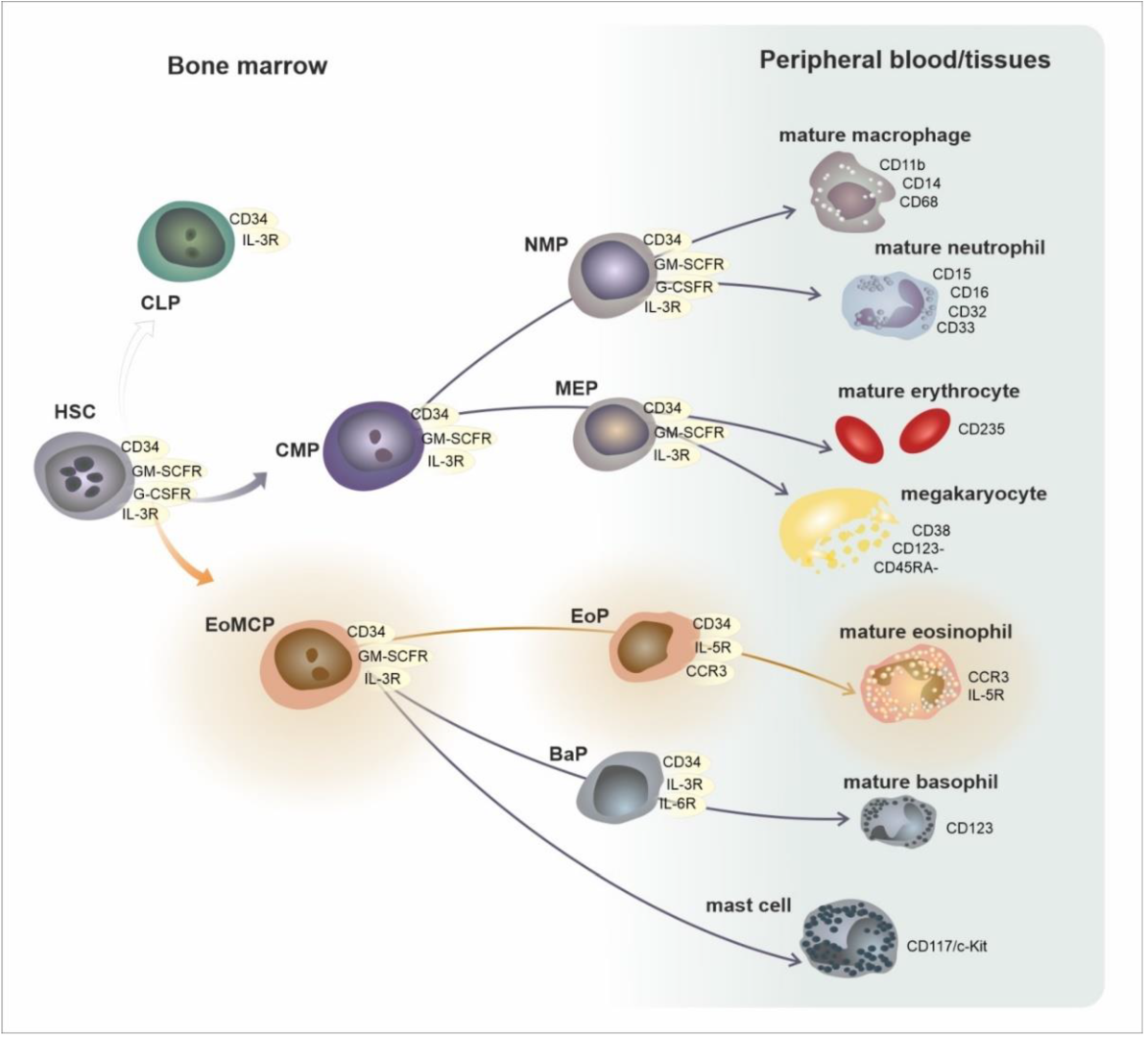
Figure 1. Development of eosinophils in the framework of general hematopoiesis. Abbreviations: HSC—hematopoietic stem cell, CLP—common lymphoid progenitor, CMP—common myeloid progenitor, EoMCP—eosinophil and mast cell progenitor, NMP—neutrophil and macrophage progenitor, MEP—megakaryocyte and erythrocyte progenitor.
Table 1. Development of eosinophils.
| Area/Subset | Developmental Cell Type | T1/2 | Phenotype |
|---|---|---|---|
| bone marrow | CMP—common myeloid progenitor EoMCP—eosinophil/mast cell progenitor |
circa 24 h | CD34, GM-SCFR, IL-3R |
| EoP—eosinophil progenitor | CD34, GM-SCFR, IL-3R, CCR3, IL-5Rα | ||
| mature eosinophil | GM-SCFR, IL-3R, CCR3, IL-5Rα, VLA-4 (CD49d/CD29), PSGL-1 (CD162) | ||
| peripheral blood | mature eosinophil | 8–24 h | GM-SCFR, IL-3R, CCR3, IL-5Rα, VLA-4 (CD49d/CD29), PSGL-1 (CD162) |
| peripheral tissues | mature eosinophil | 8 days | GM-SCFR, IL-3R, CCR3, IL-5Rα, FcεRI, VLA-4 (CD49d/CD29), PSGL-1 (CD162) |
| activated eosinophil | ? | GM-SCFR, IL-3R, CCR3, IL-5Rα, FcεRI, CD63, CD9, CD69 | |
| Mature eosinophil subsets in mice and humans | |||
| mice | hEos—homeostatic eosinophil rEos—resident eosinophil (IL-5 independent) |
lungs: 36 h, GIT: 6 days | Siglec-FmedCD62L+ CD101lo, CCR3, IL-5R, F4/80, CD11c− |
| iEos—inflammatory eosinophil (IL-5 dependent) | ? | Siglec-FhighCD62L−CD101hi, CCR3, IL-5R, CD11clow | |
| humans | hEos—homeostatic eosinophil rEos—resident eosinophil (IL-5 independent) |
? | Siglec-8+CD62L+IL-3Rlow |
| iEos—inflammatory eosinophil (IL-5 dependent) | ? | Siglec-8+CD62LlowIL-3Rhigh | |
Three central cytokines influence the maturation of eosinophils in the bone marrow—colony-stimulating factor for granulocytes and macrophages (GM-CSF), interleukin 3 (IL-3), and especially, IL-5 [11]. IL-5 also initiates the release of mature cells from the bone marrow and prolongs their survival in peripheral tissues [12]. Despite IL-5 being a key cytokine that regulates the generation and maturation of eosinophils, it is not entirely indispensable for eosinophil development, as IL-5-deficient mice and human patients receiving anti-IL-5 therapy all retain residual eosinophils [13]. Moreover, there is some level of redundancy in the IL-5 activity, since it can be produced by numerous cells (incl. Th2 lymphocytes, innate lymphoid cells (ILC)-2, NKT cells, and to a lesser extent, mast cells, epithelial cells, Reed–Sternberg cells, and EBV-transformed cells of eosinophils). It can also interact nonexclusively with eosinophils but also with basophils and possibly others [14][15].
Significant phenotypic changes occur during maturation, mainly caused by the gradual attenuation of the activity of the transcription factor FOG-1 (Friend of GATA-1) in the EoMP stage in favor of the actions of other transcription factors, GATA-1 and GATA-2 (zinc finger DNA binding proteins, that bind to the consensus DNA sequence (T/A)GATA(A/G) [16]) in EoP. Further, there is a transient increase in the activity of the transcription factors C/EBPε (CCAAT/enhancer binding protein) and finally ID-2 (DNA-binding protein inhibitor) in the mature eosinophil stage [17]. This switching leads to a gradual decrease in the expression of the surface molecule CD34 (adhesion phosphoglycoprotein specific for hematogenous stem cells) and, conversely, increases in the receptors for growth factors regulating their production and maturation, mainly the receptors for IL-3 (IL-3R) and GM-CSF (GM-CSFR) as well as the receptor for eotaxin-1 (CCR3). However, as the very first and functionally superior receptor, IL-5R is expressed on the surface of maturing eosinophils (a receptor for IL-5 consisting of the alpha subunit (IL-5Rα) and a common β chain—also described for the receptors for IL-3 and GM-CSF), the production of which is probably (at least in the EoP stage) positively regulated [18]. It has also been shown that the number of progenitors in the bone marrow (expressing CD34, CCR3, and IL-5Rα) increases 24 h after the exposure to an allergen in atopic individuals [19].
During its maturation in the bone marrow, in addition to the development of the surface receptor structure, under the influence of the transcription factors mentioned earlier (especially C/EBPε), specific (formerly secondary or α-granules) and primary (formerly β-granules, which are considered to be the immature form of specific granules) granules are gradually formed. There is also the formation of lipid bodies and so-called “sombrero vesicles” (organelles named based on their ultramicroscopy morphology). Each of these structures contains different biologically active substances, and their intentional release is a crucial element of eosinophil effector functions, both activating and inhibitory [20].
The complete surface receptor structure on the cell surface of mature cells enables the interaction of eosinophils with signals from the external environment, both activating (IL-3, GM-CSF and IL-5—through receptors IL-3R, GM-CSFR, IL-5R) and inhibitory (TGFβ—transforming growth factor β, via the TGFβR receptor or the Sialyl–Lewis X molecule (CD15s) via the Siglec-8 receptor—receptor family—Sialic acid ImmunoGlobulin-like LECtins), as well as with chemokines that attract eosinophils to the site of inflammation (eotaxin-1, 2, and 3, RANTES—Regulated upon Activation, Normal T Cell Expressed, and Presumably Secreted or PgD2 (prostaglandin D2) via CCR3, CCR1, and DP2/CRTh2 receptors, respectively). Their mutual balance represents an essential tool for controlling the production and migration of this cell population [21]. The surface immunophenotype of mature eosinophils is shown graphically in Figure 2 and Table 2.
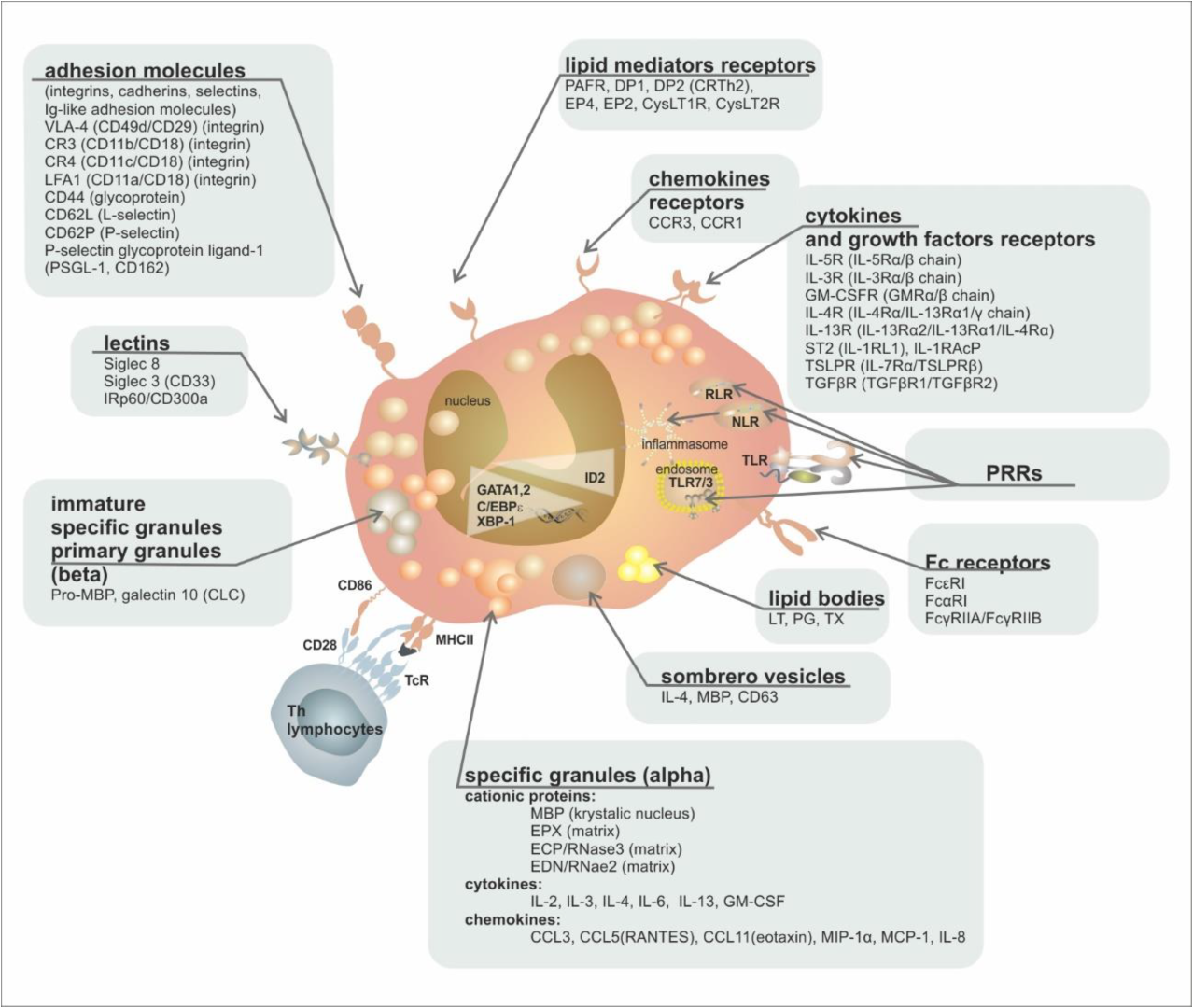
Figure 2. Eosinophil ultrastructure. Abbreviations: MBP—major basic protein, EPX—eosinophil peroxidase, ECP—eosinophil cationic protein, EDN—eosinophil-derived neurotoxin, PRRs—patterns recognising receptors, TLR—Toll-like receptor, RLR—RIG (retinoic acid-inducible gene-I)-like receptor, NLR—NOD (nucleotide-binding oligomerization domain)-like receptor, LT—leukotriene, PG—prostaglandin, TX—thromboxane, DP—D-Prostaglandin receptor, EP—E prostaglandin receptor, pro-MBP—preform of major basic protein, CLC—Charcot–Leyden crystals.
Table 2. Eosinophil receptors.
| Group of Receptors | Receptor | Ligand | Function |
|---|---|---|---|
| cytokine and growth factor receptors | IL-5R (IL-5Rα/β chain)/(CD125/CD131) | IL-5 | proliferation, growth, bone marrow escape, survival |
| IL-3R (IL-3Rα/β chain)/(CD123/CD131) | IL-3 | growth, survival | |
| GM-CSFR (GMRα/β chain)/(CD116/CD131) | GM-CSF | growth, survival | |
| IL-4R (IL-4Rα/IL-13Rα1/γ chain) | IL-4 | activation | |
| IL-13R (IL-13Rα2/IL-13Rα1/IL-4Rα) | IL-13 | activation | |
| ST2 (IL-1RL1) | IL-33 | growth, survival | |
| TSLPR (IL-7Rα/TSLPRβ) | TSLP | growth, survival | |
| TGFβR (TGFβR1/TGFβR2) | TGFβ | inhibition of survival | |
| chemokine receptors | CCR3 | Eotaxin 1, 2, 3 (CCL11/CCL24/CCL26), MCP-3, MCP-4 | chemotaxis |
| CCR1 | MIP-1α (CCL3), RANTES (CCL5) | chemotaxis | |
| lipid mediator receptors | PAFR | PAF | activation |
| DP2 (CRTh2) | PgD2 | chemotaxis | |
| DP1 | PgD1 | chemotaxis | |
| EP4 | PgE2 | activation/inactivation | |
| EP2 | PgE2 | activation/inactivation | |
| CysLT1R | LTD4, LTC4, LTE4 | activation | |
| CysLT2R | LTD4, LTC4, LTE4 | activation | |
| Pattern Recognition Receptors (PRRs) | TLR1, 2, 3, 4, 5, 6, 7, 9, 10 (Toll-Like Receptors) | PAMPs | activation, degranulation |
| NLR1, 2 (NOD-Like Receptors) | PAMPs, DAMPs | activation | |
| RLR (RIG-Like Receptors) | dsRNA | activation | |
| PAR-2 | protease-activated receptor | activation | |
| RAGE (Receptor for Advanced Glycation Products) | Advanced Glycation Products | activation | |
| Fc receptors | FcεRI | IgE | nonactivation |
| FcαRI | IgA | activation | |
| FcγRIIA/FcγRIIB | IgG | activation/inactivation | |
| MHC | MHCII (+CD80, +CD86, +CD40) | TCR + CD4 | antigen presentation |
| adhesion molecules (integrins, cadherins, selectins, Ig-like adhesion molecules) | VLA-4 (CD49d/CD29) (integrin) | VCAM, fibronectin | activation, adhesion |
| CR3 (CD11b/CD18) (integrin) | iC3b | activation, adhesion | |
| CR4 (CD11c/CD18) (integrin) | iC3b | activation, adhesion | |
| LFA1 (CD11a/CD18) (integrin) | ICAM-1, ICAM-2 | activation, adhesion | |
| CD44 (glycoprotein) | hyaluronic acid | adhesion, homing | |
| CD62L (L-selectin) | CD34, GlyCAM-1, MadCAM-1 | adhesion, homing | |
| CD62P (P-selectin) | P-selectin glycoprotein ligand-1 (PSGL-1) | activation, adhesion | |
| PSGL-1 (P-selectin glycoprotein ligand, CD162) | P-selectin (CD62P) | activation, adhesion | |
| CD34 (fosfoglykoprotein) | L-selectin | adhesion, migration | |
| lectins | Siglec-8 | Sialyl–Lewis X (CD15s) | apoptosis induction |
| Siglec-3 (CD33) | sialyl acid | apoptosis induction | |
| IRp60/CD300a | sialyl acid | inhibition of growth signals (IL-3/IL-5/GMCSF) |
However, immature progenitors of eosinophils can also leave the bone marrow and infiltrate the distant tissues (especially during the inflammatory reaction), allowing “in-situ eosinophilopoiesis”, which was also described in the mucous membrane of the respiratory tract [22] and may be associated with intensive local immune reactivity [23]. Moreover, mature eosinophils are heterogeneous cells with potentially multiple subsets. Only one subset of eosinophils is commonly detected in the blood and lungs of mice at a steady state. However, following the development of, e.g., an airway allergy to house dust mite antigens, at least two subsets of eosinophils become detectable in these animals. Similar subsets have also been detected in murine models of eosinophilic esophagitis [5]. These observations raise a question about the mechanisms leading to the endotyping of inflammatory eosinophils during eosinophilopoiesis or, instead, illustrate a picture of plasticity in the inflamed tissue. Since there are probably differences between the eosinophilopoiesis in health and in a disease state, it is likely that inflammatory signals originating from the site of inflammation, including IL-5, may modulate eosinophil development to impart functions on (subsets of) eosinophils that differ from those of their steady-state counterparts [2].
With better marker coverage by multicolor flow cytometry and single-cell level sequencing of granulocyte populations, novel phenotypes of these cells began to emerge. It is worth noting that many of these newly described subsets blend distinctions between classical myeloid lineage phenotypes. Aside from terminally differentiated eosinophils and other granulocytes (such as neutrophils) defined by conventional flow cytometry and granular protein markers, intermediate phenotypes with mixed neutrophil–eosinophil characteristics are coming to light. Two different processes may be responsible for the plasticity of granulocyte lineages: hematopoietic flexibility of granulocyte precursors and the adaptation of different subsets to local tissue and cytokine microenvironments [24].
References
- Lee, J.J.; Jacobsen, E.A.; McGarry, M.P.; Schleimer, R.P.; Lee, N.A. Eosinophils in health and disease: The LIAR hypothesis. Clin. Exp. Allergy 2010, 40, 563–575.
- Van Hulst, G.; Bureau, F.; Desmet, C.J. Eosinophils as drivers of severe eosinophilic asthma: Endotypes or plasticity? Int. J. Mol. Sci. 2021, 22, 10150.
- Loktionov, A. Eosinophils in the gastrointestinal tract and their role in the pathogenesis of major colorectal disorders. World J. Gastroenterol. 2019, 25, 3503–3526.
- Qiu, Y.; Nguyen, K.D.; Odegaard, J.I.; Cui, X.; Tian, X.; Locksley, R.M.; Palmiter, R.D.; Chawla, A. Eosinophils and type 2 cytokine signaling in macrophages orchestrate development of functional beige fat. Cell 2014, 157, 1292–1308.
- Mesnil, C.; Raulier, S.; Paulissen, G.; Xiao, X.; Birrell, M.A.; Pirottin, D.; Janss, T.; Starkl, P.; Ramery, E.; Henket, M.; et al. Lung-resident eosinophils represent a distinct regulatory eosinophil subset. J. Clin. Investig. 2016, 126, 3279–3295.
- Rothenberg, M.E. A hidden residential cell in the lung. J. Clin. Investig. 2016, 126, 3185–3187.
- Marichal, T.; Mesnil, C.; Bureau, F. Homeostatic Eosinophils: Characteristics and Functions. Front. Med. 2017, 4, 101.
- Abdala-Valencia, H.; Coden, M.E.; Chiarella, S.E.; Jacobsen, E.A.; Bochner, B.S.; Lee, J.J.; Berdnikovs, S. Shaping eosinophil identity in the tissue contexts of development, homeostasis, and disease. J. Leukoc. Biol. 2018, 104, 95–108.
- Klion, A.D.; Ackerman, S.J.; Bochner, B.S. Contributions of Eosinophils to Human Health and Disease. Annu. Rev. Pathol. Mech. Dis. 2020, 15, 179–209.
- Rühle, P.F.; Fietkau, R.; Gaipl, U.S.; Frey, B. Development of a modular assay for detailed immunophenotyping of peripheral human whole blood samples by multicolor flow cytometry. Int. J. Mol. Sci. 2016, 17, 1316.
- Rosenberg, H.F.; Phipps, S.; Foster, P.S. Eosinophil trafficking in allergy and asthma. J. Allergy Clin. Immunol. 2007, 119, 1303–1310.
- Kouro, T.; Takatsu, K. IL-5- and eosinophil-mediated inflammation: From discovery to therapy. Int. Immunol. 2009, 21, 1303–1309.
- Haldar, P.; Brightling, C.E.; Hargadon, B.; Gupta, S.; Monteiro, W.; Sousa, A.; Marshall, R.P.; Bradding, P.; Green, R.H.; Wardlaw, A.J.; et al. Mepolizumab and Exacerbations of Refractory Eosinophilic Asthma. N. Engl. J. Med. 2009, 360, 973–984.
- Roufosse, F. Targeting the interleukin-5 pathway for treatment of eosinophilic conditions other than asthma. Front. Med. 2018, 5, 49.
- Hassani, M.; Koenderman, L. Immunological and hematological effects of IL-5(Rα)-targeted therapy: An overview. Allergy Eur. J. Allergy Clin. Immunol. 2018, 73, 1979–1988.
- Gao, J.; Chen, Y.H.; Peterson, L.A.C. GATA family transcriptional factors: Emerging suspects in hematologic disorders. Exp. Hematol. Oncol. 2015, 4, 1–7.
- Bochner, B.S. The eosinophil: For better or worse, in sickness and in health. Ann. Allergy Asthma Immunol. 2018, 121, 150–155.
- Tavernier, J.; Van der Heyden, J.; Verhee, A.; Brusselle, G.; Van Ostade, X.; Vandekerckhove, J.; North, J.; Rankin, S.M.; Kay, A.B.; Robinson, D.S. Interleukin 5 regulates the isoform expression of its own receptor alpha-subunit. Blood 2000, 95, 1600–1607.
- Sehmi, R.; Wardlaw, A.J.; Cromwell, O.; Kurihara, K.; Waltmann, P.; Kay, A.B. Interleukin-5 selectively enhances the chemotactic response of eosinophils obtained from normal but not eosinophilic subjects. Blood 1992, 79, 2952–2959.
- Melo, R.C.N.; Weller, P.F. Contemporary understanding of the secretory granules in human eosinophils. J. Leukoc. Biol. 2018, 104, 85–93.
- Munitz, A.; Levi-Schaffer, F. Inhibitory receptors on eosinophils: A direct hit to a possible Achilles heel? J. Allergy Clin. Immunol. 2007, 119, 1382–1387.
- Rådinger, M.; Lötvall, J. Eosinophil progenitors in allergy and asthma—Do they matter? Pharmacol. Ther. 2009, 121, 174–184.
- Smith, S.G.; Chen, R.; Kjarsgaard, M.; Huang, C.; Oliveria, J.P.; O’Byrne, P.M.; Gauvreau, G.M.; Boulet, L.P.; Lemiere, C.; Martin, J.; et al. Increased numbers of activated group 2 innate lymphoid cells in the airways of patients with severe asthma and persistent airway eosinophilia. J. Allergy Clin. Immunol. 2016, 137, 75–86.e8.
- Berdnikovs, S. The twilight zone: Plasticity and mixed ontogeny of neutrophil and eosinophil granulocyte subsets. Semin. Immunopathol. 2021, 43, 337–346.
More
Information
Subjects:
Allergy
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.4K
Revisions:
2 times
(View History)
Update Date:
10 Apr 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





