Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jennifer Sue Kahle | -- | 2434 | 2023-04-06 05:51:30 | | | |
| 2 | Catherine Yang | + 1 word(s) | 2435 | 2023-04-06 07:16:25 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Lizama, B.N.; Kahle, J.; Catalano, S.M.; Caggiano, A.O.; Grundman, M.; Hamby, M.E. The Sigma-2 Receptor. Encyclopedia. Available online: https://encyclopedia.pub/entry/42832 (accessed on 08 February 2026).
Lizama BN, Kahle J, Catalano SM, Caggiano AO, Grundman M, Hamby ME. The Sigma-2 Receptor. Encyclopedia. Available at: https://encyclopedia.pub/entry/42832. Accessed February 08, 2026.
Lizama, Britney N., Jennifer Kahle, Susan M. Catalano, Anthony O. Caggiano, Michael Grundman, Mary E. Hamby. "The Sigma-2 Receptor" Encyclopedia, https://encyclopedia.pub/entry/42832 (accessed February 08, 2026).
Lizama, B.N., Kahle, J., Catalano, S.M., Caggiano, A.O., Grundman, M., & Hamby, M.E. (2023, April 06). The Sigma-2 Receptor. In Encyclopedia. https://encyclopedia.pub/entry/42832
Lizama, Britney N., et al. "The Sigma-2 Receptor." Encyclopedia. Web. 06 April, 2023.
Copy Citation
The sigma-2 receptor (S2R), encoded by TMEM97, is expressed in brain and retinal cells, and regulates cell functions via its co-receptor progesterone receptor membrane component 1 (PGRMC1), and through other protein–protein interactions. S2R modulates key pathways, including autophagy, trafficking, oxidative stress, and amyloid-β and α-synuclein toxicity, involved in age-related degenerative diseases of the central nervous system. Furthermore, S2R modulation can ameliorate functional deficits in cell-based and animal models of neurodegenerative disease.
S2R
TMEM97
PGRMC1
MAC30
σ2R
Alzheimer’s disease (AD)
Parkinson’s disease (PD)
Niemann-Pick's Diease type C
Dry Age-Related Macular Degeneration
Dementia with Lewy Bodies
1. History, Structure, and Function of S2R
Before describing the role of S2R in degenerative diseases, the researchers summarize the history, structure, and function of S2R (Figure 1). A clear understanding of the structure and functionality of S2R has historically been elusive. S2R was first coined meningioma-associated protein 30 (MAC30). S2R activity was initially localized to a ~20 kDa membrane protein [1][2] and first thought to be progesterone receptor membrane component 1 (PGRMC1), based on mass spectrophotometry identification following an immunoaffinity pull down with the irreversible S2R ligand WC-11, physiochemical properties, and pharmacological evidence [3][4]. In later studies by two independent groups, however, in which PGRMC1 levels were genetically manipulated through overexpression or depletion, S2R binding activity was not altered irrespective of the level of PGRMC1 expressed, as assessed using the canonical S2R ligand tritiated di-o-tolylguanidine binding in the presence of saturating (+)-pentazocine, in a human epithelial cell line or a mouse motor neuron cell line [5][6]. Moreover, full activity persisted, even in the absence of PGRMC1.
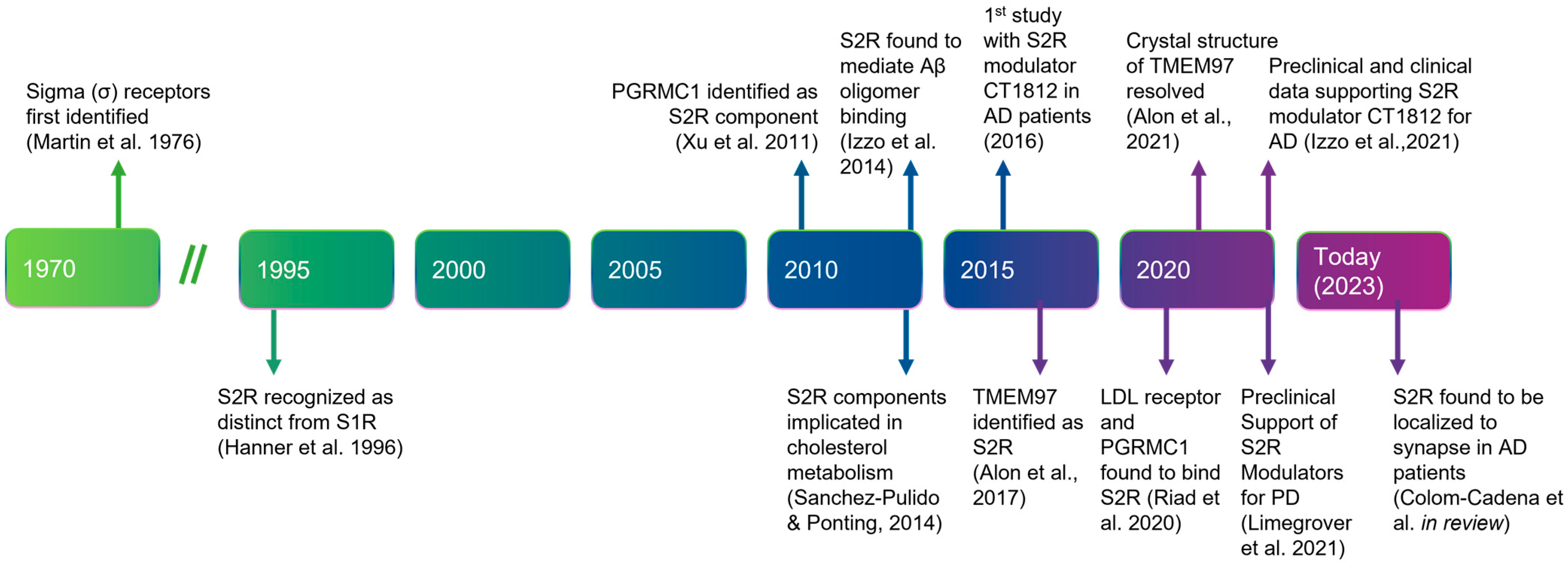
Figure 1. Timeline of the discovery and elucidation of S2R from inception to therapeutic modulation. References: Martin et al., 1976 [7]; Hanner et al., 1996 [8]; Xu et al., 2011 [9], Izzo et al., 2014 [10]; Sanchez-Pulido & Ponting, 2014 [11]; Alon et al., 2017 [12]; Riad et al., 2020 [13]; Alon et al., 2021 [14]; Izzo et al., 2021 [15]; Limegrover et al., 2021 [16]; Colom-Cadena et al., 2021 [17]. Abbreviations: AD, Alzheimer’s disease; Aβ, amyloid-β; LDL, low-density lipoprotein; PD, Parkinson’s disease; PGRMC1, progesterone receptor membrane component 1; S1R, sigma-1 receptor; S2R, sigma-2 receptor; TMEM97, transmembrane protein 97.
In 2017, the S2R was cloned [12], demonstrating that TMEM97, not PGRMC1, was the gene encoding S2R, indicating that S2R and TMEM97 are one and the same. This was confirmed in pharmacological studies assessing the activity of S2R ligands where reduction, or ablation, of S2R activity was observed when TMEM97 expression was reduced, or absent, in calf liver cells [12][18] or human cervical cancer cells [19], respectively. In further support, co-localization of TMEM97 with S2R ligands was demonstrated [20]. The identification of S2R as TMEM97 enabled a rapid advancement to the field, because it immediately merged two previously distinctly defined fields of research, enabling the application of that known about S2R pharmacology to TMEM97, and that known about TMEM97 biology to S2R (Figure 2) [12]. The crystal structure of TMEM97, although bovine, was resolved in 2021 which illuminated the docking sites of small molecule S2R ligands, PB28, and roluperidone [14], and allowed for structural enablement and docking for future medicinal chemistry efforts to identify S2R modulators that may be therapeutic candidates. This crystal structure also added additional confirmation that S2R is structurally distinct from the sigma-1 receptor [12][18][21].
2. Expression and Regulation of S2R/TMEM97
Within the brain, S2R is found in several areas, including the cerebellum, cortex, hippocampus, and substantia nigra, and is enriched in neurons as compared with glial cells in the adult brain [22]. Consistent with a role of S2R in synapses and in Alzheimer’s disease [23], S2R (TMEM97) has been demonstrated using Förster resonance energy transfer to colocalize with amyloid-β in synaptic densities [17]. In the retina, S2R is expressed in several cell types including the retinal pigment epithelium cells, photoreceptors, and retinal ganglion cells [24][25][26][27][28].
TMEM97 is a member of the EXPERA (EXPanded EBP superfamily) family of proteins along with emopamil binding protein (EBP), which is a protein domain containing four transmembrane regions and thought to have sterol isomerase activity [11]. Although TMEM97, unlike emopamil binding protein, is not thought to have isomerase activity, TMEM97 levels can be regulated by changes in cholesterol levels. More specifically, when cholesterol levels are reduced, TMEM97 expression can be induced by sterol regulatory element-binding protein-2 [29][30][31][32]. Both TMEM97 and PGRMC1 were regulated at the transcript level in skin cells by the topically applied Wnt/beta-catenin inhibitor C-82 under investigation in a clinical trial in individuals with systemic sclerosis, suggesting that Wnt signaling regulates TMEM97 [33]. Interestingly, increased expression of TMEM97 correlated with a rise in expression of genes involved in lipid metabolism, and these genes were significantly associated with the gene ontology terms cellular lipid, isoprenoid, cholesterol, and long-chain fatty-acyl-CoA biosynthetic processes [33], supporting a role of S2R in cholesterol biology. A further understanding of how TMEM97 may be regulated by different mediators and disease-relevant stressors is needed; however, in the case of Alzheimer’s disease, at least two reports suggest that TMEM97 is upregulated in synapses [17][23].
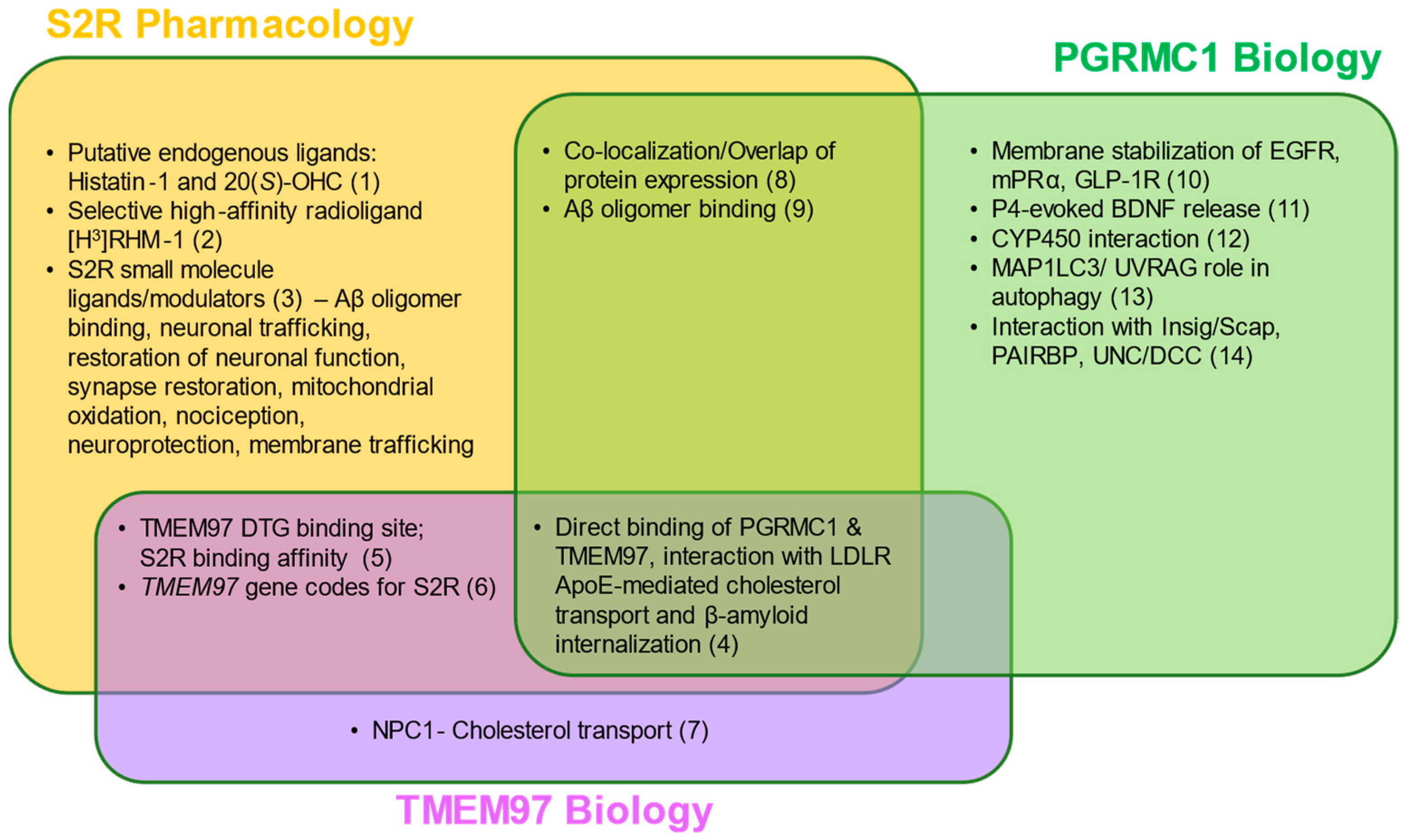
Figure 2. S2R pharmacology and the overlap with TMEM97 and PGRMC1 biology. Two putative endogenous ligands for S2R have been identified, although additional studies are needed to elucidate their roles under physiological and pathophysiological contexts. Despite this, several synthetic S2R ligands have been developed and used to investigate S2R function. Direct protein–protein interactions and functions of TMEM97 remain to be fully characterized; however, TMEM97 interacts directly with Niemann–Pick protein C1, as well as PGRMC1 and LDLR. PGRMC1 is well-identified as a hormone receptor with multiple functions, including regulation of signal transduction pathways, protein–protein interactions, membrane trafficking, and autophagy. While ongoing studies attempt to elucidate the functions of each protein, S2R, TMEM97, and PGRMC1 converge in co-localization/co-expression and their regulation of amyloid-β binding. References: (1) [34][35]; (2) [36]; (3) [14][20][27][37][38][39][40][41][42][43][44][45]; (4) [13][19]; (5) [1][46][47]; (6) [12][18]; (7) [12][48][49]; (8) [9][22][50]; (9) [10][51][52]; (10) [53][54][55]; (11) [56]; (12) [32]; (13) [57][58]; (14) [59][60][61][62] Abbreviations: Aβ, amyloid-β; Apo-E, apolipoprotein-E; BDNF, brain-derived neurotrophic factor; DTG, di-o-tolylguanidine; EGFR, epidermal growth factor receptor; GLP-1R, glucagon-like peptide-1 receptor; Insig/Scap, insulin-induced gene/sterol regulatory element-binding protein cleavage-activating protein; LDLR, low-density lipoprotein receptor; MAP1LC3/UVRAG, microtubule-associated proteins 1A/1B light chain 3/UV radiation resistance associated gene protein; mPRα, membrane progesterone receptor alpha; NPC1, Niemann–Pick protein C1; PAIRBP, plasminogen activator inhibitor 1 mRNA-binding protein; PGRMC1, progesterone receptor membrane component 1; S2R, sigma-2 receptor; TMEM97, transmembrane protein 97; UNC/DCC, UNC-40/Deleted in Colorectal Cancer.
3. Putatitve Endogenous Ligands
Until recently, the endogenous ligand of S2R was unknown, although some ligands had been historically proposed but lacked conclusive substantiation in follow-on studies [63][64]. In 2021, two studies were published that identified two putative endogenous ligands of S2R: histatin-1 [34] and the oxysterol 20(S)-OHC [35]. Histatin-1 is a salivary protein with antibacterial and antifungal activities and is known to be involved in wound healing [65][66]. Histatin-1 is also found in human tears [67], and the functional binding of histatin-1 to S2R was confirmed in human corneal epithelial cells [34]. Because histatin-1 expression is highly enriched in, and fairly limited to, salivary glands [22][68] and tears, further research is needed to clarify if histatin-1 is a ligand in the majority of tissues where S2R is expressed, including the brain and retina [35]. In contrast, the biology of 20(S)-OHC alone makes it a notable candidate. First, 20(S)-OHC is involved in Niemann–Pick C1 (NPC1)-mediated cholesterol homeostasis, including lipid membrane and sphingolipid metabolism [35]. Further, it is known that TMEM97 and NPC1 physically interact [49] and oxysterols are known to post-translationally enhance the activity of enzymes involved in sphingomyelin biosynthesis [69]. The ligand binding site described for 20(S)-OHC in this model is remarkably consistent with the ligand-bound crystal structure of S2R, recently reported for S2R in complex with synthetic S2R ligands [14]. The resolution of the crystal structure of the ligand-bound S2R will further enable the discovery of additional endogenous ligands [70]. While researchers in the field continue to investigate the endogenous ligand for S2R, there remains a clear need to elucidate definitively what, if any, previously identified putative ligands have a role in S2R signaling under both physiological and pathophysiological settings.
4. Protein Interactions Underlie the Functions of S2R
S2R couples and interacts with surrounding proteins to actuate a wide variety of cellular processes (Figure 3, Table 1). S2R has been shown to be closely associated and interact with key proteins to exert its functions, including PGRMC1 and LDLR [17][19]. PGRMC1 is a well-identified hormone receptor with multiple functions in Alzheimer’s disease [71], α-synucleinopathies [16][72], and retinal disease [73]. Moreover, PGRMC1 has been speculated to be an adaptor protein involved in regulating intracellular signal transduction and/or membrane trafficking [74][75] given its immunoreceptor tyrosine-based activation motif sequence [62][75], which enables membrane trafficking as well as protein–protein interaction [74][75]. Indeed, S2R modulators have been shown to ameliorate amyloid-β oligomer and α-synuclein oligomer-mediated deficits in neuronal trafficking [10][16][51].
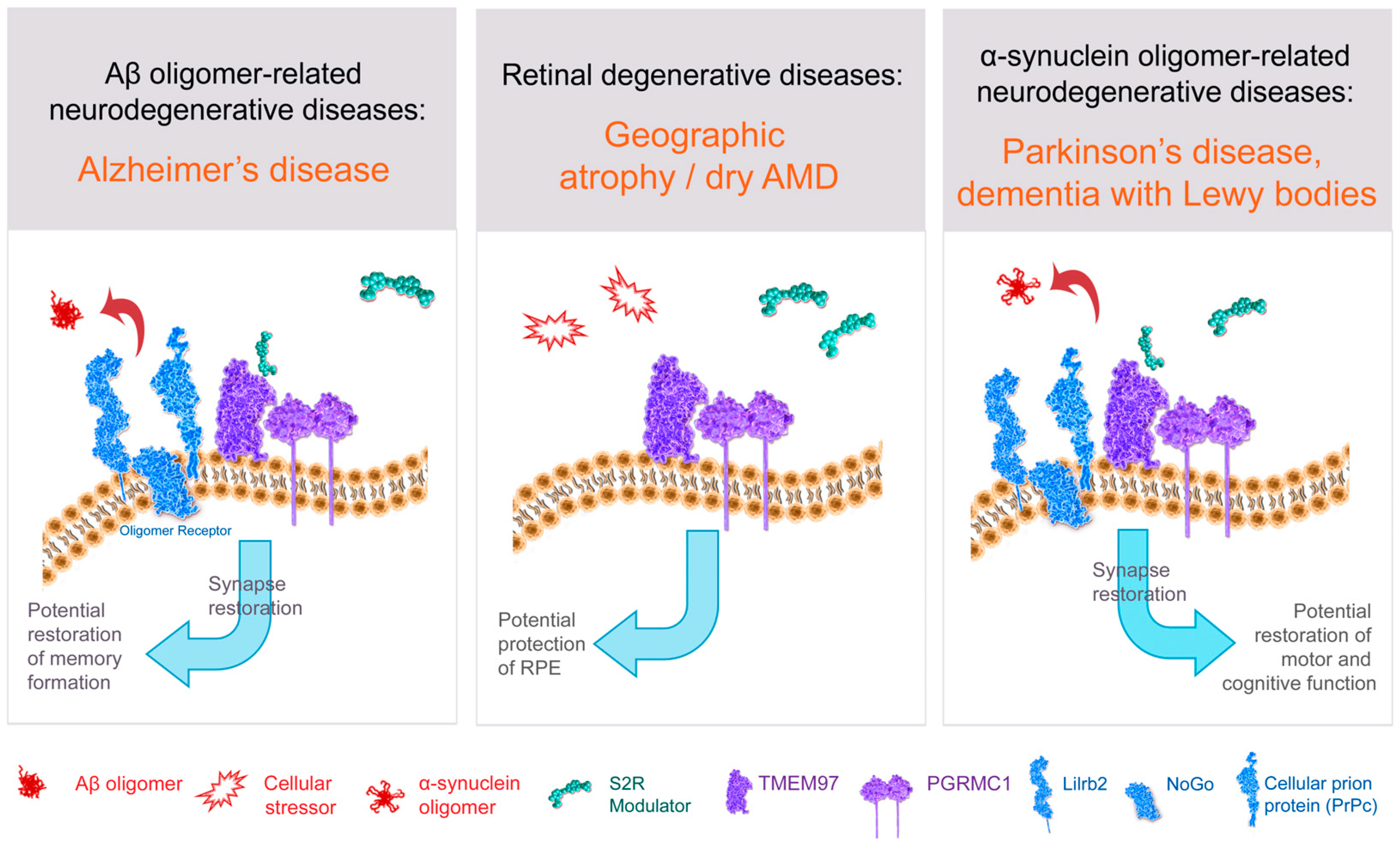
Figure 3. Structure–function of S2R and role of S2R small molecule modulators in restoring cell health and function. The amyloid-β oligomer receptor is a protein complex (light blue) that binds amyloid-β oligomers and is comprised of cellular prion protein (PrPc) along with other proteins including leukocyte immunoglobulin-like receptor subfamily B2/paired immunoglobulin-like type 2 receptor B (LilrB2), and possibly also Nogo-66 receptor 1 (Nogo). S2R small molecule modulator (green) binds to the S2R (TMEM97; purple). Abbreviations: AMD, age-related macular degeneration; Aβ, amyloid-β; PGRMC1, progesterone receptor membrane component 1; RPE, retinal pigment epithelium; S2R, sigma-2 receptor; TMEM97, transmembrane protein 97.
Table 1. Key functional roles of S2R and the molecular players involved.
| Functions of S2R | Molecules Involved | Disease Relevance |
|---|---|---|
| Blocks amyloid-β oligomers from binding neuronal synapses | TMEM97, PGRMC1 oligomer receptor |
Alzheimer’s disease |
| Blocks α-synuclein oligomers from binding neuronal synapses | TMEM97 | Dementia with Lewy bodies Parkinson’s disease |
| Mediates synaptoprotection | TMEM97, PGRMC1 mGluR5, oligomer receptor |
Alzheimer’s disease Parkinson’s disease Dementia with Lewy bodies |
| Regulates autophagy | TMEM97, PGRMC1 LAMP2A, MAP1LC3B |
Dry AMD Dementia with Lewy bodies Parkinson’s disease |
| Regulates cholesterol homeostasis | TMEM97, PGRMC1, LDLR, Apo-E, NPC1 | Alzheimer’s disease Niemann–Pick disease type C |
| Regulates membrane trafficking | TMEM97, PGRMC1, LDLR | Alzheimer’s disease Parkinson’s disease Dementia with Lewy bodies |
Abbreviations: AMD, age-related macular degeneration; Apo-E, apolipoprotein-E; LAMP2A, lysosomal-associated membrane protein 2A; LDLR, low-density lipoprotein receptor; MAP1LC3B, microtubule-associated proteins 1A/1B light chain 3; mGluR5, metabotropic glutamate receptor 5; NPC1, Niemann–Pick protein C1; PGRMC1, progesterone receptor membrane component 1; S2R, sigma-2 receptor; TMEM97, transmembrane protein 97.
Studies from multiple in vitro models have identified that both TMEM97 and PGRMC1 modulate autophagy, a quintessential cellular process needed to recycle cellular debris. PGRMC1 is a binding partner for numerous proteins in the autophagic pathway, including the key autophagic protein microtubule-associated protein 1A/1B light chain 3B (LC3) [57][58]. Treatment with S2R ligands induces processing of LC3, formation of autophagic vacuoles, and expression of downstream effectors of the mammalian Target of Rapamycin (mTOR) pathway [76]. TMEM97 knockout cell lines exhibit aberrant autophagic flux and lysosome dysfunction [77]. Some small molecule modulators of S2R, such as siramesine and SV119, trigger the induction of autophagy [76][78], while other S2R modulators such as CT2168 can restore α-synuclein oligomer-induced deficits in lysosomal-associated membrane protein 2A levels, an indirect marker of autophagy, back to healthy control levels in neurons [16]. Given that autophagy is dysregulated in neurodegenerative conditions, further elucidation of S2R in regulating autophagy in brain and retinal disease is warranted, and may foster better translation into potential neuroprotective therapies.
One important function of S2R is regulation of cholesterol homeostasis by interacting with low-density lipoprotein receptor (LDLR), an apolipoprotein-E receptor, at the plasma membrane to transport apolipoprotein E into neurons [19] and with NPC1, intracellularly, to regulate LDL cholesterol transport out of lysosomes [48][49]. Cholesterol is a key component of the cell membrane and constitutes up to 30% of total membrane lipids [79]. Cholesterol, bound to LDL, is taken up into cells via the LDLR, and endocytosed. S2R is key in facilitating the uptake of LDL, as uptake of radiolabeled LDL was significantly decreased in PGRMC1 knockout, TMEM97 knockout, and double knockout cell lines [19][48]. Using these knockout cell lines, the same group demonstrated that both PGRMC1 and TMEM97 must be present and functional for effective LDL–LDLR complex internalization [19]. Further, to understand whether S2R alters all clathrin-mediated endocytic pathways or just the uptake of LDL, radiolabeled [125I]TYR11-somatostatin and [125I] insulin uptake was assessed, but knockout of TMEM97 or PGRMC1 did not alter the uptake of either substrate, indicating a specific role of S2R regulation of trafficking through LDLR [13].
The TMEM97/PGRMC1/LDLR triad is responsible for the uptake of amyloid-β oligomers and apolipoprotein E into neurons, as knockdown of TMEM97 or modulation of TMEM97 with S2R ligands including RHM-4 and SW43 substantially decreased the uptake of both amyloid-β oligomers and apolipoprotein E, but apolipoprotein E was not required for amyloid-β oligomer uptake [13]. The lack of dependence on apolipoprotein E for synaptic uptake has been reported and discussed previously [80][81]. Apolipoprotein E also interacts with PGRMC1 in the clearance of amyloid-β from neurons. Blocking the interaction between apolipoprotein E and PGRMC1 increases amyloid-β clearance from the brain, reduces amyloid-β deposition, and reduces related pathology in Alzheimer animal models [82]. This is consistent with results showing that specific small-molecule ligands that bind to S2R displace endogenous β-amyloid oligomers from human brain samples from individuals with Alzheimer’s disease, displace β-amyloid oligomer binding to primary cultured neurons, and reverse cognitive deficits in Alzheimer model mice [10][51].
There is a host of direct and indirect evidence linking S2R to the amyloid-β oligomer receptor. Amyloid-β oligomers bind to a protein complex comprised of cellular prion protein (PrPc) along with other proteins including leukocyte immunoglobulin-like receptor subfamily B2/paired immunoglobulin-like type 2 receptor B (LilrB2), and possibly also Nogo-66 receptor 1 (Nogo-1), low-affinity nerve growth factor (NGF) receptor [83] (Figure 3), and the metabotropic glutamate receptor 5 [84]. In the brain, the expressions of oligomer receptor constituents are enriched in neuronal synapses [84]. Several lines of evidence indicate a physical and/or functional interaction of S2R with this protein complex that binds amyloid-β oligomers. Amyloid-β oligomer binding results in aberrant neuronal signaling and the endocytosis of the dysfunctional synapse into the cell [84][85]. Amyloid-β oligomer binding to neuronal synapses is abrogated by knockdown of PGRMC1 [51], PrPc, and with S2R modulators in neurons [10][51], an effect that leads to restoration of neuronal health and function. Furthermore, treatment with a metabotropic glutamate receptor 5 modulator, which displaces amyloid-β oligomers, effectively prevented synaptotoxicity and restored functional synapses in a mouse model of Alzheimer’s disease [85]. Conditional deletion of the oligomer receptor PrPc in Alzheimer transgenic mice rescues cognitive and synaptic deficits [86]. Lastly, S2R modulators restore cognitive deficits in Alzheimer transgenic mice [10][15]. In sum, substantial evidence linking S2R biology to that of the protein complex that binds amyloid-β oligomers supports the pursuit of small-molecule modulators of S2R to prevent amyloid-β oligomer-mediated toxicity.
References
- Hellewell, S.B.; Bowen, W.D. A Sigma-like Binding Site in Rat Pheochromocytoma (PC12) Cells: Decreased Affinity for (+)-Benzomorphans and Lower Molecular Weight Suggest a Different Sigma Receptor Form from That of Guinea Pig Brain. Brain Res. 1990, 527, 244–253.
- Hellewell, S.B.; Bruce, A.; Feinstein, G.; Orringer, J.; Williams, W.; Bowen, W.D. Rat Liver and Kidney Contain High Densities of Sigma 1 and Sigma 2 Receptors: Characterization by Ligand Binding and Photoaffinity Labeling. Eur. J. Pharmacol. 1994, 268, 9–18.
- Zeng, C.; Garg, N.; Mach, R.H. The PGRMC1 Protein Level Correlates with the Binding Activity of a Sigma-2 Fluorescent Probe (SW120) in Rat Brain Cells. Mol. Imaging Biol. 2016, 18, 172–179.
- Nicholson, H.; Mesangeau, C.; McCurdy, C.R.; Bowen, W.D. Sigma-2 Receptors Play a Role in Cellular Metabolism: Stimulation of Glycolytic Hallmarks by CM764 in Human SK-N-SH Neuroblastomas. J. Pharmacol. Exp. Ther. 2016, 356, 232–243.
- Abate, C.; Niso, M.; Infantino, V.; Menga, A.; Berardi, F. Elements in Support of the ‘ Non-Identity ’ of the PGRMC1 Protein with the σ 2 Receptor. Eur. J. Pharmacol. 2015, 758, 16–23.
- Chu, U.B.; Mavlyutov, T.A.; Chu, M.; Yang, H.; Schulman, A.; Mesangeau, C.; McCurdy, C.R.; Guo, L.; Ruoho, A.E. The Sigma-2 Receptor and Progesterone Receptor Membrane Component 1 Are Different Binding Sites Derived From Independent Genes. EBioMedicine 2015, 2, 1806–1813.
- Martin, W.R.; Eades, C.G.; Thompson, J.A.; Huppler, R.E.; Gilbert, P.E. The Effects of Morphine- and Nalorphine-like Drugs in the Nondependent and Morphine-Dependent Chronic Spinal Dog. J. Pharmacol. Exp. Ther. 1976, 197, 517–532.
- Hanner, M.; Moebius, F.F.; Flandorfer, A.; Knaus, H.G.; Striessnig, J.; Kempner, E.; Glossmann, H. Purification, Molecular Cloning, and Expression of the Mammalian Sigma1-Binding Site. Proc. Natl. Acad. Sci. USA 1996, 93, 8072–8077.
- Xu, J.; Zeng, C.; Chu, W.; Pan, F.; Rothfuss, J.M.; Zhang, F.; Tu, Z.; Zhou, D.; Zeng, D.; Vangveravong, S.; et al. Identification of the PGRMC1 Protein Complex as the Putative Sigma-2 Receptor Binding Site. Nat. Commun. 2011, 2, 380.
- Izzo, N.J.; Staniszewski, A.; To, L.; Fa, M.; Teich, A.F.; Saeed, F.; Wostein, H.; Walko, T.; Vaswani, A.; Wardius, M.; et al. Alzheimer’s Therapeutics Targeting Amyloid Beta 1–42 Oligomers I: Abeta 42 Oligomer Binding to Specific Neuronal Receptors Is Displaced by Drug Candidates That Improve Cognitive Deficits. PLoS ONE 2014, 9, e111898.
- Sanchez-Pulido, L.; Ponting, C.P. TM6SF2 and MAC30, New Enzyme Homologs in Sterol Metabolism and Common Metabolic Disease. Front. Genet. 2014, 5, 439.
- Alon, A.; Schmidt, H.R.; Wood, M.D.; Sahn, J.J.; Martin, S.F.; Kruse, A.C. Identification of the Gene That Codes for the σ 2 Receptor. Proc. Natl. Acad. Sci. USA 2017, 114, 7160–7165.
- Riad, A.; Lengyel-Zhand, Z.; Zeng, C.; Weng, C.-C.; Lee, V.M.-Y.; Trojanowski, J.Q.; Mach, R.H. The Sigma-2 Receptor/TMEM97, PGRMC1, and LDL Receptor Complex Are Responsible for the Cellular Uptake of Aβ42 and Its Protein Aggregates. Mol. Neurobiol. 2020, 57, 3803–3813.
- Alon, A.; Lyu, J.; Braz, J.M.; Tummino, T.A.; Craik, V.; O’Meara, M.J.; Webb, C.M.; Radchenko, D.S.; Moroz, Y.S.; Huang, X.; et al. Structures of the Σ2 Receptor Enable Docking for Bioactive Ligand Discovery. Nature 2021, 600, 759–764.
- Izzo, N.J.; Yuede, C.M.; LaBarbera, K.M.; Limegrover, C.S.; Rehak, C.; Yurko, R.; Waybright, L.; Look, G.; Rishton, G.; Safferstein, H.; et al. Preclinical and Clinical Biomarker Studies of CT1812: A Novel Approach to Alzheimer’s Disease Modification. Alzheimer’s Dement. 2021, 17, 1365–1382.
- Limegrover, C.S.; Yurko, R.; Izzo, N.J.; LaBarbera, K.M.; Rehak, C.; Look, G.; Rishton, G.; Safferstein, H.; Catalano, S.M. Sigma-2 Receptor Antagonists Rescue Neuronal Dysfunction Induced by Parkinson’s Patient Brain-derived A-synuclein. J. Neurosci. Res. 2021, 99, 1161–1176.
- Colom-Cadena, M.; Tulloch, J.; Jackson, R.J.; Catterson, J.H.; Rose, J.; Davies, C.; Hooley, M.; Anton-Fernandez, A.; Dunnett, S.; Tempelaar, R.; et al. TMEM97 Increases in Synapses and Is a Potential Synaptic Aβ Binding Partner in Human Alzheimer’s Disease. bioRxiv 2021.
- Schmidt, H.R.; Kruse, A.C. The Molecular Function of σ Receptors: Past, Present, and Future. Trends Pharmacol. Sci. 2019, 40, 636–654.
- Riad, A.; Zeng, C.; Weng, C.-C.C.; Winters, H.; Xu, K.; Makvandi, M.; Metz, T.; Carlin, S.; Mach, R.H. Sigma-2 Receptor/TMEM97 and PGRMC-1 Increase the Rate of Internalization of LDL by LDL Receptor through the Formation of a Ternary Complex. Sci. Rep. 2018, 8, 16845.
- Cantonero, C.; Camello, P.J.; Abate, C.; Berardi, F.; Salido, G.M.; Rosado, J.A.; Redondo, P.C. NO1, a New Sigma 2 Receptor/TMEM97 Fluorescent Ligand, Downregulates SOCE and Promotes Apoptosis in the Triple Negative Breast Cancer Cell Lines. Cancers 2020, 12, 257.
- Langa, F.; Codony, X.; Tovar, V.; Lavado, A.; Gimenez, E.; Cozar, P.; Cantero, M.; Dordal, A.; Hernandez, E.; Perez, R.; et al. Generation and Phenotypic Analysis of Sigma Receptor Type I (Sigma1) Knockout Mice. Eur. J. Neurosci. 2003, 18, 2188–2196.
- Karlsson, M.; Zhang, C.; Méar, L.; Zhong, W.; Digre, A.; Katona, B.; Sjöstedt, E.; Butler, L.; Odeberg, J.; Dusart, P.; et al. A Single–Cell Type Transcriptomics Map of Human Tissues. Sci. Adv. 2021, 7, eabh2169.
- Hesse, R.; Hurtado, M.L.; Jackson, R.J.; Eaton, S.L.; Herrmann, A.G.; Colom-Cadena, M.; Tzioras, M.; King, D.; Rose, J.; Tulloch, J.; et al. Comparative Profiling of the Synaptic Proteome from Alzheimer’s Disease Patients with Focus on the APOE Genotype. Acta Neuropathol. Commun. 2019, 7, 214.
- Wang, J.-H.; Urrutia-Cabrera, D.; Mora, S.M.; Nguyen, T.; Hung, S.; Hewitt, A.W.; Edwards, T.L.; Wong, R.C.B. Functional Study of the AMD-Associated Gene TMEM97 in Retinal Pigmented Epithelium Using CRISPR Interference. bioRxiv 2020, 2020-07.
- Shanmugam, A.K.; Mysona, B.A.; Wang, J.; Zhao, J.; Tawfik, A.; Sanders, A.; Markand, S.; Zorrilla, E.; Ganapathy, V.; Bollinger, K.E.; et al. Progesterone Receptor Membrane Component 1 (PGRMC1) Expression in Murine Retina. Curr. Eye Res. 2016, 41, 1105–1112.
- Ratnapriya, R.; Sosina, O.A.; Starostik, M.R.; Kwicklis, M.; Kapphahn, R.J.; Fritsche, L.G.; Walton, A.; Arvanitis, M.; Gieser, L.; Pietraszkiewicz, A.; et al. Retinal Transcriptome and EQTL Analyses Identify Genes Associated with Age-Related Macular Degeneration. Nat. Genet. 2019, 51, 606–610.
- Wang, H.; Peng, Z.; Li, Y.; Sahn, J.J.; Hodges, T.R.; Chou, T.H.; Liu, Q.; Zhou, X.; Jiao, S.; Porciatti, V.; et al. Σ2R/TMEM97 in Retinal Ganglion Cell Degeneration. Sci. Rep. 2022, 12, 20753.
- Kim, H.Y.; Lee, J.Y.; Hsieh, C.-J.; Riad, A.; Izzo, N.J.; Catalano, S.M.; Graham, T.J.A.; Mach, R.H. Screening of σ 2 Receptor Ligands and In Vivo Evaluation of 11C-Labeled 6,7-Dimethoxy-2--1,2,3,4-Tetrahydroisoquinoline for Potential Use as a σ2 Receptor Brain PET Tracer. J. Med. Chem. 2022, 65, 6261–6272.
- Ahmed, I.S.A.; Chamberlain, C.; Craven, R.J. S2R(Pgrmc1): The Cytochrome-Related Sigma-2 Receptor That Regulates Lipid and Drug Metabolism and Hormone Signaling. Expert Opin. Drug Metab. Toxicol. 2012, 8, 361–370.
- Lee, S.R.; Kwon, S.W.; Kaya, P.; Lee, Y.H.; Lee, J.G.; Kim, G.; Lee, G.-S.; Baek, I.-J.; Hong, E.-J. Loss of Progesterone Receptor Membrane Component 1 Promotes Hepatic Steatosis via the Induced de Novo Lipogenesis. Sci. Rep. 2018, 8, 15711.
- Cai, H.L.; Tan, Q.Y.; Jiang, P.; Dang, R.L.; Xue, Y.; Tang, M.M.; Xu, P.; Deng, Y.; Li, H.D.; Yao, J.K. A Potential Mechanism Underlying Atypical Antipsychotics-Induced Lipid Disturbances. Transl. Psychiatry 2015, 5, e661.
- Hughes, A.L.; Powell, D.W.; Bard, M.; Eckstein, J.; Barbuch, R.; Link, A.J.; Espenshade, P.J. Dap1/PGRMC1 Binds and Regulates Cytochrome P450 Enzymes. Cell Metab. 2007, 5, 143–149.
- Lafyatis, R.; Mantero, J.C.; Gordon, J.; Kishore, N.; Carns, M.; Dittrich, H.; Spiera, R.; Simms, R.W.; Varga, J. Inhibition of β-Catenin Signaling in the Skin Rescues Cutaneous Adipogenesis in Systemic Sclerosis: A Randomized, Double-Blind, Placebo-Controlled Trial of C-82. J. Investig. Dermatol. 2017, 137, 2473–2483.
- Son, K.; Lee, H.; Shah, D.; Kalmodia, S.; Miller, R.C.; Ali, M.; Balasubramaniam, A.; Cologna, S.M.; Kong, H.; Shukla, D.; et al. Histatin-1 Is an Endogenous Ligand of the Sigma-2 Receptor. FEBS J. 2021, 288, 6815–6827.
- Cheng, Y.; Zhang, T.; Ma, X.; Pratuangtham, S.; Zhang, G.C.; Ondrus, A.A.; Mafi, A.; Lomenick, B.; Jones, J.J.; Ondrus, A.E. A Proteome-Wide Map of 20(S)-Hydroxycholesterol Interactors in Cell Membranes. Nat. Chem. Biol. 2021, 17, 1271–1280.
- Mach, R.H.; Wheeler, K.T. Development of Molecular Probes for Imaging Sigma-2 Receptors in Vitro and in Vivo. Cent. Nerv. Syst. Agents Med. Chem. 2009, 9, 230–245.
- Jin, J.; Arbez, N.; Sahn, J.J.; Lu, Y.; Linkens, K.T.; Hodges, T.R.; Tang, A.; Wiseman, R.; Martin, S.F.; Ross, C.A. Neuroprotective Effects of σ 2 R/TMEM97 Receptor Modulators in the Neuronal Model of Huntington’s Disease. ACS Chem. Neurosci. 2022, 13, 2852–2862.
- Turgutalp, B.; Bhattarai, P.; Ercetin, T.; Luise, C.; Reis, R.; Gurdal, E.E.; Isaak, A.; Biriken, D.; Dinter, E.; Sipahi, H.; et al. Discovery of Potent Cholinesterase Inhibition-Based Multi-Target- Directed Lead Compounds for Synaptoprotection in Alzheimer’s Disease. J. Med. Chem. 2022, 65, 12292–12318.
- Mavlyutov, T.A.; Li, J.; Liu, X.; Shen, H.; Yang, H.; McCurdy, C.R.; Pattnaik, B.; Guo, L.W. Retinal Photoreceptor Protection in an AMD-Related Mouse Model by Selective Sigma-1 or Sigma-2 Receptor Modulation. Genes 2022, 13, 2386.
- Vilner, B.J.; Bowen, W.D. Modulation of Cellular Calcium by Sigma-2 Receptors: Release from Intracellular Stores in Human SK-N-SH Neuroblastoma Cells. J. Pharmacol. Exp. Ther. 2000, 292, 900–911.
- Cassano, G.; Gasparre, G.; Contino, M.; Niso, M.; Berardi, F.; Perrone, R.; Colabufo, N.A. The Sigma-2 Receptor Agonist PB28 Inhibits Calcium Release from the Endoplasmic Reticulum of SK-N-SH Neuroblastoma Cells. Cell Calcium 2006, 40, 23–28.
- Guo, L.; Zhen, X. Sigma-2 Receptor Ligands: Neurobiological Effects. Curr. Med. Chem. 2015, 22, 989–1003.
- Mach, R.H.; Zeng, C.; Hawkins, W.G. The σ2 Receptor: A Novel Protein for the Imaging and Treatment of Cancer. J. Med. Chem. 2013, 56, 7137–7160.
- Zeng, C.; Weng, C.-C.C.; Schneider, M.E.; Puentes, L.; Riad, A.; Xu, K.; Makvandi, M.; Jin, L.; Hawkins, W.G.; Mach, R.H. TMEM97 and PGRMC1 Do Not Mediate Sigma-2 Ligand-Induced Cell Death. Cell Death Discov. 2019, 5, 58.
- Pati, M.L.; Hornick, J.R.; Niso, M.; Berardi, F.; Spitzer, D.; Abate, C.; Hawkins, W. Sigma-2 Receptor Agonist Derivatives of 1-Cyclohexyl-4-Piperazine (PB28) Induce Cell Death via Mitochondrial Superoxide Production and Caspase Activation in Pancreatic Cancer. BMC Cancer 2017, 17, 51.
- Quirion, R.; Bowen, W.D.; Itzhak, Y.; Junien, J.L.; Musacchio, J.M.; Rothman, R.B.; Su, T.P.; Tam, S.W.; Taylor, D.P. A Proposal for the Classification of Sigma Binding Sites. Trends Pharmacol. Sci. 1992, 13, 85–86.
- Vilner, B.J.; John, C.S.; Bowen, W.D. Sigma-1 and Sigma-2 Receptors Are Expressed in a Wide Variety of Human and Rodent Tumor Cell Lines. Cancer Res. 1995, 55, 408–413.
- Bartz, F.; Kern, L.; Erz, D.; Zhu, M.; Gilbert, D.; Meinhof, T.; Wirkner, U.; Erfle, H.; Muckenthaler, M.; Pepperkok, R.; et al. Identification of Cholesterol-Regulating Genes by Targeted RNAi Screening. Cell Metab. 2009, 10, 63–75.
- Ebrahimi-Fakhari, D.; Wahlster, L.; Bartz, F.; Werenbeck-Ueding, J.; Praggastis, M.; Zhang, J.; Joggerst-Thomalla, B.; Theiss, S.; Grimm, D.; Ory, D.S.; et al. Reduction of TMEM97 Increases NPC1 Protein Levels and Restores Cholesterol Trafficking in Niemann-Pick Type C1 Disease Cells. Hum. Mol. Genet. 2015, 25, 3588–3599.
- Intlekofer, K.A.; Petersen, S.L. Distribution of MRNAs Encoding Classical Progestin Receptor, Progesterone Membrane Components 1 and 2, Serpine MRNA Binding Protein 1, and Progestin and ADIPOQ Receptor Family Members 7 and 8 in Rat Forebrain. Neuroscience 2011, 172, 55–65.
- Izzo, N.J.; Xu, J.; Zeng, C.; Kirk, M.J.; Mozzoni, K.; Silky, C.; Rehak, C.; Yurko, R.; Look, G.; Rishton, G.; et al. Alzheimer’s Therapeutics Targeting Amyloid Beta 1–42 Oligomers II: Sigma-2/PGRMC1 Receptors Mediate Abeta 42 Oligomer Binding and Synaptotoxicity. PLoS ONE 2014, 9, e111899.
- Catalano, S.M.; Mozzoni, K.; Rehak, C.; Waybright, L.; Sadlek, K.; Safferstein, H.; Watto, E.; Izzo, N.J.; Grundman, M.; Dekosky, S.; et al. CT1812 Demonstrates Evidence of Synapse Preservation in Alzheimer’s Disease Patients and Abeta Oligomer Displacement in Preclinical Models; Presentation 200.15/B19; Society for Neuroscience: Chicago, IL, USA, 2019.
- Ahmed, I.S.; Rohe, H.J.; Twist, K.E.; Mattingly, M.N.; Craven, R.J. Progesterone Receptor Membrane Component 1 (Pgrmc1): A Heme-1 Domain Protein That Promotes Tumorigenesis and Is Inhibited by a Small Molecule. J. Pharmacol. Exp. Ther. 2010, 333, 564–573.
- Thomas, P.; Pang, Y.; Dong, J. Enhancement of Cell Surface Expression and Receptor Functions of Membrane Progestin Receptor α (MPRα) by Progesterone Receptor Membrane Component 1 (PGRMC1): Evidence for a Role of PGRMC1 as an Adaptor Protein for Steroid Receptors. Endocrinology 2014, 155, 1107–1119.
- Zhang, M.; Robitaille, M.; Showalter, A.D.; Huang, X.; Liu, Y.; Bhattacharjee, A.; Willard, F.S.; Han, J.; Froese, S.; Wei, L.; et al. Progesterone Receptor Membrane Component 1 Is a Functional Part of the GLP-1 Receptor Complex in Pancreatic Beta Cells. Mol. Cell. Proteom. 2014, 1, 3049–3062.
- Su, C.; Cunningham, R.L.; Rybalchenko, N.; Singh, M. Progesterone Increases the Release of Brain-Derived Neurotrophic Factor from Glia via Progesterone Receptor Membrane Component 1 (Pgrmc1)-Dependent ERK5 Signaling. Endocrinology 2012, 153, 4389–4400.
- Mir, S.U.R.; Schwarze, S.R.; Jin, L.; Zhang, J.; Friend, W.; Miriyala, S.; Clair, D.S.; Craven, R.J.; St Clair, D.; Craven, R.J. Progesterone Receptor Membrane Component 1/ Sigma-2 Receptor Associates with MAP1LC3B and Promotes Autophagy. Autophagy 2013, 9, 1566–1578.
- Behrends, C.; Sowa, M.E.; Gygi, S.P.; Harper, J.W. Network Organization of the Human Autophagy System. Nature 2010, 466, 68–76.
- Rohe, H.J.; Ahmed, I.S.; Twist, K.E.; Craven, R.J. PGRMC1 (Progesterone Receptor Membrane Component 1): A Targetable Protein with Multiple Functions in Steroid Signaling, P450 Activation and Drug Binding. Pharmacol. Ther. 2009, 121, 14–19.
- Peluso, J.J.; Yuan, A.; Liu, X.; Lodde, V. Plasminogen Activator Inhibitor 1 RNA-Binding Protein Interacts with Progesterone Receptor Membrane Component 1 to Regulate Progesterone’s Ability to Maintain the Viability of Spontaneously Immortalized Granulosa Cells and Rat Granulosa Cells. Biol. Reprod. 2013, 88, 20.
- Runko, E.; Kaprielian, Z. Expression of Vema in the Developing Mouse Spinal Cord and Optic Chiasm. J. Comp. Neurol. 2002, 451, 289–299.
- Runko, E.; Kaprielian, Z. Caenorhabditis Elegans VEM-1, a Novel Membrane Protein, Regulates the Guidance of Ventral Nerve Cord-Associated Axons. J. Neurosci. 2004, 24, 9015–9026.
- Contreras, P.C.; Dimaggio, D.A.; O’Donohue, T.L. An Endogenous Ligand for the Sigma Opioid Binding Site. Synapse 1987, 1, 57–61.
- Connor, M.A.; Chavkin, C. Ionic Zinc May Function as an Endogenous Ligand for the Haloperidol-Sensitive Sigma 2 Receptor in Rat Brain. Mol. Pharmacol. 1992, 42, 471–479.
- Oudhoff, M.J.; Bolscher, J.G.M.; Nazmi, K.; Kalay, H.; Hof, W.; Amerongen, A.V.N.; Veerman, E.C.I. Histatins Are the Major Wound-closure Stimulating Factors in Human Saliva as Identified in a Cell Culture Assay. FASEB J. 2008, 22, 3805–3812.
- Torres, P.; Hernández, N.; Mateluna, C.; Silva, P.; Reyes, M.; Solano, L.; Venegas, S.; Criollo, A.; Nazmi, K.; Bikker, F.J.; et al. Histatin-1 Is a Novel Osteogenic Factor That Promotes Bone Cell Adhesion, Migration, and Differentiation. J. Tissue Eng. Regen. Med. 2021, 15, 336–346.
- Kalmodia, S.; Son, K.N.; Cao, D.; Lee, B.S.; Surenkhuu, B.; Shah, D.; Ali, M.; Balasubramaniam, A.; Jain, S.; Aakalu, V.K. Presence of Histatin-1 in Human Tears and Association with Aqueous Deficient Dry Eye Diagnosis: A Preliminary Study. Sci. Rep. 2019, 9, 10304.
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419.
- Ridgway, N.D. 25-Hydroxycholesterol Stimulates Sphingomyelin Synthesis in Chinese Hamster Ovary Cells. J. Lipid Res. 1995, 36, 1345–1358.
- Xu, K.; Hsieh, C.-J.; Lee, J.Y.; Riad, A.; Izzo, N.J.; Look, G.; Catalano, S.; Mach, R.H. Exploration of Diazaspiro Cores as Piperazine Bioisosteres in the Development of σ2 Receptor Ligands. Int. J. Mol. Sci. 2022, 23, 8259.
- Xu, X.; Ruan, X.; Ju, R.; Wang, Z.; Yang, Y.; Cheng, J.; Gu, M.; Mueck, A.O. Progesterone Receptor Membrane Component-1 May Promote Survival of Human Brain Microvascular Endothelial Cells in Alzheimer’s Disease. Am. J. Alzheimers. Dis. Other Demen. 2022, 37.
- Kline, R.A.; Kaifer, K.A.; Osman, E.Y.; Carella, F.; Tiberi, A.; Ross, J.; Pennetta, G.; Lorson, C.L.; Murray, L.M. Comparison of Independent Screens on Differentially Vulnerable Motor Neurons Reveals Alpha-Synuclein as a Common Modifier in Motor Neuron Diseases. PLoS Genet. 2017, 13, e1006680.
- Wyse Jackson, A.C.; Roche, S.L.; Byrne, A.M.; Ruiz-Lopez, A.M.; Cotter, T.G. Progesterone Receptor Signalling in Retinal Photoreceptor Neuroprotection. J. Neurochem. 2016, 136, 63–77.
- Cahill, M.A.; Jazayeri, J.A.; Catalano, S.M.; Toyokuni, S.; Kovacevic, Z.; Richardson, D.R. The Emerging Role of Progesterone Receptor Membrane Component 1 (PGRMC1) in Cancer Biology. Biochim. Biophys. Acta 2016, 1866, 339–349.
- Cahill, M.A.; Jazayeri, J.A.; Kovacevic, Z.; Richardson, D.R. PGRMC1 Regulation by Phosphorylation: Potential New Insights in Controlling Biological Activity! Oncotarget 2016, 67, 1079–1082.
- Zeng, C.; Rothfuss, J.; Zhang, J.; Chu, W.; Vangveravong, S.; Tu, Z.; Pan, F.; Chang, K.C.; Hotchkiss, R.; Mach, R.H. Sigma-2 Ligands Induce Tumour Cell Death by Multiple Signalling Pathways. Br. J. Cancer 2012, 106, 693–701.
- Shen, H.; Li, J.; Heisler-Taylor, T.; Makin, R.; Yang, H.; Mavlyutov, T.A.; Gelfand, B.; Cebulla, C.M.; Guo, L.W. TMEM97 Ablation Aggravates Oxidant-Induced Retinal Degeneration. Cell Signal. 2021, 86, 110078.
- Ostenfeld, M.S.; Høyer-Hansen, M.; Bastholm, L.; Fehrenbacher, N.; Olsen, O.D.; Groth-Pedersen, L.; Puustinen, P.; Kirkegaard-Sørensen, T.; Nylandsted, J.; Farkas, T.; et al. Anti-Cancer Agent Siramesine Is a Lysosomotropic Detergent That Induces Cytoprotective Autophagosome Accumulation. Autophagy 2008, 4, 487–499.
- Krause, M.R.; Regen, S.L. The Structural Role of Cholesterol in Cell Membranes: From Condensed Bilayers to Lipid Rafts. Acc. Chem. Res. 2014, 47, 3512–3521.
- Gylys, K.H.; Fein, J.A.; Tan, A.M.; Cole, G.M. Apolipoprotein E Enhances Uptake of Soluble but Not Aggregated Amyloid-β Protein into Synaptic Terminals. J. Neurochem. 2003, 84, 1442–1451.
- Fernandez, C.G.; Hamby, M.E.; McReynolds, M.L.; Ray, W.J. The Role of ApoE4 in Disrupting the Homeostatic Functions of Astrocytes and Microglia in Aging and Alzheimer’s Disease. Front. Aging Neurosci. 2019, 10, 14.
- Wisniewski, T.; Drummond, E. APOE-Amyloid Interaction: Therapeutic Targets. Neurobiol. Dis. 2020, 138, 104784.
- Smith, L.M.; Strittmatter, S.M. Binding Sites for Amyloid-β Oligomers and Synaptic Toxicity. Cold Spring Harb. Perspect. Med. 2017, 7, a024075.
- Um, J.W.; Kaufman, A.C.; Kostylev, M.; Heiss, J.K.; Stagi, M.; Takahashi, H.; Kerrisk, M.E.; Vortmeyer, A.; Wisniewski, T.; Koleske, A.J.; et al. Metabotropic Glutamate Receptor 5 Is a Coreceptor for Alzheimer Aβ Oligomer Bound to Cellular Prion Protein. Neuron 2013, 79, 887–902.
- Spurrier, J.; Nicholson, L.; Fang, X.T.; Stoner, A.J.; Toyonaga, T.; Holden, D.; Sieger, T.R.; Laird, W.; Allnutt, M.A.; Chiasseu, M.; et al. Reversal of Synapse Loss in Alzheimer Mouse Models by Targeting MGluR5 to Prevent Synaptic Tagging by C1Q. Sci. Transl. Med. 2022, 14, eabi8593.
- Salazar, S.V.; Gallardo, C.; Kaufman, A.C.; Herber, C.S.; Haas, L.T.; Robinson, S.; Manson, J.C.; Lee, M.K.; Strittmatter, S.M. Conditional Deletion of Prnp Rescues Behavioral and Synaptic Deficits after Disease Onset in Transgenic Alzheimer’s Disease. J. Neurosci. 2017, 37, 9207–9221.
More
Information
Subjects:
Clinical Neurology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.9K
Entry Collection:
Neurodegeneration
Revisions:
2 times
(View History)
Update Date:
06 Apr 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




