
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ivana Viola | -- | 1684 | 2023-03-31 18:18:28 | | | |
| 2 | Jessie Wu | + 138 word(s) | 1822 | 2023-04-03 05:19:49 | | |
Video Upload Options
TEOSINTE BRANCHED1, CYCLOIDEA, PROLIFERATING CELL FACTOR 1 and 2 (TCP) proteins constitute a plant-specific transcription factors family exerting effects on multiple aspects of plant development, such as germination, embryogenesis, leaf and flower morphogenesis, and pollen development, through the recruitment of other factors and the modulation of different hormonal pathways. They are divided into two main classes, I and II. The roles of class TCPs from class I in plant growth and development, as well as the modulation of their activity through interaction with other proteins and redox interconversions, proteolytic processing, or intra- or intercellular movement are discussed. Additionally, the function of these proteins in response to different environmental conditions is discussed.
1. Introduction
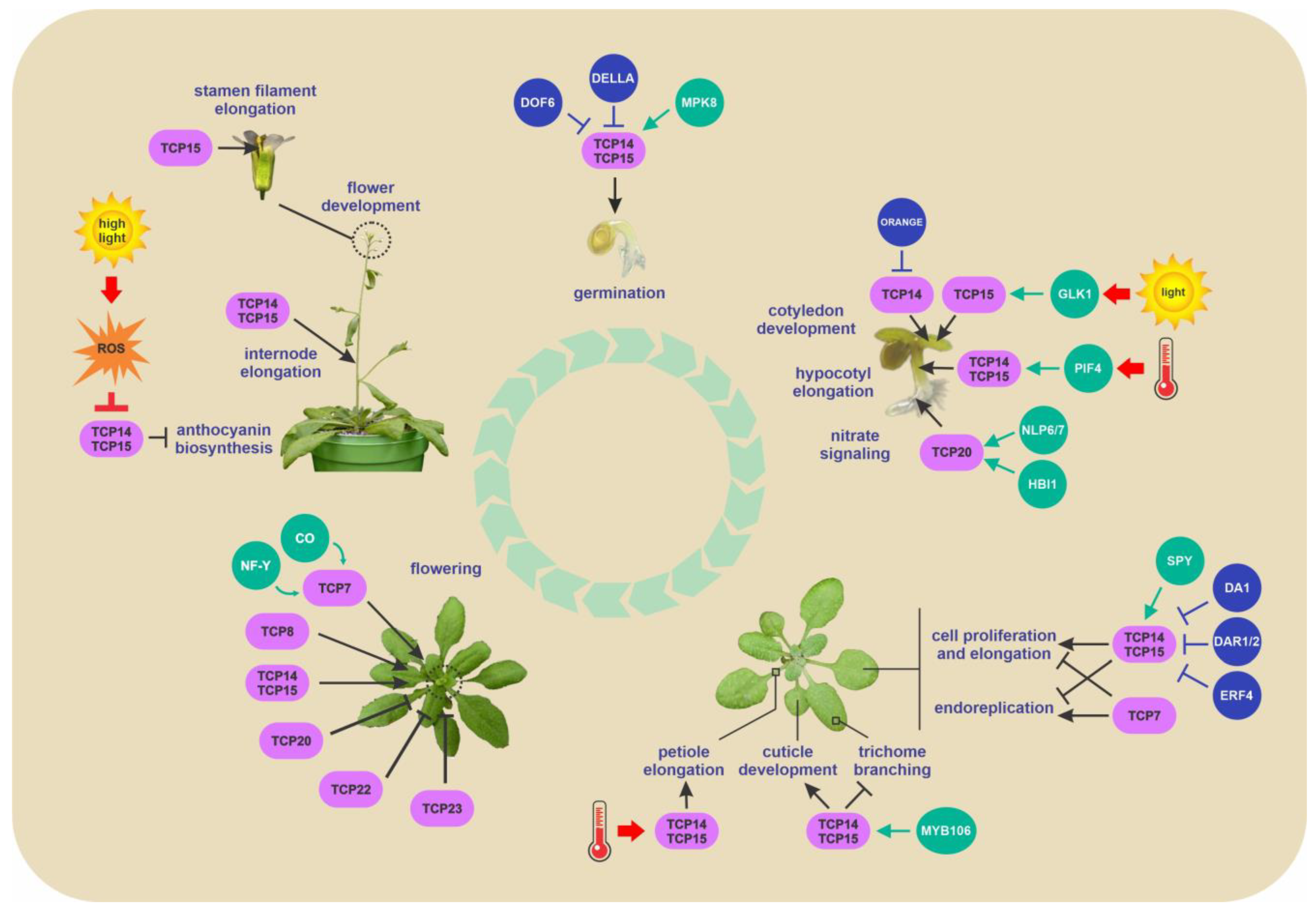
2. Interaction with Non-TEOSINTE BRANCHED1, CYCLOIDEA, PROLIFERATING CELL FACTOR 1 and 2 Transcriptional Regulators
3. Class I TEOSINTE BRANCHED1, CYCLOIDEA, PROLIFERATING CELL FACTOR 1 and 2 Proteins in Redox Signaling
4. Subcellular Distribution of Class I TEOSINTE BRANCHED1, CYCLOIDEA, PROLIFERATING CELL FACTOR 1 and 2 Proteins
References
- Yang, L.; Teixeira, P.J.P.L.; Biswas, S.; Finkel, O.M.; He, Y.; Salas-Gonzalez, I.; English, M.E.; Epple, P.; Mieczkowski, P.; Dangl, J.L. Pseudomonas Syringae Type III Effector HopBB1 Promotes Host Transcriptional Repressor Degradation to Regulate Phytohormone Responses and Virulence. Cell Host Microbe 2017, 21, 156–168.
- Peng, Y.; Chen, L.; Lu, Y.; Wu, Y.; Dumenil, J.; Zhu, Z.; Bevan, M.W.; Lia, Y. The Ubiquitin Receptors DA1, DAR1, and DAR2 Redundantly Regulate Endoreduplication by Modulating the Stability of TCP14/15 in Arabidopsis. Plant Cell 2015, 27, 649–662.
- Dong, H.; Dumenil, J.; Lu, F.H.; Na, L.; Vanhaeren, H.; Naumann, C.; Klecker, M.; Prior, R.; Smith, C.; McKenzie, N.; et al. Ubiquitylation Activates a Peptidase That Promotes Cleavage and Destabilization of Its Activating E3 Ligases and Diverse Growth Regulatory Proteins to Limit Cell Proliferation in Arabidopsis. Genes Dev. 2017, 31, 197–208.
- Steiner, E.; Efroni, I.; Gopalraj, M.; Saathoff, K.; Tseng, T.S.; Kieffer, M.; Eshed, Y.; Olszewski, N.; Weiss, D. The Arabidopsis O-Linked N-Acetylglucosamine Transferase SPINDLY Interacts with Class I TCPs to Facilitate Cytokinin Responses in Leaves and Flowers. Plant Cell 2012, 24, 96–108.
- Steiner, E.; Livne, S.; Kobinson-Katz, T.; Tal, L.; Pri-Tal, O.; Mosquna, A.; Tarkowská, D.; Mueller, B.; Tarkowski, P.; Weiss, D. The Putative O-Linked N-Acetylglucosamine Transferase SPINDLY Inhibits Class I TCP Proteolysis to Promote Sensitivity to Cytokinin. Plant Physiol. 2000, 171, 1485–1494.
- Steiner, E.; Triana, M.R.; Kubasi, S.; Blum, S.; Paz-Ares, J.; Rubio, V.; Weiss, D. KISS ME DEADLY F-Box Proteins Modulate Cytokinin Responses by Targeting the Transcription Factor TCP14 for Degradation. Plant Physiol. 2021, 185, 1495–1499.
- Zentella, R.; Sui, N.; Barnhill, B.; Hsieh, W.P.; Hu, J.; Shabanowitz, J.; Boyce, M.; Olszewski, N.E.; Zhou, P.; Hunt, D.F.; et al. The Arabidopsis O-Fucosyltransferase SPINDLY Activates Nuclear Growth Repressor Della. Nat. Chem. Biol. 2017, 13, 479–485.
- Wang, Y.; He, Y.; Su, C.; Zentella, R.; Sun, T.; Wang, L. Nuclear Localized O-Fucosyltransferase SPY Facilitates PRR5 Proteolysis to Fine-Tune the Pace of Arabidopsis Circadian Clock. Mol. Plant 2020, 13, 446–458.
- Liang, L.; Wang, Q.; Song, Z.; Wu, Y.; Liang, Q.; Wang, Q.; Yang, J.; Bi, Y.; Zhou, W.; Fan, L.M. O-Fucosylation of CPN20 by SPINDLY Derepresses Abscisic Acid Signaling During Seed Germination and Seedling Development. Front. Plant Sci. 2021, 12, 724144.
- Zhang, W.; Cochet, F.; Ponnaiah, M.; Lebreton, S.; Matheron, L.; Pionneau, C.; Boudsocq, M.; Resentini, F.; Huguet, S.; Blázquez, M.; et al. The MPK8-TCP14 Pathway Promotes Seed Germination in Arabidopsis. Plant J. 2019, 100, 677–692.
- Mo, W.; Zhang, J.; Zhang, L.; Yang, Z.; Yang, L.; Yao, N.; Xiao, Y.; Li, T.; Li, Y.; Zhang, G.; et al. Arabidopsis Cryptochrome 2 Forms Photobodies with TCP22 under Blue Light and Regulates the Circadian Clock. Nat. Commun. 2022, 13, 2631.
- Wang, Z.; Cui, D.; Liu, C.; Zhao, J.; Liu, J.; Liu, N.; Tang, D.; Hu, Y. TCP transcription factors interact with ZED1-related kinases as components of the temperature-regulated immunity. Plant Cell Environ. 2019, 42, 2045–2056.
- Wang, K.; Zhang, N.; Fu, X.; Zhang, H.; Liu, S.; Pu, X.; Wang, X.; Si, H. StTCP15 Regulates Potato Tuber Sprouting by Modulating the Dynamic Balance between Abscisic Acid and Gibberellic Acid. Front. Plant Sci. 2022, 13, 1009552.
- Xu, S.L.; Chalkley, R.J.; Maynard, J.C.; Wang, W.; Ni, W.; Jiang, X.; Shin, K.; Cheng, L.; Savage, D.; Hühmer, A.F.R.; et al. Proteomic Analysis Reveals O-GlcNAc Modification on Proteins with Key Regulatory Functions in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E1536–E1543.
- Purayannur, S.; Kumar, K.; Kaladhar, V.C.; Verma, P.K. Phylogenomic Analysis of MKKs and MAPKs from 16 Legumes and Detection of Interacting Pairs in Chickpea Divulge MAPK Signalling Modules. Sci. Rep. 2017, 7, 5026.
- Zhang, N.; Wang, Z.; Bao, Z.; Yang, L.; Wu, D.; Shu, X.; Hua, J. MOS1 Functions Closely with TCP Transcription Factors to Modulate Immunity and Cell Cycle in Arabidopsis. Plant J. 2018, 93, 66–78.
- Li, X.; Zhang, G.; Liang, Y.; Hu, L.; Zhu, B.; Qi, D.; Cui, S.; Zhao, H. TCP7 Interacts with Nuclear Factor-Ys to Promote Flowering by Directly Regulating SOC1 in Arabidopsis. Plant J. 2021, 108, 1493–1506.
- Alem, A.L.; Ariel, F.D.; Cho, Y.; Hong, J.C.; Gonzalez, D.H.; Viola, I.L. TCP15 Interacts with GOLDEN2-LIKE 1 to Control Cotyledon Opening in Arabidopsis. Plant J. 2022, 110, 748–763.
- Ma, Y.-N.; Xu, D.-B.; Li, L.; Zhang, F.; Fu, X.-Q.; Shen, Q.; Lyu, X.-Y.; Wu, Z.-K.; Pan, Q.-F.; Shi, P.; et al. Jasmonate Promotes Artemisinin Biosynthesis by Activating the TCP14-ORA Complex in Artemisia annua. Sci. Adv. 2018, 4, eaas9357.
- Wu, J.-F.; Tsai, H.-L.; Joanito, I.; Wu, Y.-C.; Chang, C.-W.; Li, Y.-H.; Wang, Y.; Hong, J.C.; Chu, J.-W.; Hsu, C.-P.; et al. LWD–TCP Complex Activates the Morning Gene CCA1 in Arabidopsis. Nat. Commun. 2016, 7, 13181.
- Li, M.; Chen, H.; Chen, J.; Chang, M.; Palmer, I.A.; Gassmann, W.; Liu, F.; Fu, Z.Q. Tcp Transcription Factors Interact with Npr1 and Contribute Redundantly to Systemic Acquired Resistance. Front. Plant Sci. 2018, 9, 1153.
- Chu, X.; Li, M.; Zhang, S.; Fan, M.; Han, C.; Xiang, F.; Li, G.; Wang, Y.; Xiang, C.; Wang, J.; et al. HBI1-TCP20 Interaction Positively Regulates the CEPs-mediated Systemic Nitrate Acquisition. J. Integr. Plant Biol. 2021, 63, 902–912.
- Resentini, F.; Felipo-Benavent, A.; Colombo, L.; Blázquez, M.A.; Alabadí, D.; Masiero, S. TCP14 and TCP15 Mediate the Promotion of Seed Germination by Gibberellins in Arabidopsis thaliana. Mol. Plant 2015, 8, 482–485.
- Ding, A.-M.; Xu, C.-T.; Xie, Q.; Zhang, M.-J.; Yan, N.; Dai, C.-B.; Lv, J.; Cui, M.-M.; Wang, W.-F.; Sun, Y.-H. ERF4 Interacts with and Antagonizes TCP15 in Regulating Endoreduplication and Cell Growth in Arabidopsis. J. Integr. Plant Biol. 2022, 64, 1673–1689.
- Sun, T.; Zhou, F.; Huang, X.-Q.; Chen, W.-C.; Kong, M.-J.; Zhou, C.-F.; Zhuang, Z.; Li, L.; Lu, S. ORANGE Represses Chloroplast Biogenesis in Etiolated Arabidopsis Cotyledons via Interaction with TCP14. Plant Cell 2019, 31, 2996–3014.
- Wang, Q.; Xu, G.; Zhao, X.; Zhang, Z.; Wang, X.; Liu, X.; Xiao, W.; Fu, X.; Chen, X.; Gao, D.; et al. Transcription Factor TCP20 Regulates Peach Bud Endodormancy by Inhibiting DAM5/DAM6 and Interacting with ABF2. J. Exp. Bot. 2020, 71, 1585–1597.
- Wu, J.; Wu, W.; Liang, J.; Jin, Y.; Gazzarrini, S.; He, J.; Yi, M. GhTCP19 Transcription Factor Regulates Corm Dormancy Release by Repressing GhNCED Expression in Gladiolus. Plant Cell Physiol. 2019, 60, 52–62.
- Masuda, H.P.; Cabral, L.M.; De Veylder, L.; Tanurdzic, M.; De Almeida Engler, J.; Geelen, D.; Inzé, D.; Martienssen, R.A.; Ferreira, P.C.G.; Hemerly, A.S. ABAP1 Is a Novel Plant Armadillo BTB Protein Involved in DNA Replication and Transcription. EMBO J. 2008, 27, 2746–2756.
- Pruneda-Paz, J.L.; Breton, G.; Para, A.; Kay, S.A. A Functional Genomics Approach Reveals CHE as a Component of the Arabidopsis Circadian Clock. Science 2009, 323, 1481–1485.
- Liu, Y.J.; An, J.P.; Gao, N.; Wang, X.; Chen, X.X.; Wang, X.F.; Zhang, S.; You, C.X. MdTCP46 interacts with MdABI5 to negatively regulate ABA signalling and drought response in apple. Plant Cell Environ. 2022, 45, 3233–3248.
- Chen, G.-H.; Sun, J.-Y.; Liu, M.; Liu, J.; Yang, W.-C. SPOROCYTELESS Is a Novel Embryophyte-Specific Transcription Repressor That Interacts with TPL and TCP Proteins in Arabidopsis. J. Genet. Genom. 2014, 41, 617–625.
- Viola, I.L.; Camoirano, A.; Gonzalez, D.H. Redox-Dependent Modulation of Anthocyanin Biosynthesis by the TCP Transcription Factor TCP15 during Exposure to High Light Intensity Conditions in Arabidopsis. Plant Physiol. 2016, 170, 74–85.
- Viola, I.L.; Güttlein, L.N.; Gonzalez, D.H. Redox Modulation of Plant Developmental Regulators from the Class I TCP Transcription Factor Family. Plant Physiol. 2013, 162, 1434–1447.
- Busch, A.; Deckena, M.; Almeida-Trapp, M.; Kopischke, S.; Kock, C.; Sch€ Ussler, E.; Tsiantis, M.; Mith€ Ofer, A.; Zachgo, S. MpTCP1 Controls Cell Proliferation and Redox Processes in Marchantia Polymorpha. New Phytol. 2019, 224, 1627–1641.
- Valsecchi, I.; Guittard-Crilat, E.; Maldiney, R.; Habricot, Y.; Lignon, S.; Lebrun, R.; Miginiac, E.; Ruelland, E.; Jeannette, E.; Lebreton, S. The Intrinsically Disordered C-Terminal Region of Arabidopsis Thaliana TCP8 Transcription Factor Acts Both as a Transactivation and Self-Assembly Domain. Mol. Biosyst. 2013, 9, 2282.
- Willig, J.J.; Guarneri, N.; van Steenbrugge, J.J.M.; de Jong, W.; Chen, J.; Goverse, A.; Lozano Torres, J.L.; Sterken, M.G.; Bakker, J.; Smant, G. The Arabidopsis Transcription Factor TCP9 Modulates Root Architectural Plasticity, Reactive Oxygen Species-Mediated Processes, and Tolerance to Cyst Nematode Infections. Plant J. 2022, 112, 1070–1083.
- Liu, H.; Gao, Y.; Wu, M.; Shi, Y.; Wang, H.; Wu, L.; Xiang, Y. TCP10, a TCP Transcription Factor in Moso Bamboo (Phyllostachys edulis), Confers Drought Tolerance to Transgenic Plants. Environ. Exp. Bot. 2020, 172, 104002.
- Xu, Y.; Liu, H.; Gao, Y.; Xiong, R.; Wu, M.; Zhang, K.; Xiang, Y. The TCP Transcription Factor PeTCP10 Modulates Salt Tolerance in Transgenic Arabidopsis. Plant Cell Rep. 2021, 40, 1971–1987.
- Schippers, J.H.; Foyer, C.H.; van Dongen, J.T. Redox Regulation in Shoot Growth, SAM Maintenance and Flowering. Curr. Opin. Plant Biol. 2016, 29, 121–128.
- Eljebbawi, A.; del Guerrero, Y.C.R.; Dunand, C.; Estevez, J.M. Highlighting Reactive Oxygen Species as Multitaskers in Root Development. iScience 2021, 24, 101978.
- Kim, S.H.; Son, G.H.; Bhattacharjee, S.; Kim, H.J.; Nam, J.C.; Nguyen, P.D.T.; Hong, J.C.; Gassmann, W. The Arabidopsis Immune Adaptor SRFR1 Interacts with TCP Transcription Factors That Redundantly Contribute to Effector-Triggered Immunity. Plant J. 2014, 78, 978–989.
- Mazur, M.J.; Spears, B.J.; Djajasaputra, A.; Van Der Gragt, M.; Vlachakis, G.; Beerens, B.; Gassmann, W.; Van Den Burg, H.A. Arabidopsis TCP Transcription Factors Interact with the SUMO Conjugating Machinery in Nuclear Foci. Front. Plant Sci. 2017, 8, 1–18.
- Hammani, K.; Gobert, A.; Hleibieh, K.; Choulier, L.; Small, I.; Giegé, P. An Arabidopsis Dual-Localized Pentatricopeptide Repeat Protein Interacts with Nuclear Proteins Involved in Gene Expression Regulation. Plant Cell 2011, 23, 730–740.
- Spears, B.J.; McInturf, S.A.; Collins, C.; Chlebowski, M.; Cseke, L.J.; Su, J.; Mendoza-Cózatl, D.G.; Gassmann, W. Class I TCP Transcription Factor AtTCP8 Modulates Key Brassinosteroid-Responsive Genes. Plant Physiol. 2022, 190, 1457–1473.
- Camoirano, A.; Alem, A.L.; Gonzalez, D.H.; Viola, I.L. Arabidopsis Thaliana TCP15 Interacts with the MIXTA-like Transcription Factor MYB106/NOECK. Plant Signal. Behav. 2021, 16, 1938432.
- Ferrero, L.V.; Viola, I.L.; Ariel, F.D.; Gonzalez, D.H. Class I TCP Transcription Factors Target the Gibberellin Biosynthesis Gene GA20ox1 and the Growth-Promoting Genes HBI1 and PRE6 during Thermomorphogenic Growth in Arabidopsis. Plant Cell Physiol. 2019, 60, 1633–1645.
- Perez, M.; Guerringue, Y.; Ranty, B.; Pouzet, C.; Jauneau, A.; Robe, E.; Mazars, C.; Galaud, J.P.; Aldon, D. Specific TCP Transcription Factors Interact with and Stabilize PRR2 within Different Nuclear Sub-Domains. Plant Sci. 2019, 287, 110197.
- Karaaslan, E.S.; Wang, N.; Faiß, N.; Liang, Y.; Montgomery, S.A.; Laubinger, S.; Berendzen, K.W.; Berger, F.; Breuninger, H.; Liu, C. Marchantia TCP Transcription Factor Activity Correlates with Three-Dimensional Chromatin Structure. Nat. Plants 2020, 6, 1250–1261.
- Guan, P.; Ripoll, J.J.; Wang, R.; Vuong, L.; Bailey-Steinitz, L.J.; Ye, D.; Crawford, N.M. Interacting TCP and NLP Transcription Factors Control Plant Responses to Nitrate Availability. Proc. Natl. Acad. Sci. USA 2017, 114, 2419–2424.




