
Video Upload Options
Rice is one of the most economically important staple food crops in the world. Soil salinization and drought seriously restrict sustainable rice production. Drought aggravates the degree of soil salinization, and, at the same time, increased soil salinity also inhibits water absorption, resulting in physiological drought stress. Salt tolerance in rice is a complex quantitative trait controlled by multiple genes. The recent research developments on salt stress impact on rice growth, rice salt tolerance mechanisms, the identification and selection of salt-tolerant rice resources, and strategies to improve rice salt tolerance are discussed. The increased cultivation of water-saving and drought-resistance rice (WDR) has shown great application potential in alleviating the water resource crisis and ensuring food and ecological security. An innovative germplasm selection strategy of salt-tolerant WDR is presented, using a population that is developed by recurrent selection based on dominant genic male sterility. The efficient genetic improvement can be referenced and germplasm innovation of complex traits (drought and salt tolerance) that can be translated into breeding all economically important cereal crops.
1. Salt Tolerance Mechanisms
1.1. Osmotic Adjustment
1.2. Ionic Homeostasis Mechanisms
1.3. Resistance to Oxidative Stress
1.4. Signal Molecules
2. Strategies for Salt Tolerance Improvement in Rice
2.1. Conventional Breeding
2.2. Molecular Marker-Assisted Selection (MAS)
2.3. Genome Editing
3. Breeding of Salt-Tolerant Drought-Resistance Rice
For most plant breeders and physiologists, yield loss under stress is the most meaningful measurement of drought resistance or salt tolerance. Rice plants may adapt to drought or salinity through complex physiological or molecular mechanisms. These are very important for identifying specific traits related to drought resistance or salt tolerance and developing the appropriate screening techniques in breeding programs [57].
Drought resistance is a complex quantitative trait, including at least two mechanisms—drought avoidance and drought tolerance. Drought avoidance refers to plants’ ability to absorb or reduce water loss in water-deficient environments. This process requires water absorption through a sophisticated root system and water transport to the aboveground tissues of plants. Water consumption can also be reduced by moderately closed stomata or thickened cuticles. Drought tolerance refers to the ability of plants to maintain physiological and metabolic activities under water stress when leaf water potential decreases. Drought-tolerant plants actively accumulate osmolytes in the cells to increase their osmotic adjustment capacity and maintain high turgor pressure. In addition, they can also improve the ability to remove harmful compounds that are accumulating, and thus resist oxidative stress under drought conditions [58]. In fact, the drought resistance of plants is often a combined effect of the above properties. Drought avoidance performs a major role, and drought tolerance is considered the second line of defense for drought resistance [59].
Drought resistance is the result of a systemic network of interactions between multiple drought-resistance genes rather than the effect of a single drought-resistance gene. As mentioned in Pennisi [60], among numerous candidate drought-resistance genes revealed by genomics studies, almost none has significant impact on crop performance under field conditions, indicating that the research progress of molecular breeding on rice drought-resistance is slow. Therefore, selecting a breeding strategy that aggregates genes with different drought-resistance mechanisms is necessary. WDR is a newly cultivated rice type developed by introducing upland rice’s water-saving and drought-resistance characteristics based on technological progress in rice breeding science [58]. Wei et al. [61] also confirmed that drought-resistance genes and their networks could be transmitted in the offspring through conventional hybridization combined with high-intensity stress selections in the target environment. By using this strategy of crossing lowland rice and upland rice combined with selection under severe stress in the target environment, more than 20 new varieties of WDR have been generated. These WDR varieties are planted in dry and irrigated fields in China. Several varieties are successfully cultivated in Asian and African countries, and the planting area in China exceeds one million hectares [62][63]. The successful breeding of WDR provides a feasible solution for rice drought-resistance breeding.
The molecular and physiological mechanisms of plants’ responses to drought and salt stress exhibit similarities. Salt stress can cause ion and metabolic imbalance due to plant ion toxicity. Another salinity component is high osmotic stress, which leads to a water deficit similar to that caused by drought stress. Plants generally counteract the negative effects of salinity and drought by activating biochemical reactions, such as osmolytes synthesis and accumulation, intracellular ion homeostasis maintenance, and reactive oxygen species scavenging (Figure 1) [64][65]. Osmolytes performs important roles in plant response to abiotic stress. Enhancing the biosynthesis of trehalose can increase yield potential in marker-free transgenic rice under drought and salt conditions [66]. Heat shock proteins (Hsps) were also involved in osmotic adjustment. Overexpression of OsHsp17.0 and OsHsp23.7 enhances drought and salt tolerance in rice [67]. A novel C2H2 zinc finger transcription factor, DST (drought and salt tolerance), that controls stomatal aperture under drought and salt stress in rice. DST contributes to stomata movement via regulation of genes involved in ROS homeostasis [54]. OsRLCK241 improved salt and drought tolerance in rice is mainly due to improved ROS detoxification, increased accumulation of osmolytes, and altered expression of stress-responsive genes [68].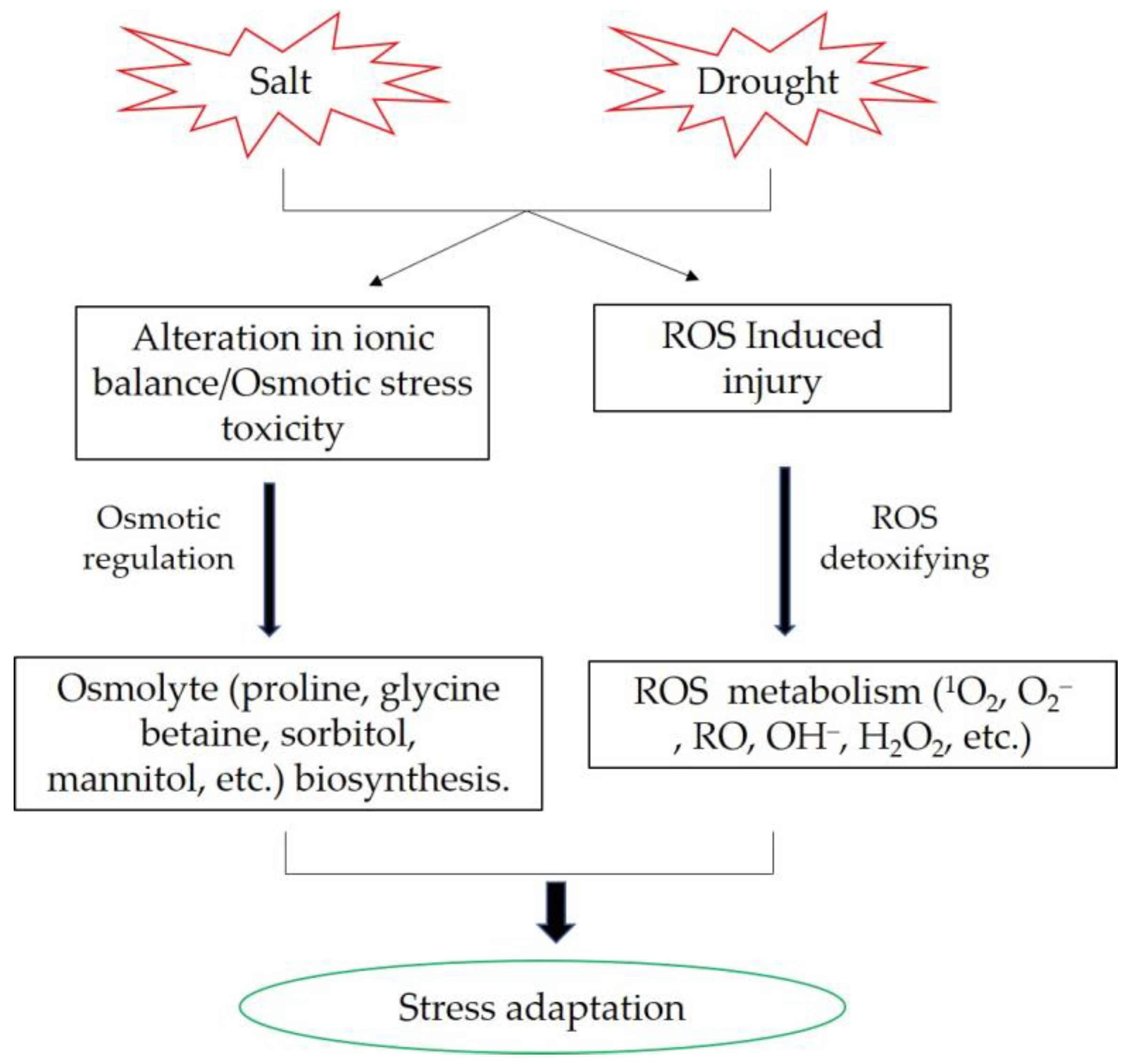
Traditional cross or backcross breeding involves only a few parents are involved and the opportunities for excellent gene recombination are limited, resulting in a narrow genetic base of the developed cultivars [69]. As drought and salt tolerance are highly complex and quantitative traits that are governed by multiple genes/QTLs, and there are lots of cryptic beneficial alleles [70]. It is difficult to use and pyramid all these genes through limited cross breeding.

- The dominant genic male sterile WDR rice lines can be generated by crossing more than five rounds of backcrossing with the dominant genic male sterile lines (female parents) and the core WDR parents (male parents).
- F1 seeds are obtained by crossing the WDR genic male sterile lines (female parents) with genotypes, such as ‘Haidao 86’, ‘Dongxiang wild rice’, and salt-tolerant donors.
- F1 seeds are mixed and sowed, the rice plants are pollinated during the heading stage, and the seeds are harvested at maturity from the male sterile plants with decent agronomic traits to obtain the first-generation recurrent selection population.
- The first-generation recurrent selection population is planted in a dry land, relying only on rainfall throughout the growth period, without artificial irrigation. At the heading stage, artificial pollination is carried out. The seeds from the elite male sterile plants are harvested at the maturity stage to obtain the generation recurrent selection population.
- The second generation of the recurrent selection population is planted in saline-alkali land or coastal land, and the seeds from the elite male sterile plants are harvested to obtain the third generation of the recurrent selection population.
- According to the breeding objectives and selection pressure, different selection environments can be set to obtain recurrent selection populations, or fertile individual plants that meet the breeding objectives can be selected to conduct conventional breeding programs.
4. Conclusions and Prospects
Soil salt stress threatens rice production and global food security. Cultivating salt-tolerant rice varieties is the most effective approach to overcome this environmental hurdle. However, rice salt tolerance mechanisms are highly complex, involving various physiological and biochemical pathways. It is challenging to improve quantitative traits controlled by multiple genes through traditional breeding methods; thus, the current progress is slow. MAS and genetic engineering approaches can accelerate the process of breeding salt-tolerant rice varieties. However, most existing research has focused on transferring a single salt-tolerance QTL—Saltol, resulting in a simple genetic basis of salt tolerance in the bred varieties. Thus, it is difficult for these varieties to adapt to different types of saline land environments, which limits their large-scale cultivation. In addition, obtaining salt-tolerant varieties that can be used in field production by introducing a single gene or a few related genes is challenging.
Therefore, the effective generation of salt-tolerant rice varieties translated to increased field productivity may require the simultaneous introduction of multiple key genes, which allows the genetic improvement on multiple pathways of the salt-tolerance regulatory networks. This requires a breakthrough in breeding methods. The development of genome editing technologies makes it a routine operation to mutate multiple target genes simultaneously. Studies have shown that genes negatively regulating salt tolerance in rice involve different salt-tolerance pathways. Therefore, simultaneously knocking out multiple genes negatively regulating salt tolerance can integrate multiple salt-tolerance genes. This process is expected to stack the effects of different salt-tolerance pathways and achieve the generation of rice lines with stronger salt tolerance, suitable for cultivation in the field.
Researchers further introduced the development of WDR by crossing lowland rice and upland rice combined with selection under severe stress conditions in the target environment. The successful development of WDR provides a feasible technical scheme and concept for the improvement of complex traits (drought resistance and salt tolerance) in rice cultivars. The breeding approaches towards the generation of salt-tolerant and WDR lines show that recurrent selection is an effective method for crop population improvement and the generation of lines with novel traits, especially regarding the simultaneous improvement of multiple complex agronomic traits.References
- Lin, H.; Zhu, M.; Gao, G.; Liang, Z.; Yano, M.; Su, W.; Hu, X.; Ren, Z.; Chao, D. QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theor. Appl. Genet. 2004, 108, 253–260.
- Garg, A.K. Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc. Natl. Acad. Sci. USA 2003, 99, 15898–15903.
- Vinocur, B.; Altman, A. Recent advances in engineering plant tolerance to abiotic stress: Achievements and limitations. Curr. Opin. Biotechnol. 2005, 16, 123–132.
- Sripinyowanich, S.; Klomsaku, L.P.; Boonburapong, B.; Bangyeekhun, T.; Asami, T.; Gu, H.; Buaboocha, T.; Chadchawan, E. Exogenous ABA induces salt tolerance in indica rice (Oryza sativa L.): The role of Os P5CS1 and Os P5CR gene expression during salt stress. Environ. Exp. Bot. 2013, 86, 94–105.
- Yu, J.; Li, Y.; Tang, W.; Liu, J.; Lu, B.; Liu, Y. The Accumulation of Glycine Betaine Is Dependent on Choline Monooxygenase (OsCMO), Not on Phosphoethanolamine N-Methyltransferase (OsPEAMT1), in Rice (Oryza sativa L. ssp. japonica). Plant. Mol. Biol. Rep. 2014, 32, 916–922.
- Hasthanasombut, S.; Supaibulwatana, K.; Mii, M.; Nakamura, I. Genetic manipulation of Japonica rice using the OsBADH1 gene from Indica rice to improve salinity tolerance. Plant Cell 2011, 104, 79–89.
- Li, H.; Zang, B.; Deng, X.; Wang, X. Overexpression of the trehalose-6-phosphate synthase gene OsTPS1 enhances abiotic stress tolerance in rice. Planta 2011, 234, 1007–1018.
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.; Bohnert, H.J. Plant Cellular and Molecular Responses To High Salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 463–499.
- Sun, J.; Chen, S.; Dai, S.; Wang, R.; Li, N.; Shen, X.; Zhou, X.; Lu, C.; Zheng, X.; Hu, Z.; et al. Ion flux profiles and plant ion homeostasis control under salt stress. Plant Signal Behav. 2009, 4, 261–264.
- Hauser, F.; Horie, T. A conserved primary salt tolerance mechanism mediated by HKT transporters: A mechanism for sodium exclusion and maintenance of high K+/Na+ ratio in leaves during salinity stress. Plant Cell Environ. 2010, 33, 552–565.
- Ren, Z.; Gao, J.; Li, L.; Cai, X.; Huang, W.; Chao, D.; Zhu, M.; Wang, Z.; Luan, S.; Lin, H. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 2005, 37, 1141–1146.
- Wang, H.; Zhang, M.; Guo, R.; Shi, D.; Liu, B.; Lin, X.; Yang, C. Effects of salt stress on ion balance and nitrogen metabolism of old and young leaves in rice (Oryza sativa L.). BMC Plant Biol. 2012, 12, 194.
- Martínez-Atienza, J.; Jiang, X.; Garciadeblas, B.; Mendoza, I.; Zhu, J.; Pardo, J.M.; Quintero, F.J. Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 2007, 143, 1001–1012.
- Quintero, F.J.; Martinez-Atienza, J.; Villalta, I.; Jiang, X.; Kim, W.Y.; Ali, Z.; Fujii, H.; Mendoza, I.; Yun, D.; Zhu, J.; et al. Activation of the plasma membrane Na/H antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc. Natl. Acad. Sci. USA 2011, 108, 2611–2616.
- Bassil, E.; Coku, A.; Blumwald, E. Cellular ion homeostasis: Emerging roles of intracellular NHX Na+/H+ antiporters in plant growth and development. J. Exp. Bot. 2012, 63, 5727–5740.
- Liu, S.; Zheng, L.; Xue, Y.; Zhang, Q.; Wang, L.; Shuo, H. Overexpression of OsVP1 and OsNHX1 increases tolerance to drought and salinity in rice. J. Plant Biol. 2010, 53, 444–452.
- Fukuda, A.; Nakamura, A.; Hara, N.; Toki, S.; Tanaka, Y. Molecular and functional analyses of rice NHX-type Na+/H+ antiporter genes. Planta 2011, 233, 175–188.
- Ashraf, M. Biotechnological approach of improving plant salt tolerance using antioxidants as markers. Biotechnol. Adv. 2009, 27, 84–93.
- Shigeoka, S.; Ishikawa, T.; Tamoi, M.; Miyagawa, Y.; Takeda, T.; Yabuta, Y.; Yoshimura, K. Regulation and function of ascorbate peroxidase isoenzymes. J. Exp. Bot. 2002, 53, 1305–1319.
- Wu, T.; Lin, W.; Kao, C.; Hong, C. Gene knockout of glutathione reductase 3 results in increased sensitivity to salt stress in rice. Plant Mol. Biol. 2015, 87, 555–564.
- Zhang, Z.; Zhang, Q.; Wu, J.; Zheng, X.; Zheng, S.; Sun, X.; Qiu, Q.; Lu, T. Gene knockout study reveals that cytosolic ascorbate peroxidase 2 (OsAPX2) plays a critical role in growth and reproduction in rice under drought, salt and cold stresses. PLoS ONE 2013, 8, e57472.
- Dodd, A.N.; Kudla, J.; Sanders, D. The language of calcium signaling. Annu. Rev. Plant Biol. 2010, 61, 593–620.
- Jiang, Z.; Zhou, X.; Tao, M.; Yuan, F.; Liu, L.; Wu, F.; Wu, X.; Xiang, Y.; Niu, Y.; Liu, F.; et al. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature 2019, 572, 341–346.
- Ma, L.; Ye, J.; Yang, Y.; Lin, H.; Yue, L.; Luo, J.; Long, Y.; Fu, H.; Liu, X.; Zhang, Y.; et al. The SOS2-SCaBP8 complex generates and finetunes an AtANN4-dependent calcium signature under salt stress. Dev. Cell 2019, 48, 697–709.
- Yang, Z.; Wang, C.; Xue, Y.; Liu, X.; Chen, S.; Song, C.; Yang, Y.; Guo, Y. Calcium-activated 14-3-3 proteins as a molecular switch in salt stress tolerance. Nat. Commun. 2019, 10, 1199.
- Lou, L.; Yu, F.; Tian, M.; Liu, G.; Wu, Y.; Wu, Y.; Xia, R.; Pardo, J.; Guo, Y.; Xie, Q. ESCRT-I component VPS23A sustains salt tolerance by strengthening the SOS module in Arabidopsis. Mol. Plant 2020, 13, 1134–1148.
- Li, J.; Zhou, H.; Zhang, Y.; Li, Z.; Yang, Y.; Guo, Y. The GSK3-like kinase BIN2 is a molecular switch between the salt stress response and growth recovery in Arabidopsis thaliana. Dev. Cell 2020, 55, 367–380.
- Chen, T.; Shabala, S.; Niu, Y.; Chen, Z.; Shabala, L.; Meinke, H.; Venkataraman, G.; Pareek, A.; Xu, J.; Zhou, M. Molecular mechanisms of salinity tolerance in rice. Crop J. 2021, 9, 506–520.
- Song, Y.; Miao, Y.; Song, C. Behind the scenes: The roles of reactive oxygen species in guard cells. New Phytol. 2014, 201, 1121–1140.
- Das, P.; Nutan, K.K.; Singla-Pareek, S.L.; Pareek, A. Understanding salinity responses and adopting ‘omics-based’ approaches to generate salinity tolerant cultivars of rice. Front. Plant Sci. 2015, 6, 712.
- Yuan, L. Saline-Alkaline Tolerance Rice Breeding Technology; Shandong Technology Press: Jinan, China, 2019; pp. 121–131.
- Das, P.; Mishra, M.; Lakra, N.; Singla-Pareek, S.L.; Pareek, A. Mutation breeding: A powerful approach for obtaining abiotic stress tolerant crops and upgrading food security for human nutrition. In Mutagenesis: Exploring Novel Genes and Pathways; Tomlekova, N.B., Kozgar, M.I., Wani, M.R., Eds.; Wageningen Academic Publisher: Wageningen, The Netherlands, 2014; pp. 615–621.
- Mustafa, G.; Soomro, A.M.; Baloch, A.W.; Siddiqui, K.A. “Shua-92,” a new cultivar of rice (Oryza sativa L.) developed through fast neutrons irradiation. Mutat. Breed. Newsl. 1997, 43, 35–36.
- Saleem, M.Y.; Mukhtar, Z.; Cheema, A.A.; Atta, B.M. Induced mutation and in vitro techniques as a method to induce salt tolerance in Basmati rice (Oryza sativa L.). Int. J. Environ. Sci. Technol. 2005, 2, 141–145.
- Takagi, H.; Tamiru, M.; Abe, A.; Yoshida, K.; Uemura, A.; Yaegashi, H.; Obara, T.; Oikawa, K.; Utsushi, H.; Kanzaki, E.; et al. MutMap accelerates breeding of a salt-tolerant rice cultivar. Nat. Biotechnol. 2015, 33, 445–449.
- Ashraf, M.Y.; Wu, L. Breeding for salinity tolerance in plants. Crit. Rev. Plant Sci. 1994, 13, 17–42.
- Deng, P.; Jiang, D.; Dong, Y.; Shi, X.; Jing, W.; Zhang, W. Physiological characterization and fine mapping of salt-tolerant mutant in rice (Oryza sativa). Funct. Plant Biol. 2015, 42, 1026–1035.
- Ganapati, R.K.; Naveed, S.A.; Zafar, S.; Wang, W.; Xu, J. Saline-Alkali Tolerance in Rice: Physiological Response, Molecular Mechanism, and QTL Identification and Application to Breeding. Rice Sci. 2022, 29, 412–434.
- Singh, R.; Singh, Y.; Xalaxo, S.; Verulkar, S.; Yadav, N.; Singh, S.; Singh, N.; Prasad, K.S.N.; Kondayya, K.; Rao, P.V.R.; et al. From QTL to variety-harnessing the benefits of QTLs for drought, flood and salt tolerance in mega rice varieties of India through a multi-institutional network. Plant Sci. 2016, 242, 278–287.
- Vinod, K.K.; Krishnan, S.G.; Babu, N.N.; Nagarajan, M.; Singh, A.K. Improving salt tolerance in rice: Looking beyond the conventional. In Salt Stress in Plants: Signalling, Omics and Adaptations; Ahmad, P., Azooz, M.M., Prasas, M.N., Eds.; Springer: New York, NY, USA, 2013; pp. 219–260.
- Fan, X.; Jiang, H.; Meng, L.; Chen, J. Gene Mapping, Cloning and Association Analysis for Salt Tolerance in Rice. Int. J. Mol. Sci. 2021, 22, 11674.
- Gregorio, G.B.; Senadhira, D. Genetic analysis of salinity tolerance in rice. Theor. Appl. Genet. 1993, 86, 333–338.
- Qin, H.; Li, Y.; Huang, R. Advances and challenges in the breeding of salt-tolerant rice. Int. J. Mol. Sci. 2020, 21, 8385.
- Huyen, L.T.N.; Cuc, L.M.; Ismail, A.M.; Ham, L.H. Introgression the Salinity Tolerance QTLs Saltol into AS996, the Elite Rice Variety of Vietnam. Am. J. Plant Sci. 2012, 3, 981–987.
- Bimpong, I.K.; Manneh, B.; Sock, M.; Diaw, F.; Amoah, N.K.A.; Ismail, A.M.; Gregorio, G.; Singh, R.K.; Wopereis, M. Improving salt tolerance of lowland rice cultivar ‘Rassi’ through marker-aided backcross breeding in West Africa. Plant Sci. 2016, 242, 288–299.
- Singh, A.K.; Gopalakrishnan, S.; Singh, V.P.; Prabhu, K.V.; Mohapatra, T.; Singh, N.K.; Sharma, T.R.; Nagarajan, M.; Ellur, R.K.; Singh, A.; et al. Marker assisted selection: A paradigm shift in Basmati breeding. Indian J. Genet. Plant Breed. 2011, 71, 120.
- Geetha, S.; Vasuki, A.; Selvam, P.J.; Saraswathi, R.; Krishnamurthy, S.L.; Dhasarathan, M.; Thamodharan, G.; Baskar, M. Development of sodicity tolerant rice varieties through marker assisted backcross breeding. Electron. J. Plant Breed. 2017, 8, 1013.
- Chukwu, S.C.; Rafii, M.Y.; Ramlee, S.I.; Ismail, S.I.; Oladosu, Y.; Okporie, E.; Onyishi, G.; Utobo, E.; Ekwu, L.; Swaray, S.; et al. Marker-assisted selection and gene pyramiding for resistance to bacterial leaf blight disease of rice (Oryza sativa L.). Biotechnol. Biotechnol. Equip. 2019, 33, 440–455.
- Shailani, A.; Joshi, R.; Singla-Pareek, S.L.; Pareek, A. Stacking for future: Pyramiding genes to improve drought and salinity tolerance in rice. Physiol. Plant. 2021, 172, 1352–1362.
- Wu, F.; Yang, J.; Yu, D.; Xu, P. Identification and validation a major QTL from“Sea Rice 86” seedlings conferred salt tolerance. Agronomy 2020, 10, 410.
- Li, J.; Sun, Y.; Du, J.; Zhao, Y.; Xia, L. Generation of Targeted Point Mutations in Rice by a Modified CRISPR/Cas9 System. Mol. Plant 2017, 10, 526–529.
- Mishra, R.; Joshi, R.K.; Zhao, K. Genome Editing in Rice: Recent Advances, Challenges, and Future Implications. Front. Plant Sci. 2018, 9, 1361.
- Ponce, K.S.; Meng, L.; Guo, L.; Leng, Y.; Ye, G. Advances in sensing, response and regulation mechanism of salt tolerance in rice. Int. J. Mol. Sci. 2021, 22, 2254.
- Huang, X.; Chao, D.; Gao, J.; Zhu, M.; Shi, M.; Lin, H. A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 2009, 23, 1805–1817.
- Santosh-Kumar, V.V.; Verma, R.K.; Yadav, S.K.; Yadav, P.; Watts, A.; Rao, M.V.; Chinnusamy, V. CRISPR-Cas9 mediated genome editing of drought and salt tolerance (OsDST) gene in indica mega rice cultivar MTU1010. Physiol. Mol. Biol. Plants 2020, 26, 1099–1110.
- Zhang, A.; Liu, Y.; Wang, F.; Li, T.; Chen, Z.; Kong, D.; Bi, J.; Zhang, F.; Luo, X.; Wang, J.; et al. Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breed. 2019, 39, 47.
- Li, Z.K.; Xu, J.L. Breeding for Drought and Salt Tolerant Rice (Oryza Sativa L.): Progress and Perspectives. In Advances in Molecular Breeding Toward Drought and Salt Tolerant Crops; Jenks, M.A., Hasegawa, P.M., Jain, S.M., Eds.; Springer: Dordrecht, The Netherlands, 2007; pp. 531–564.
- Luo, L. Breeding for water-saving and drought-resistance rice (WDR) in China. J. Exp. Bot. 2010, 61, 3509–3517.
- Blum, A. Drought resistance, water-use efficiency, and yield potential–are they compatible, dissonant, or mutually exclusive? Aust. J. Agric. Res. 2005, 25, 1159–1168.
- Pennisi, E. The blue revolution, drop by drop, gene by gene. Science 2008, 320, 171–173.
- Wei, H.; Feng, F.; Lou, Q.; Xia, H.; Ma, X.; Liu, Y.; Xu, K.; Yu, X.; Mei, H.; Luo, L. Genetic determination of the enhanced drought resistance of rice maintainer HuHan2B by pedigree breeding. Sci. Rep. 2016, 6, 37302.
- Luo, L.; Mei, H.; Yu, X.; Xia, H.; Chen, L.; Liu, H.; Zhang, A.; Xu, K.; Wei, H.; Liu, G.; et al. Water-saving and drought-resistance rice: From the concept to practice and theory. Mol. Breed. 2019, 39, 145.
- Xia, H.; Zhang, X.; Liu, Y.; Bi, J.; Ma, X.; Zhang, A.; Liu, H.; Chen, L.; Zhou, S.; Gao, H.; et al. Blue revolution for food security under carbon neutrality: A case from the water-saving and drought-resistance rice. Mol. Plant 2022, 15, 1401–1404.
- Flowers, T.J. Improving crop salt tolerance. J. Exp. Bot. 2004, 55, 307–319.
- Ashraf, M.; Akram, N.A. Improving salinity tolerance of plants through conventional breeding and genetic engineering: An analytical comparison. Biotechnol. Adv. 2009, 27, 744–752.
- Joshi, R.; Sahoo, K.; Singh, A.; Anwar, K.; Pundir, P.; Gautam, R.; Krishnamurthy, S.; Sopory, S.; Pareek, A.; Singla-Pareek, S. Enhancing trehalose biosynthesis improves yield potential in marker-free transgenic rice under drought, saline, and sodic conditions. J. Exp. Bot. 2020, 71, 653–668.
- Zou, J.; Liu, C.; Liu, A.; Zou, D.; Chen, X. Overexpression of OsHsp17.0 and OsHsp23.7 enhances drought and salt tolerance in rice. J. Plant Physiol. 2012, 169, 628–635.
- Zhang, H.; Zhai, N.; Ma, X.; Zhou, H.; Cui, Y.; Wang, C.; Xu, G. Overexpression of OsRLCK241 confers enhanced salt and drought tolerance in transgenic rice (Oryza sativa L.). Gene 2021, 768, 145278.
- Li, Z.; Zhang, F. Rice breeding in the post-genomics era: From concept to practice. Curr. Opin. Plant Biol. 2013, 16, 261–269.
- Ali, A.; Xu, J.; Ismail, A.; Fu, B.; Vijaykumar, C.; Gao, Y.; Domingo, J.; Maghirang, R.; Yu, S.; Gregorio, G.; et al. Hidden diversity for abiotic and biotic stress tolerances in the primary gene pool of rice revealed by a large backcross breeding program. Field Crops Res. 2006, 97, 66–76.
- Zhang, A.; Wang, F.; Luo, X.; Liu, Y.; Zhang, F.; Liu, G.; Yu, X.; Luo, L. Germplasm enhancement of water-saving and drought-resistance rice based on recurrent selection facilitated by dominant nucleus male sterility. Acta Agric. 2022, 38, 91–95.




