Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Daniela Minerdi | -- | 1268 | 2023-03-27 14:25:13 | | | |
| 2 | Conner Chen | Meta information modification | 1268 | 2023-03-29 05:51:50 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Minerdi, D.; Savoi, S.; Sabbatini, P. The Role of Cytochromes P450 in Plants. Encyclopedia. Available online: https://encyclopedia.pub/entry/42562 (accessed on 01 March 2026).
Minerdi D, Savoi S, Sabbatini P. The Role of Cytochromes P450 in Plants. Encyclopedia. Available at: https://encyclopedia.pub/entry/42562. Accessed March 01, 2026.
Minerdi, Daniela, Stefania Savoi, Paolo Sabbatini. "The Role of Cytochromes P450 in Plants" Encyclopedia, https://encyclopedia.pub/entry/42562 (accessed March 01, 2026).
Minerdi, D., Savoi, S., & Sabbatini, P. (2023, March 27). The Role of Cytochromes P450 in Plants. In Encyclopedia. https://encyclopedia.pub/entry/42562
Minerdi, Daniela, et al. "The Role of Cytochromes P450 in Plants." Encyclopedia. Web. 27 March, 2023.
Copy Citation
Cytochromes P450 are ancient enzymes diffused in organisms belonging to all kingdoms of life, including viruses, with the largest number of P450 genes found in plants. The functional characterization of cytochromes P450 has been extensively investigated in mammals, where these enzymes are involved in the metabolism of drugs and in the detoxification of pollutants and toxic chemicals.
grapevine
cytochrome P450 enzyme
1. Cytochromes P450 Enzymes: General Features
Cytochromes P450 (P450s or CYPs) are heme–thiolate terminal monooxygenases that transfer one atom from molecular oxygen to X–H bonds, where X may be the -C, -N, or S of a substrate, with a concomitant reduction in the remaining oxygen atom to water [1]. The CYP-mediated molecular oxygen activation gives rise to ketones, alcohols, aldehydes, epoxides, and carboxylic acids. Radicals or reactive oxygenated intermediates generated by CYPs can also lead to the formation of oxidative C–C bond cleavage [2], dealkylation [3], desaturation [4], dehydration [5], the oxidative rearrangement of carbon skeletons [6], and decarboxylation [7]. CYPs are a large superfamily of enzymes that show an absorbance peak at 450 nm when their heme is reduced and complexed with carbon monoxide. These enzymes were discovered in 1958, while studying the spectrophotometric properties of pigments in a microsomal fraction of rat livers [8]. A few years later, it was demonstrated that this pigment is a heme-containing protein, and it was called “P-450” (P for pigment) [9]. The heme group plays essential roles in catalysis, providing the P450s’ carbon monoxide-binding ability; moreover, the signature sequence FxxGxxxCxG in the heme-binding domain functions as the fifth ligand to the heme iron. CYPs are ancient enzymes that are extensively diffused in the organisms of all kingdoms of life (Figure 1), including viruses [10], with the largest number of P450 genes being present in plants [11]. Plant P450s are classified into the A-type group and are involved in plant-specific biochemical pathways [12]. The non-A-type P450 enzymes [13] form distinct clades and are more closely related to non-plant P450 enzymes than to the A-type. CYPs are classified into families on the basis of the sequence identity of their amino acid sequences, and the symbol CYP is the root followed by an Arabic numeral representing family therefore, the subfamily is indicated by a letter and the gene is represented by a number [14]. The features and functions of P450 enzymes have been studied from bacteria to mammals (see http://drnelson.uthsc.edu/CytochromeP450.html, accessed on 15 November 2022) [14]. Human P450 enzymes metabolize drugs and synthesize endogenous compounds essential for human physiology. Often, alterations in specific P450s affect the biological processes that they mediate, leading to a disease [15].
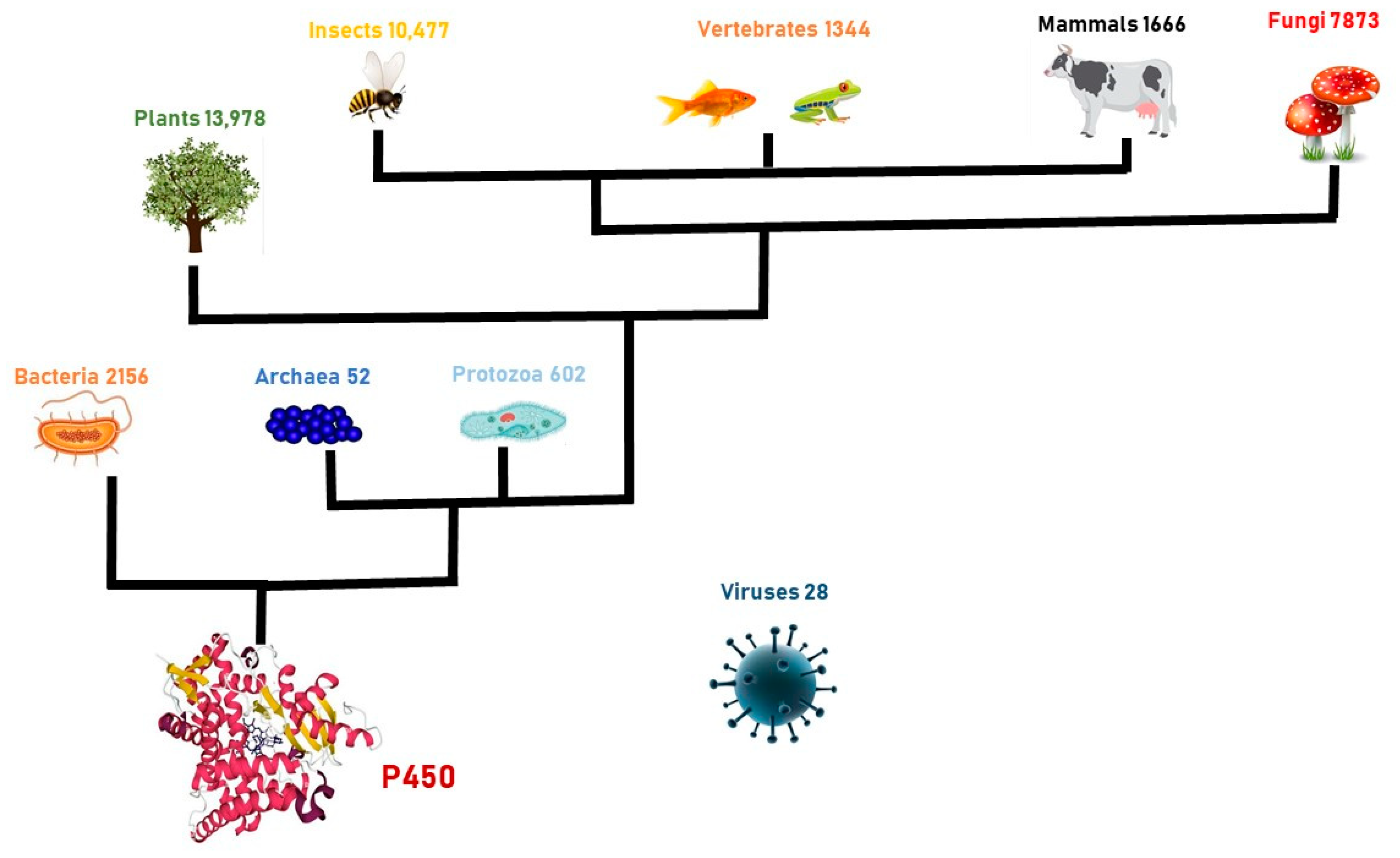
Figure 1. Diffusion of cytochrome P450 enzyme in the kingdom of Life.
2. Cytochrome P450 Reactivity
CYPs, through the activation of molecular oxygen, are able to monooxygenate the substrate, followed by the insertion of a single atom of oxygen into an organic substrate and a reduction in the another oxygen atom to water according to the following scheme:
RH + O2 + NAD(P)H + H+ → ROH + H2O + NAD(P)+
NAD(P)H supplies electron equivalents through an electron transfer chain that involves different redox partners, the nature of which is utilized to classify cytochromes P450 into 10 classes [16]. The organic substrate (R) binds to the heme group of the enzyme and this binding allows the transfer of an electron from NADPH through cytochrome P450 reductase (CPR) to the hem domain that reduces the iron (Fe) from the ferric state (Fe3+) to the ferrous state (Fe2+). The molecular oxygen binds to ferrous P450, forming a ferrous CYP–dioxygen complex, and the second electron is transferred from CPR to the ferrous CYP–dioxygen complex, forming the peroxo complex, and this complex is rapidly protonated twice to form one molecule of water and an iron–oxo complex. The oxidized reaction product (RO) is formed during the last step of the catalytic cycle when the atom of oxygen in the iron–oxo complex binds to the organic substrate (R) [17] (Figure 2).
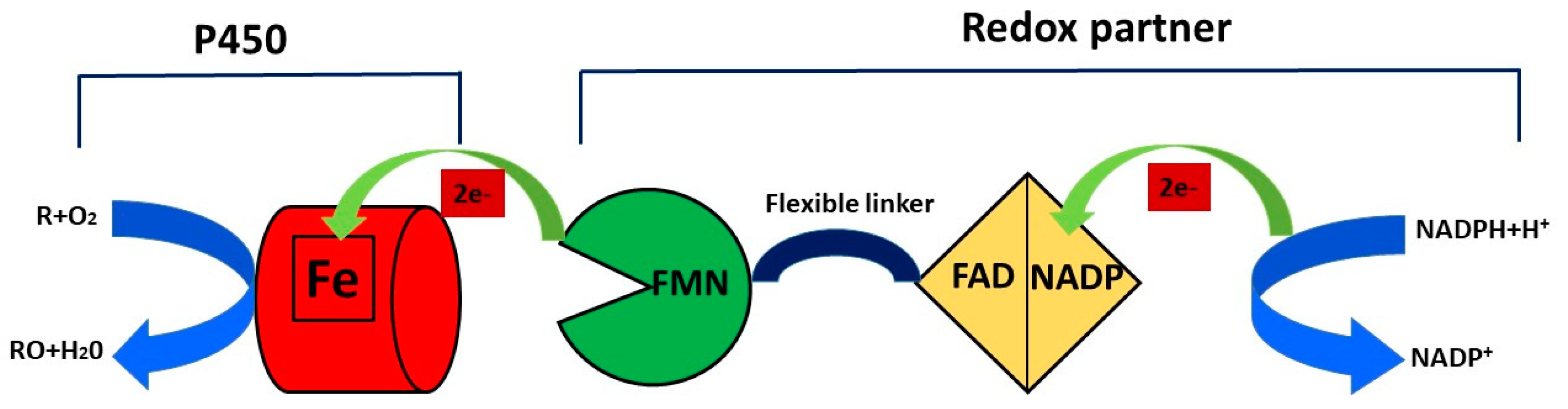
Figure 2. Catalytic cycle of cytochrome P450 enzyme.
3. The Role of Cytochromes P450 in Plants
In plant cytochromes, P450 enzymes are present in several organelles and organs, namely, the endoplasmic reticulum, mitochondria, and chloroplast [18]. They are also found in shoot, bulbs, hypocotyl, roots, and the endosperm of germinating seeds. In both plants and animals, CYPs have an absorbance range from 447 to 452 nm and constitute the largest family of enzymes in the metabolism of a plant, where they are grouped into ten clans [19][20]. The most studied family is CYP51, with a pivotal role in regulating the synthesis of sterols [21] and triterpenes [22]. CYP71 impacts shikimate products and intermediates [23][24]. The CYP72 clan has a role in the catabolism of isoprenoid hormones [25], and the CYP74 synthetizes oxylipin derivatives and allene oxide in the octadecanoid and jasmonate pathways [26][27]. CYP85 participates in the metabolism of cyclic terpenes and sterols in the brassinosteroid and in abscisic acid and gibberellin pathways [28]. The CYP86 hydroxylates fatty acids [29], the CYP97 hydroxylates carotenoids [30], and CYP710 control the synthesis of sterol C-22 desaturase [5]. The CYP450 family in plants has a molecular mass between 45 and 62 kDa [4][31][32]. In plants, P450 enzymes are involved in the biosynthesis of structural polymers, defense against pathogen infection, communication with other organisms, hormonal signaling, herbicide resistance, and stress tolerance (Figure 3).
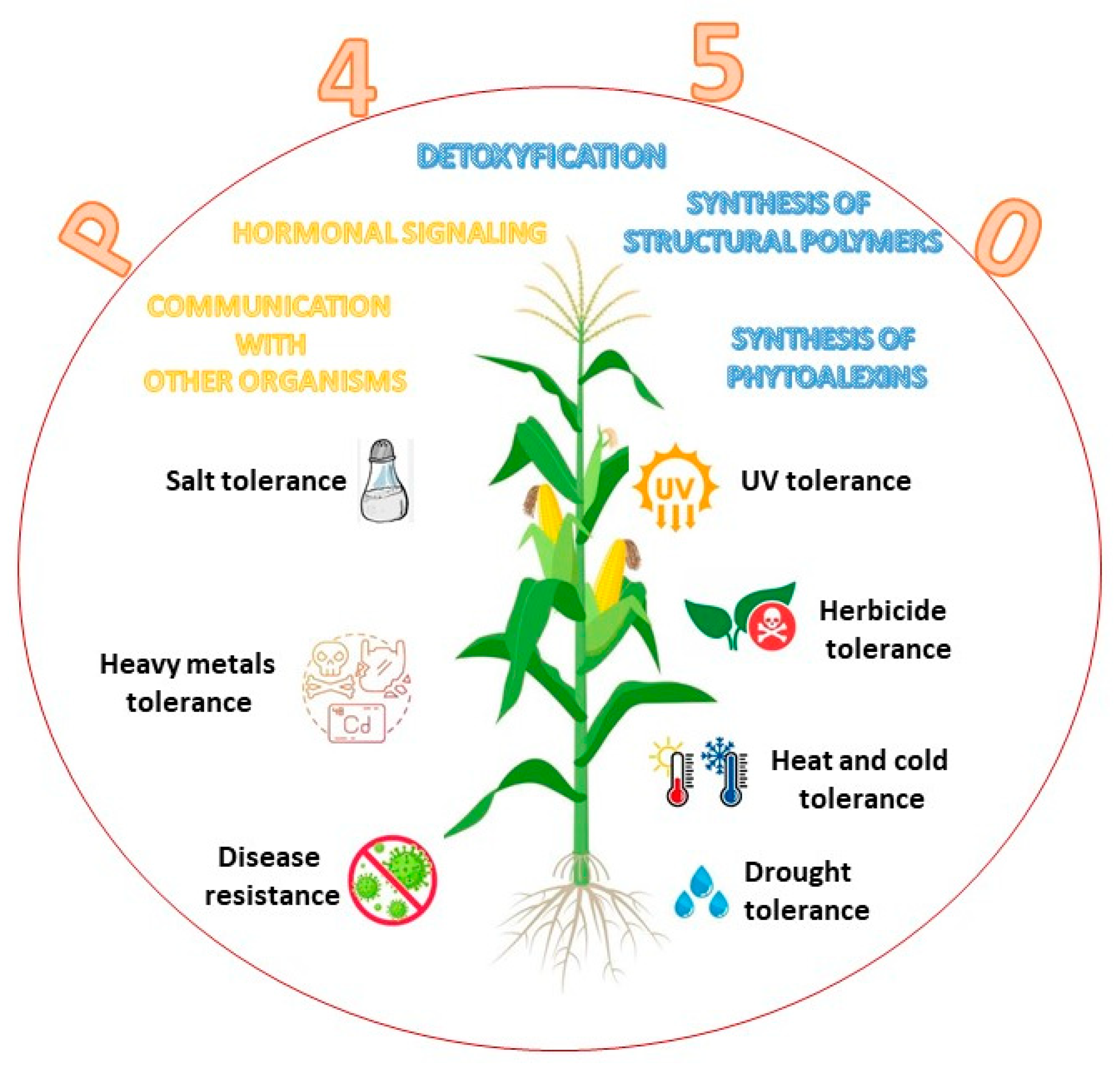
Figure 3. Role of cytochrome P450 enzyme in plants.
Plant hormone metabolism is also regulated by cytochrome P450 enzymes that control cell division and cell expansion, vascular differentiation, fruit growth, root development, and flower formation. Cytochromes P450 protect plants from dehydration [3], UV stress [33], and drought [34]. Moreover, P450s play a role in physiological process, such as detoxification, adaptation responses to heavy metals, salts, and chemicals (e.g., herbicides) [35][36]. In addition, many P450s participate in the biosynthesis of cell wall components [37]. CYP97 engages in the biosynthetic pathway of xanthophylls, hydroxylating the beta and gamma rings of carotenoids [38] (Table 1).
Table 1. Role of cytochrome P450 enzyme in crop plants.
| Plant | P450 | Function | Reference |
|---|---|---|---|
| Solanum lycopersicum | CYP789A | Fruit size Plant architecture |
[39] |
| Solanum tuberosum | CYP72A208 CYP188 |
Glycolakaloid biosynthesis | [40] |
| Oryza sativa | CYP703A3 | Male fertility | [41] |
| Cucumis sativus | CYP88L2 CYP81Q58 CYP78D20 |
Synthesis of cucurbitacin | [42][43] |
| Maesa lanceolata | CYP72A CYP76A |
Synthesis of triterpenes | [22][44] |
| Olea europea | CYP72 | Synthesis of secoxy-iridoids | [45] |
| Malus domestica | CYP716A CYP175 |
Synthesis of triterpenic acids | [46] |
| Sorghum bicolor | CYP97C1 CYPP97A3 |
Synthesis of lutein | [32] |
In Solanum lycopersicum, the CYP78 subfamily modulates the number and length of the lateral shoots and fruit ripening time [39]. In Solanum tuberosum, CYP72 is involved in the biosynthesis of two steroidal glycoalkaloids that catalyze the 26- and 22-hydroxylation steps. The knockdown plants of CYP72 are sterile, and tubers do not sprout during storage.
Tuber sprouting and the biosynthesis of glycoalkaloid in potatoes, two traits that significantly impact potato breeding and are important for the industry, can be controlled by tools provided by the functional analyses of the CYP72 family [40]. The cuticle of anther and the pollen exine are protective envelopes of the male gametophyte and the pollen grain. The fatty acid hydroxylase CYP703 is essential for male fertility in rice because the CYP703A3-2 mutant shows that pollen exine is defective and that the anthers exhibit a reduced level of cutin monomers and wax components [41]. Cucumber and melon contain cucurbitacins, highly oxygenated triterpenoids responsible for the bitter taste [47]. Cucurbitacins derive from the tetracyclic triterpenoid cucurbitadienol that is further oxidized to cucurbitacin at different positions by CYP81 and CYP87 [42][43][48] and CYP72 and CYP71 [22][44]. In Olea europaea, CYP72 catalyzes the oxidative C–C bond cleavage in the biosynthesis of secoxy-iridoids, a series of phenolic compounds that are essential for the flavor and quality of olive oil [45]. CYP716 is involved in the biosynthesis of the triterpenic acids present in apple fruit [46], and CYP97 is involved in the biosynthetic pathway of lutein in Sorghum [32].
References
- Garfinkel, D. Studies on Pig Liver Microsomes. I. Enzymic and Pigment Composition of Different Microsomal Fractions. Arch. Biochem. Biophys. 1958, 77, 493–509.
- Irmler, S.; Schröder, G.; St-Pierre, B.; Crouch, N.P.; Hotze, M.; Schmidt, J.; Strack, D.; Matern, U.; Schröder, J. Indole Alkaloid Biosynthesis in Catharanthus Roseus: New Enzyme Activities and Identification of Cytochrome P450 CYP72A1 as Secologanin Synthase. Plant J. 2000, 24, 797–804.
- Bak, S.; Kahn, R.A.; Olsen, C.E.; Halkier, B.A. Cloning and Expression in Escherichia Coli of the Obtusifoliol 14α-demethylase of Sorghum bicolor (L.) Moench, a Cytochrome P450 Orthologous to the Sterol 14α-demethylases (CYP51) from Fungi and Mammals. Plant J. 1997, 11, 191–201.
- Morikawa, T.; Mizutani, M.; Aoki, N.; Watanabe, B.; Saga, H.; Saito, S.; Oikawa, A.; Suzuki, H.; Sakurai, N.; Shibata, D. Cytochrome P450 CYP710A Encodes the Sterol C-22 Desaturase in Arabidopsis and Tomato. Plant Cell 2006, 18, 1008–1022.
- Hansen, C.C.; Sørensen, M.; Veiga, T.A.M.; Zibrandtsen, J.F.S.; Heskes, A.M.; Olsen, C.E.; Boughton, B.A.; Møller, B.L.; Neilson, E.H.J. Reconfigured Cyanogenic Glucoside Biosynthesis in Eucalyptus Cladocalyx Involves a Cytochrome P450 CYP706C55. Plant Physiol. 2018, 178, 1081–1095.
- Nasomjai, P.; Reed, D.W.; Tozer, D.J.; Peach, M.J.G.; Slawin, A.M.Z.; Covello, P.S.; O’Hagan, D. Mechanistic Insights into the Cytochrome P450-mediated Oxidation and Rearrangement of Littorine in Tropane Alkaloid Biosynthesis. ChemBioChem 2009, 10, 2382–2393.
- Koch, B.M.; Sibbesen, O.; Halkier, B.A.; Svendsen, I.; Møller, B.L. The Primary Sequence of Cytochrome P450tyr, the MultifunctionalN-Hydroxylase Catalyzing the Conversion OfL-Tyrosine Top-Hydroxyphenylacetaldehyde Oxime in the Biosynthesis of the Cyanogenic Glucoside Dhurrin in Sorghum bicolor (L.) Moench. Arch. Biochem. Biophys. 1995, 323, 177–186.
- Klingenberg, M. Pigments of Rat Liver Microsomes. Arch. Biochem. Biophys. 1958, 75, 376–386.
- Omura, T.; Sato, R. The Carbon Monoxide-Binding Pigment of Liver Microsomes: II. Solubilization, Purification, and Properties. J. Biol. Chem. 1964, 239, 2379–2385.
- Lamb, D.C.; Lei, L.; Warrilow, A.G.S.; Lepesheva, G.I.; Mullins, J.G.L.; Waterman, M.R.; Kelly, S.L. The First Virally Encoded Cytochrome P450. J. Virol. 2009, 83, 8266–8269.
- Paquette, S.M.; Bak, S.; Feyereisen, R. Intron–Exon Organization and Phylogeny in a Large Superfamily, the Paralogous Cytochrome P450 Genes of Arabidopsis Thaliana. DNA Cell Biol. 2000, 19, 307–317.
- Durst, F.; Nelson, D.R. Diversity and Evolution of Plant P450 and P450-Reductases. Drug Metab. Drug Interact. 1995, 12, 189–206.
- Feldmann, K.A. Cytochrome P450s as Genes for Crop Improvement. Curr. Opin. Plant Biol. 2001, 4, 162–167.
- Nelson, D.R. The Cytochrome P450 Homepage. Hum. Genom. 2009, 4, 59.
- Pikuleva, I.A.; Waterman, M.R. Cytochromes P450: Roles in Diseases. J. Biol. Chem. 2013, 288, 17091–17098.
- Hannemann, F.; Bichet, A.; Ewen, K.M.; Bernhardt, R. Cytochrome P450 Systems—Biological Variations of Electron Transport Chains. Biochim. Et Biophys. Acta (BBA)-Gen. Subj. 2007, 1770, 330–344.
- Guengerich, F.P. Mechanisms of Cytochrome P450-Catalyzed Oxidations. ACS Catal. 2018, 8, 10964–10976.
- Morant, M.; Bak, S.; Møller, B.L.; Werck-Reichhart, D. Plant Cytochromes P450: Tools for Pharmacology, Plant Protection and Phytoremediation. Curr. Opin. Biotechnol. 2003, 14, 151–162.
- Laffaru Singpho, N.; Sharma, J.G. Importance of Cytochrome P450 Gene Family from Metabolite Biosynthesis to Stress Tolerance: A Review. In Proceedings of the IOP Conference Series: Earth and Environmental Science, Jakarta, Indonesia, 25–26 September 2021; Volume 775, p. 012012.
- Nelson, D.; Werck-Reichhart, D. A P450-centric View of Plant Evolution. Plant J. 2011, 66, 194–211.
- Jain, K.S.; Khedkar, V.M.; Arya, N.; Rane, P.V.; Chaskar, P.K.; Coutinho, E.C. Design, Synthesis & Evaluation of Condensed 2H-4-Arylaminopyrimidines as Novel Antifungal Agents. Eur. J. Med. Chem. 2014, 77, 166–175.
- Geisler, K.; Hughes, R.K.; Sainsbury, F.; Lomonossoff, G.P.; Rejzek, M.; Fairhurst, S.; Olsen, C.-E.; Motawia, M.S.; Melton, R.E.; Hemmings, A.M. Biochemical Analysis of a Multifunctional Cytochrome P450 (CYP51) Enzyme Required for Synthesis of Antimicrobial Triterpenes in Plants. Proc. Natl. Acad. Sci. USA 2013, 110, E3360–E3367.
- Chen, J.; Wu, X.T.; Xu, Y.Q.; Zhong, Y.; Li, Y.X.; Chen, J.K.; Li, X.; Nan, P. Global transcriptome analysis profiles metabolic pathways in traditional herb Astragalus membranaceus Bge. var. mongolicus (Bge.) Hsiao. BMC Genom. 2015, 16, S15.
- Williams, D.; De Luca, V. Plant cytochrome P450s directing monoterpene indole alkaloid (MIA) and benzylisoquinoline alkaloid (BIA) biosynthesis. Phytochem. Rev. 2022, 1–30.
- Magome, H.; Nomura, T.; Hanada, A.; Takeda-Kamiya, N.; Ohnishi, T.; Shinma, Y.; Katsumata, T.; Kawaide, H.; Kamiya, Y.; Yamaguchi, S. CYP714B1 and CYP714B2 Encode Gibberellin 13-Oxidases That Reduce Gibberellin Activity in Rice. Proc. Natl. Acad. Sci. USA 2013, 110, 1947–1952.
- Hoffmann, I.; Jernerén, F.; Oliw, E.H. Expression of Fusion Proteins of Aspergillus Terreus Reveals a Novel Allene Oxide Synthase. J. Biol. Chem. 2013, 288, 11459–11469.
- Yoeun, S.; Rakwal, R.; Han, O. Dual Positional Substrate Specificity of Rice Allene Oxide Synthase-1: Insight into Mechanism of Inhibition by Type II Ligand Imidazole. BMB Rep. 2013, 46, 151.
- Ohnishi, T.; Watanabe, B.; Sakata, K.; Mizutani, M. CYP724B2 and CYP90B3 Function in the Early C-22 Hydroxylation Steps of Brassinosteroid Biosynthetic Pathway in Tomato. Biosci. Biotechnol. Biochem. 2006, 70, 2071–2080.
- Delventhal, R.; Falter, C.; Strugala, R.; Zellerhoff, N.; Schaffrath, U. Ectoparasitic Growth of Magnaporthe on Barley Triggers Expression of the Putative Barley Wax Biosynthesis Gene CYP96B22 Which Is Involved in Penetration Resistance. BMC Plant Biol. 2014, 14, 26.
- Cui, H.; Yu, X.; Wang, Y.; Cui, Y.; Li, X.; Liu, Z.; Qin, S. Evolutionary Origins, Molecular Cloning and Expression of Carotenoid Hydroxylases in Eukaryotic Photosynthetic Algae. BMC Genom. 2013, 14, 457.
- Vasav, A.P.; Barvkar, V.T. Phylogenomic Analysis of Cytochrome P450 Multigene Family and Their Differential Expression Analysis in Solanum Lycopersicum L. Suggested Tissue Specific Promoters. BMC Genom. 2019, 20, 116.
- Kim, J.-E.; Cheng, K.M.; Craft, N.E.; Hamberger, B.; Douglas, C.J. Over-Expression of Arabidopsis Thaliana Carotenoid Hydroxylases Individually and in Combination with a β-Carotene Ketolase Provides Insight into in Vivo Functions. Phytochemistry 2010, 71, 168–178.
- Sasaki, T.; Mogi, S.; Kaneko, T.; Kojima, H.; Katoh, S.; Sano, A.; Kojima, S. Relationship between Tissue Hydroxyl Radical and Oxidatively Modified Macromolecule Levels. Geriatr. Gerontol. Int. 2014, 14, 498–507.
- Mao, G.; Seebeck, T.; Schrenker, D.; Yu, O. CYP709B3, a Cytochrome P450 Monooxygenase Gene Involved in Salt Tolerance in Arabidopsis thaliana. BMC Plant Biol. 2013, 13, 169.
- Iquebal, M.A.; Soren, K.R.; Gangwar, P.; Shanmugavadivel, P.S.; Aravind, K.; Singla, D.; Jaiswal, S.; Jasrotia, R.S.; Chaturvedi, S.K.; Singh, N.P. Discovery of Putative Herbicide Resistance Genes and Its Regulatory Network in Chickpea Using Transcriptome Sequencing. Front. Plant Sci. 2017, 8, 958.
- Dimaano, N.G.; Iwakami, S. Cytochrome P450-mediated Herbicide Metabolism in Plants: Current Understanding and Prospects. Pest Manag. Sci. 2021, 77, 22–32.
- Grausem, B.; Widemann, E.; Verdier, G.; Nosbüsch, D.; Aubert, Y.; Beisson, F.; Schreiber, L.; Franke, R.; Pinot, F. CYP 77 A 19 and CYP 77 A 20 Characterized from Solanum Tuberosum Oxidize Fatty Acids in Vitro and Partially Restore the Wild Phenotype in an A Rabidopsis Thaliana Cutin Mutant. Plant Cell Environ. 2014, 37, 2102–2115.
- Fiore, A.; Dall’Osto, L.; Cazzaniga, S.; Diretto, G.; Giuliano, G.; Bassi, R. A Quadruple Mutant of Arabidopsis Reveals a β-Carotene Hydroxylation Activity for LUT1/CYP97C1 and a Regulatory Role of Xanthophylls on Determination of the PSI/PSII Ratio. BMC Plant Biol. 2012, 12, 50.
- Chakrabarti, M.; Zhang, N.A.; Sauvage, C.; Muños, S.; Blanca, J.; Cañizares, J.; Diez, M.J.; Schneider, R.; Mazourek, M.; McClead, J. A Cytochrome P450 Regulates a Domestication Trait in Cultivated Tomato. Proc. Natl. Acad. Sci. USA 2013, 110, 17125–17130.
- Umemoto, N.; Nakayasu, M.; Ohyama, K.; Yotsu-Yamashita, M.; Mizutani, M.; Seki, H.; Saito, K.; Muranaka, T. Two Cytochrome P450 Monooxygenases Catalyze Early Hydroxylation Steps in the Potato Steroid Glycoalkaloid Biosynthetic Pathway. Plant Physiol. 2016, 171, 2458–2467.
- Yang, X.; Wu, D.I.; Shi, J.; He, Y.I.; Pinot, F.; Grausem, B.; Yin, C.; Zhu, L.; Chen, M.; Luo, Z. Rice CYP703A3, a Cytochrome P450 Hydroxylase, Is Essential for Development of Anther Cuticle and Pollen Exine. J. Integr. Plant Biol. 2014, 56, 979–994.
- Shang, Y.; Ma, Y.; Zhou, Y.; Zhang, H.; Duan, L.; Chen, H.; Zeng, J.; Zhou, Q.; Wang, S.; Gu, W. Biosynthesis, Regulation, and Domestication of Bitterness in Cucumber. Science 2014, 346, 1084–1088.
- Zhou, Y.; Ma, Y.; Zeng, J.; Duan, L.; Xue, X.; Wang, H.; Lin, T.; Liu, Z.; Zeng, K.; Zhong, Y. Convergence and Divergence of Bitterness Biosynthesis and Regulation in Cucurbitaceae. Nat. Plants 2016, 2, 16183.
- Moses, T.; Pollier, J.; Faizal, A.; Apers, S.; Pieters, L.; Thevelein, J.M.; Geelen, D.; Goossens, A. Unraveling the Triterpenoid Saponin Biosynthesis of the African Shrub Maesa Lanceolata. Mol. Plant 2015, 8, 122–135.
- Rodríguez-López, C.E.; Hong, B.; Paetz, C.; Nakamura, Y.; Koudounas, K.; Passeri, V.; Baldoni, L.; Alagna, F.; Calderini, O.; O’Connor, S.E. Two Bi-functional Cytochrome P450 CYP72 Enzymes from Olive (Olea europaea) Catalyze the Oxidative C-C Bond Cleavage in the Biosynthesis of Secoxy-iridoids–Flavor and Quality Determinants in Olive Oil. New Phytol. 2021, 229, 2288–2301.
- Andre, C.M.; Legay, S.; Deleruelle, A.; Nieuwenhuizen, N.; Punter, M.; Brendolise, C.; Cooney, J.M.; Lateur, M.; Hausman, J.; Larondelle, Y. Multifunctional Oxidosqualene Cyclases and Cytochrome P450 Involved in the Biosynthesis of Apple Fruit Triterpenic Acids. New Phytol. 2016, 211, 1279–1294.
- Takase, S.; Kera, K.; Nagashima, Y.; Mannen, K.; Hosouchi, T.; Shinpo, S.; Kawashima, M.; Kotake, Y.; Yamada, H.; Saga, Y. Allylic Hydroxylation of Triterpenoids by a Plant Cytochrome P450 Triggers Key Chemical Transformations That Produce a Variety of Bitter Compounds. J. Biol. Chem. 2019, 294, 18662–18673.
- Zhang, J.; Dai, L.; Yang, J.; Liu, C.; Men, Y.; Zeng, Y.; Cai, Y.; Zhu, Y.; Sun, Y. Oxidation of Cucurbitadienol Catalyzed by CYP87D18 in the Biosynthesis of Mogrosides from Siraitia Grosvenorii. Plant Cell Physiol. 2016, 57, 1000–1007.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.3K
Revisions:
2 times
(View History)
Update Date:
29 Mar 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





