1. Bioluminescence-Induced Optical Biosensors for Ion Sensing
Calcium ions (Ca
2+), being universal intracellular secondary messengers, regulate abundant physiological processes. To achieve precise control over Ca
2+ signaling, a variety of optogenetic probes have been crafted by coupling light-sensitive domains with intracellular signaling proteins to enable remote control of intracellular Ca
2+ signaling both in cellulo and in vivo
[1][2]. Bioluminescence-based Ca
2+ imaging is a perfect strategy that is highly compatible with optogenetic actuators for imaging and monitoring of multiple cellular Ca
2+ dynamics
[3]. Undesirable consequences resulting from functional crosstalk between optogenetic light-sensitive domains can be avoided as bioluminescent indicators have self-luminescent excitation. The bioluminescent optogenetic Ca
2+ sensors are helpful for the further study of Ca
2+-related physiology, pathophysiology, and drug screening.
It should be noted that a bioluminescent optogenetic opsin named bMCOII has been reported
[4]. bMCOII was created by fusing the C-terminus of mutated opsins from algae (Chlamydomonas) with the N-terminus of a bioluminescent protein GeNL made of NanoLuc and GFP. The calmodulin-M13 domain fused with GeNL is located on the cytosolic side of the membrane and just beneath the bMCOII-TM actuator domain, which allows GeNL to bind to Ca
2+ flowing into the cell when the actuator is evoked. Upon light stimulation, the transmembrane opsin channel is opened, allowing Ca
2+ to enter the transfected cells, which can be detected and reported by the Ca
2+-GeNL sensor immediately. This is a hybrid optogenetic actuator and a bioluminescence Ca
2+ sensor, which enables simultaneous optical modulation and bioluminescence imaging of cortical activities in vitro and in vivo. Moreover, Johnson’s group developed a BRET-based Ca
2+ sensor that can be partnered with optogenetic probes (
Figure 1a). This BRET system consisted of bright NanoLuc luciferase in conjunction with the fluorescent protein Venus. The Ca
2+ flux elicited by the optogenetic ChR2 domains and melanopsin in response to light in hippocampal and fibroblast neurons was imaged and quantified successfully by use of this Ca
2+ sensor
[5]. A dual-purpose probe named LuCID has recently been crafted to provide real-time dynamics and the transcriptional record of Ca
2+ [6] (
Figure 1b). The LuCID probe is constructed by inserting NanoBiT fusions into the GI/FKF1 system. In the presence of high Ca
2+ and furimazine, BRET from NanoBiT activates the blue-light-induced protein dimerization system through the LOV domain in FKF1
[7], which leads to the transcription of reporter genes. The real-time readout of Ca
2+ activation is reflected via reconstitution of NanoBiT, while the steady recording of past Ca
2+ activity is provided in the form of reporter expression controlled by the GI/FKF1 system, enabling LuCID to function in both Ca
2+ indicator and transcriptional Ca
2+ integrator capacities in living cells. In addition to Ca
2+ sensors, Inagaki and co-workers developed a bioluminescent NanoLuc protein-based voltage indicator, called LOTUS-V, which was combined with the optogenetic proteins ChR2 and eNpHR3.0 to allow long-term membrane voltage imaging in an in vitro cardiomyocyte model
[8] and in vivo imaging of brain activity
[9]. This is a useful live imaging tool with multipurpose applications, such as for studying cardiomyocyte behavior
[8] and recording brain activity in multiple socially interactive animals
[9].
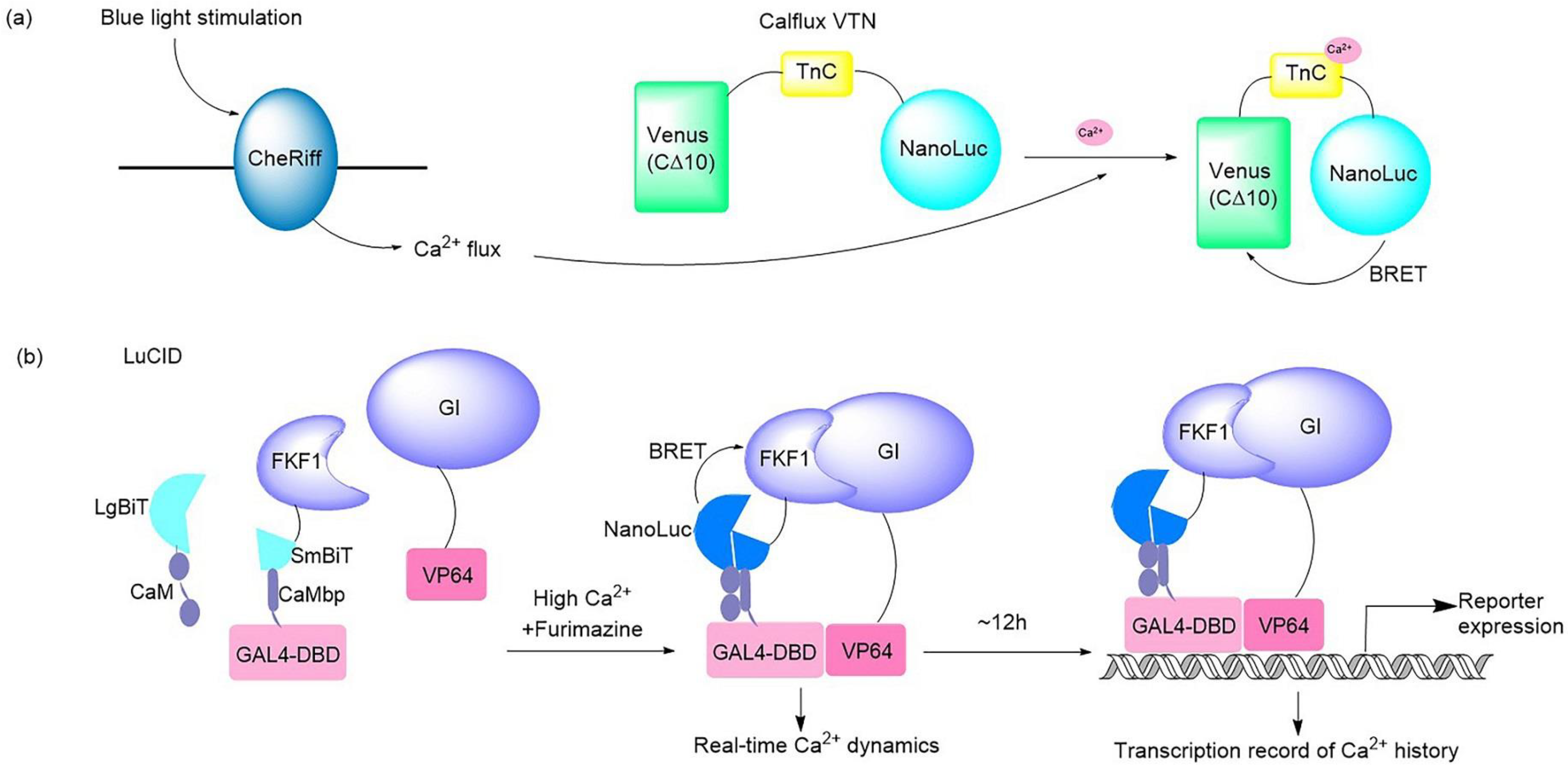
Figure 1. Illustrations of typical bioluminescent optogenetic Ca
2+ sensors. (
a) Schematic of BRET-based Ca
2+ sensor Calflux VTN. Calflux VTN was designed by inserting troponin C domain between NanoLuc and Venus. The Ca
2+ sensitive troponin undergoes a conformational change in response to binding Ca
2+, which brings NanoLuc closer to Venus so that BRET can occur with a spectral shift
[5]. (
b) Schematic of dual Ca
2+ indicator LuCID. In the presence of high Ca
2+ and furimazine, BRET from NanoLuc activates the GI/FKF1 system, leading to the transcription of reporter genes. The real-time readout of Ca
2+ activation is reflected via reconstitution of NanoBiT, while the steady recording of past Ca
2+ activity is provided in the form of reporter expression controlled by the GI/FKF1 system
[6].
The cooperation mode of the optogenetic component and luciferase sensor varies in different systems, and is mainly divided into two typical principles: in one case, the optical channel is triggered by light stimulation, resulting in Ca
2+ current, which is detected and reported by the designed bioluminescent Ca
2+-sensitive protein
[4][5]. In the other case, the bioluminescence generated by the Ca
2+ indicator binding with Ca
2+ is responsible for stimulating optogenetic proteins, leading to a series of downstream reactions to record and display the dynamics of Ca
2+ [6].
Regarding the experimental protocols for the application of typical bioluminescent optogenetic Ca
2+ sensors, the bioluminescent optogenetic Ca
2+ sensors may be a fusion-type protein constructed by optogenetic domains and bioluminescent Ca
2+ indicators
[4][6], or the two partners may be expressed separately in target cells by co-transfection
[5][8][9]. The primary intended use of bioluminescent optogenetic sensors is for the imaging and recording of biological activities over a long period and in a noninvasive manner, as well as avoiding the undesirable effects of fluorescence irradiation such as tissue autofluorescence, excitation-induced tissue photodamage, and poor tissue penetration. The luciferase NanoLuc is commonly involved in bioluminescence Ca
2+ sensors: its unique luciferin furimazine should be prepared for a substrate solution as a bioluminescence trigger
[4][5][6][8][9]. Apart from this, other basic routes include cell preparation and transfection, virus preparation, virus injection to generate animal models, cranial surgery, brain slice preparation, imaging and recording by an EMCCD camera, and patch clamp/voltage clamp recording. Considering that Ca
2+ signaling is not only important in neuronal functions but also plays a role in other biological processes such as cardiocyte activities, bioluminescent optogenetic Ca
2+ sensors may allow a wide range of imaging and the quantification of live Ca
2+ fluxes in multiple physiological and pathophysiological conditions.
In addition to the above, many bioluminescent indicators have been generated and have potential to be a partner of optogenetic probes. The Renilla luciferase and its improved variant Rluc8, NanoLuc, and the bioluminescent aequorin protein have been used in the construction of bioluminescent Ca
2+ indicators. For example, a series of multicolor nanolanterns that resulted from the coupling of Rluc8 or NanoLuc luciferase with a fluorescent protein were demonstrated to image Ca
2+ dynamics in the cytosol, nucleus, and mitochondria
[10][11], as well as in the sarco/endoplasmic reticulum (SR/ER)
[12]. Moreover, a high-affinity fluorescence/bioluminescence bimodal indicator that fused the Ca
2+ sensing tool GCaMP6f to a NanoLuc-derived reporter NanoBiT was demonstrated to be useful for imaging cytosolic Ca
2+ dynamics. Another low-affinity bimodal Ca
2+ indicator variant that coupled the Ca
2+ sensing tool R-CEPIA1er with NanoBiT was created to target Ca
2+ imaging of ER
[13]. A bioluminescent Ca
2+ indicator, LUCI-GECO1, based on the coupling of BRET from the NanoLuc luciferase to a topological variant of GCaMP6s is reported to be able to image Ca
2+ changes in cultured cells and primary neurons
[14]. Tricoire and co-workers engineered a bioluminescent Ca
2+ probe that consisted of a fusion of aequorin and GFP and could be used in whole-field bioluminescence imaging of neuronal network dynamics
[15]. The function of bioluminescent indicators is not limited to ion sensing. For example, a stereospecific bioluminescent luciferase probe has been engineered to detect endogenous D-cysteine quantitatively in mammals. D-cysteine combines with 2-cyano-6-hydroxybenzothiazole (CHBT) after the addition of base and reducing agent to form D-luciferin, which can be used as the substrate of luciferase to make it emit light, allowing a noninvasive method to monitor D-cysteine and further demonstrate its function
[16]. Based on BRET, Park and co-workers created an ERα dimerization assay and subsequent ERβ dimerization assay in human cells to elucidate the ER dimerization potential of estrogenic compounds
[17][18]. Furthermore, NanoBiT subunits LgBiT and SmBiT were respectively fused to the human androgen receptor (hAR) to construct a system that images the activation state of ARs. When testosterone binds to the AR, a series of conformational changes cause dimerization of the hAR and the reconstruction of NanoBiT, which in turn will yield bioluminescence in the presence of the furimazine
[19]. Accordingly, these bioluminescent indicators may represent maximal biocompatibility and the best signal-to-noise ratios, avoiding the interference from crosstalk between the optogenetic tools and fluorescent indicators when they are applied with optogenetic probes. However, a major limitation is that the consumption of the substrates could gradually reduce signal brightness
[20], which results in partially sacrificing the spatial resolution
[1][21].
Although optogenetic proteins have been widely used in eukaryotic systems, especially in neuroscience, a variety of photosensitive proteins have been discovered from microorganisms
[22]. The renaissance of optogenetic applications in prokaryotes has been underway since synthetic sensors for light-induced gene expression were created with the use of photoreceptor Cph1 and the histidine kinase EnvZ in 2005
[23]. Optogenetic tools have been developed to manipulate molecular processes, interbacterial interactions, and cell-to-environment interactions in microbiology. A bacterial sensor for Hg
2+ based on a lux bioluminescence-triggered photoswitchable magnet system was developed, which provides a new strategy for whole-cell biosensors
[24]. When the Hg
2+ binds to the transcriptional regulator MerR, the lux operon undergoes expression to emit bioluminescence, which leads to the activation of photoswitchable proteins that are displayed on bacterial surfaces. Thus, this bioluminescent optogenetic tool with the ability to control bacterial aggregation provides an alternative tool in the regulation of bacterial consortia.
2. Bioluminescence-Aided Optical Tools for Reprogramming Cellular Activities
According to the BRET mechanism, a luciferase could be engineered as a bioluminescent–fluorescent protein that emits light from cyan to red across the visible spectrum. Hence, the BRET strategy has the ability to alter the spectrum of a bioluminescent protein, which is beneficial to match more light-sensitive domains to the design of sophisticated bioluminescent optogenetic tools.
The BRET strategy has been applied in optogenetics to generate optogenetic probes for Ca
2+ sensing
[5] as mentioned above, and engineer inhibitory luminopsins
[25] for controlling neuron activities. Recently, a series of novel and delicate BRET-based optogenetic tools have been reported (
Figure 2). NanoLuc is the preferred luciferase as a bioluminescence donor due to its ability to activate blue-to-green light-sensitive proteins via BRET, which results in downstream events. In 2018, Komatsu and co-workers developed a BRET–FRET hybrid biosensor named hyBRET that consists of luciferase Rluc8, a yellow fluorescent protein, and a cyan fluorescent protein for intramolecular ERK activity. This biosensor is compatible with the optogenetic protein CRY2 for imaging and monitoring ERK activation in live cells and in vivo. In this case, a prolonged half-life substrate diacetyl CTZ h was used to induce bioluminescence for longer in vivo imaging. It is a promising platform for the visualization of the multiscale dynamics of cell signaling, and even for the visualization of pharmacodynamics in living animals, which is helpful for drug development
[26].
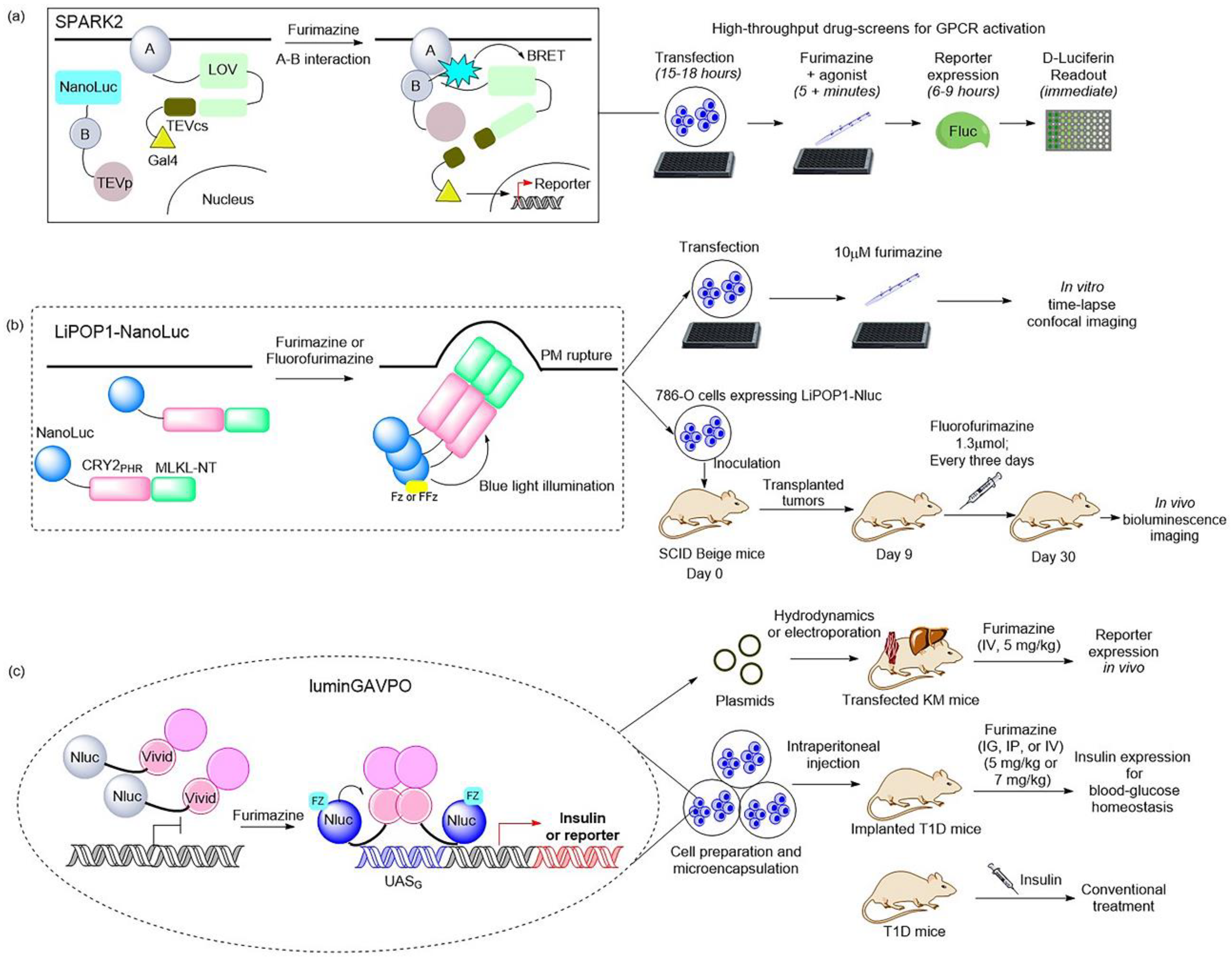
Figure 2. Illustrations of classic BRET-based photosensory probes. (
a) Schematic of SPARK2 and its general flow for high-throughput GPCR agonist screening
[21][27]. SPARK2 contains two modules: One is the fusion of target protein A, an LOV domain, and a transcription factor with a peptide linker including the TEVcs domain. The other is the fusion of target protein B, NanoLuc, and the TEV protease. When the target proteins are close enough to interact and the luciferin is provided, the BRET activates the LOV domain to expose TEVcs for protease cleavage in order to induce the release of transcription factors. (
b) Schematic of LiPOP1 and its experimental procedure for controlling nonapoptotic cell death in vitro and in vivo
[28]. LiPOP1–NanoLuc was generated by inserting NanoLuc into LiPOP1 between mCherry and CRY2. With the addition of the FFz, the blue bioluminescence catalyzed by NanoLuc stimulates CRY2 to undergo oligomerization, which causes MLKL to expose the N-terminal helix bundle domain and destroy the membrane. (
c) Schematic of luminGAVPO and its application for pulsatile activation of transgene expression in mice
[29]. luminGAVPO is the fusion of NanoLuc and a light-switchable transcription factor GAVPO. Light stemmed from the NanoLuc–furimazine reaction activates the VVD domain in the transactivator in order to cause dimerization and binding of luminGAVPO to the related promoter, thus leading to downstream gene transcription.
Moreover, a BRET-based optogenetic transcription reporter, named SPARK2, used for the quantitative detection of protein–protein interactions, has been reported
[21][27] (
Figure 3a). This transcription reporter has the ability to switch gates between external light and the addition of a luciferin for temporal specificity. NanoLuc was employed as a light-emitting moiety to control the blue-light-sensing LOV domain from Avena sativa (asLOV2), which yielded a lower background, leading to improved protein–protein interaction specificity. As regards the generality of SPARK2′s design, it contains two modules. One is the fusion of a target protein, an LOV domain, and a transcription factor with a peptide linker including the TEVcs domain for protease recognition. The other is the protein fused by a target protein, NanoLuc, and the TEVp protease. When the target proteins are close enough to interact and the luciferin is provided, the BRET activates the light-sensing LOV domain to expose the TEVcs site for protease cleavage in order to induce the release of transcription factors. Then, the downstream gene expression is regulated by the released transcription factors. This is the AND logic of SPARK2 for target protein–protein interaction. This tool has been applied in high-throughput screening for GPCR agonists and the study of transcellular interactions. For example, when SPARK2 was applied to detect the PPI interaction between the GPCR beta-2 adrenergic receptor (β2AR) and its cytosolic effector arrestin, the NanoLuc was fused to arrestin–TEVp and the LOV-sensing domain component was fused to β2AR; these two constructs were co-expressed with the gene reporter of citrine in HEK293T cells. With the addition of the β2AR agonist isoetharine and 10 μM furimazine for 15 min, robust citrine expression was observed. SPARK2 displayed furimazine- and agonist-dependent expression of citrine when it was designed for other GPCR/arrestin pairs, such as arginine vasopressin receptor 2 (AVPR2) and dopamine receptor type I (DRD1). Another similar BRET-activated optogenetic paradigm, BEACON, has been developed with the use of self-illuminating bioluminescent–fluorescent proteins (LumiFluor)
[30] that were generated by fusing the N-terminus of NanoLuc to the C-terminus of fluorescent proteins eGFP or mCerulean3 with a flexible linker. The reaction of NanoLuc and furimazine resulted in biological light, and BRET was carried out to control a diverse variety of blue–green light-sensitive proteins, including CRY2, LOV, and VVD, which provided a novel strategy for achieving rapid and robust activation toward a variety of commonly used optogenetic systems in a spatiotemporally restricted manner. For example, the light emitted from LumiFluor upon the addition of 10 μM furimazine activated the cryptochrome-based CRY2–CIBN system, the LOV-based FKF1–GI and iLID system, and the VVD-based pMagnet system, then gene expression or protein co-localization was achieved. However, further studies are required to determine whether bioluminescence activates BEACON to trigger downstream processes in vivo.
Recently, NanoLuc and a novel furimazine analog, fluorofurimazine (FFz), were utilized to emit bioluminescence, which provided a biocompatible approach by which to effectively activate a novel optogenetic tool, LiPOP1, based on photosensitive domain CRY2 to achieve wireless control of tumor lysis in living animals
[28] (
Figure 2b). LiPOP1 is a fusion protein created by fusing the N-terminal of mCherry-CRY2 to the C-terminal of MLKL that plays a role in the execution of necroptosis. A hybrid fusion protein, LiPOP1–NanoLuc, was generated by inserting NanoLuc into LiPOP1 between mCherry and CRY2 in order to validate the bioluminescence-mediated optogenetic stimulation to activate LiPOP1 via CRY2 oligomerization. With the addition of the substrate FFz, the blue bioluminescence catalyzed by NanoLuc stimulates CRY2 to undergo monomer-to-oligomer transition, which also leads to the oligomerization of MLKL. Activated MLKL exposes the N-terminal helix bundle domain (HBD) and migrates to the plasma membrane, destroying the membrane and causing necroptosis. Additionally, LiPOP1–NanoLuc was demonstrated to be able to induce PM translocation and necroptosis in three cancer cells lines, including HeLa cells, B16 cells, and 786-O cells. Furthermore, human 786-O cells were engineered to express LiPOP1–NanoLuc, and these were subcutaneously inoculated into mice to generate a mouse xenograft model. The mice were treated with FFz (1.3 μmol/25 g) by intratumoral injection every three days for a total of seven injections, after which they displayed a significant reduction in tumor size. This tool will likely act as a synthetic light-switchable gene that allows selective killing of abnormal cells and the elimination of therapeutic cells after treatment in the future.
Furthermore, the description of NanoLuc-driven bioluminescence directly activating the light-switchable transcription factor that fused to NanoLuc was reported
[29] (
Figure 2c). This is a bioluminescence transcription factor, termed luminGAVPO, in which NanoLuc was fused to a light-switchable transcription factor GAVPO
[31]. NanoLuc is close enough to VVD in this fusion protein to allow BRET to occur. Light stemmed from the NanoLuc–furimazine reaction and the external blue laser activated the light-sensitive VVD domain in the transactivator in order to cause dimerization of luminGAVPO and binding of luminGAVPO to the related promoter, thus leading to downstream gene transcription. When the bioluminescence was dimmed due to the consumption of luciferin, the luminGAVPO dimer gradually dissociated from the promoter, thereby stopping the transcription of the target gene. The downstream gene expression reached the highest level and was sustained for 2–3 h when the furimazine concentration was 2.5 μM in vitro. The pulse amplitude and duration of the transgene expression were consistent with the furimazine concentration, which is suitable for studying the pulsing dynamics of signaling proteins. Moreover, this tool was activated in a pulsatile manner by the administration of furimazine for target gene expression in transfected mice. As shown in a publication, the synthetic BRET-induced transgene expression system (LuminON) based on luminGAVPO for gene therapy was successfully used to mediate blood glucose homeostasis in a type 1 diabetic mouse model (T1D) in a pulsatile fashion. The stable cell line that expressed luminGAVPO-mediated insulin was engineered and microencapsulated into coherent, semipermeable, and immunoprotective alginate-poly-(L-lysine)-alginate beads. The T1D mice were intraperitoneally injected with microencapsulated transgenic cells and treated with 5 mg/kg furimazine, which resulted in significant restoration of blood glucose levels and enhanced glucose homeostasis. This study provided a promising BRET-based optogenetic tool for the precise control and a better understanding of the pulsing behaviors in pharmacological studies.
Another example is the light-inducible transcription factor EL222, which was directly controlled by its fused partner, Gaussia luciferase
[32]. Unlike luminGAVPO used for gene therapy, this kind of bioluminescence- or BRET-induced transcription factor was successfully applied to achieve communication between synthetic cells and natural cells. Schroeder’s group engineered the high-level Gaussia luciferase-expressing synthetic cell to activate sensitive proteins such as retinal rhodopsins in order to induce photoconidation in the fungus Trichoderma atroviride. The bioluminescence generated in synthetic cells with the addition of 5 μM CTZ enabled intercellular signaling between a synthetic cell and a natural cell. Then, they utilized the light-dependent transcription mechanism mediated by the transcription factor EL222 and the BRET strategy to demonstrate intercellular signaling between light-producing synthetic cells and light-responsive natural cells. The synthetic cells were first engineered to contain the self-activating fusion proteins composed of Gluc on their N-terminal end connected through a flexible peptide linker to the light-inducible bacterial transcription factor EL222 on their C-terminal end. Native CTZ (0.2 nmol) was added every 30 min to the synthetic cell culture in a 384-well microplate to produce and maintain bioluminescence to activate the downstream transcription of the RFP protein. Then, they engineered another fusion protein: N-terminal Gluc fused to C-terminal iLID with a linker peptide and an N-terminal his-tag. The addition of CTZ led to a reaction with the Gluc being localized in the synthetic cells’ membranes to emit bioluminescence, which stimulated iLID and then activated membrane recruitment of sspB-tagged proteins to Gluc-iLID-labeled synthetic cells. Additionally, these BRET-based optogenetic fusion proteins could be imaged simultaneously during the activation process with the use of a microscope, providing spatial information about the activation. Their study to facilitate communication between synthetic cells and natural cells holds promise for deploying synthetic cells as a tunable and embeddable tool by which to control engineered processes inside tissues.
Additionally, a bioluminescence-activated optogenetic tool, bPAC–nLuc for cAMP synthesis, was generated by fusing a light-activated adenylyl cyclase from Beggiatoa with a myc tag to NanoLuc
[33]. This tool can be precisely activated by furimazine and blue light to regulate cAMP production temporally and spatially in cellulo, which means that it is adaptable in imitating physiological levels and maintains cAMP synthesis to control downstream processes in living cells. For instance, the PCCL3 rat thyroid cell stably expressing bPAC–nLuc was engineered to verify this probe’s function and downstream signaling. Upon the addition of Fz or its analog Fz-4377 with long-lasting luminescence and lower toxicity, bPAC–nLuc is activated and sustained to continuously synthesize cAMP localized in the cytosol or nucleus, which leads to cAMP-dependent thyroid cell proliferation. Considering that aberrant cAMP signaling is linked to a variety of diseases such as cancer and heart disease, this tool may play a role in the study of cAMP-involved biology and pharmacological interventions.
Although the cellular activities regulated by these systems are different, their control principles can all be attributed to the optogenetic programming of living organisms using genetically encoded bioluminescence systems. When the substrate is added, the bioluminescence catalyzed by luciferase triggers the light-switchable module to undergo conformational change or expose the cleavage site, leading to the reprogramming of downstream signal transduction and gene expression.
The general procedure for the application of the tools in this section essentially includes cell culture and transfection, luciferin solution preparation, estimation of energy transfer efficiencies, compatibility tests with optogenetic tools, bioluminescence activation and imaging in vitro, tests for specific signaling molecules, animal preparation, and bioluminescence activation and imaging in vivo. In general, researchers have designed these tools as fusion proteins consisting of luciferase and light-switchable domains. Most of these BRET-optogenetic probes and platforms discussed in this section are not designed for neuronal control, but are fit for the study of protein–protein interaction and drug screening (e.g., hyBRET, SPARK2, and BEACON), gene therapy (e.g., LiPOP–NanoLuc and luminGAVPO), and synthetic biological activities (reference
[32] and bPAC–nLuc). Their application could be carried out using plate readers and an imaging system with a CCD camera. When they are applied in animals, surgery protocols such as craniotomy are not needed, but transgenic cell line-derived xenograft models are often generated. Considering that most of them can be expressed in many cell lines using common transfection methods, and that their “on–off” switch can be induced by chemical substrates more conveniently, they may be adapted for the high-throughput screening method of target molecules and can expand the boundaries of gene therapy in future applications.
3. Luminopsins: Bioluminescent Optogenetics Probes in Neuroscience
Luminopsins are a fusion of light-emitting luciferases and light-sensing opsins, which results in optical and chemical control in one molecule. The advantages of utilizing luminopsins are obvious: they allow tracking of the signaling with spatial and temporal precision in a noninvasive manner, which bypasses the major challenge of implanting an external light source. To date, luminopsins have been successfully applied in the modulation of neuronal activity and intracellular signaling at different temporal and spatial resolutions. Some typical luminopsins and their applications are shown as follows (Figure 3).
In 2013, the Hochgeschwender group first reported the development of the fusion of Gaussia luciferase and channelrhodopsin as a luminopsin for use in the manipulation of neuronal activity. The light-activated ion channel, channelrhodopsin-2 from Chlamydomonas (ChR2), and Gaussia luciferase (Gluc), a secreted form of luciferase from the marine copepod Gaussia princeps, were chosen to be fused for the creation of the luminopsin LMO. The addition of coelenterazine (CTZ), the substrate of luciferase Gluc, results in sufficient bioluminescence to activate the coupled ChR2, which in turn leads to the influx of a large number of extracellular cations to produce action potential
[34][35]. Another more efficient bioluminescence-aided optogenetic probe, LMO2, that consists of the red-shifted and more sensitive Volvox channelrhodopsin-1 (VChR1) and Gaussia luciferase, was further developed
[34]. The latter was able to modulate the intrinsic excitability of neurons. Then, this bioluminescent-driven optogenetic tool, LMO2, was applied in noninvasive in vivo imaging and modulation of neuronal activity in mice
[36]. When the luciferin CTZ was intravenously administrated and reached the brain, it reacted with Gluc to produce bioluminescence, which allowed imaging of the neurons and illuminated channelrhodopsin to alter neuronal activity.
Additionally, the Gross group reported an inhibitory luminopsin (iLMO) series that consisted of Renilla luciferase (Rluc) and Natronomonas halorhodopsin (NpHR) and its means of suppressing neural activity in vitro and in vivo
[25]. The luciferases were coupled to light-sensitive opsin NpHR to generate iLMO1 and iLMO2, respectively. Their study showed that iLMO2 had the robust effect of suppressing action potential firing and synchronous bursting in the entire neural network in vitro and in vivo. This inhibitory luminopsin was found to have the ability to modulate neural activity in freely behaving animals and target specific sites in the brain in a barely invasive manner. Further, iLMO2 was utilized in the optogenetic modulation of multiple nodes in an epileptic network for noninvasive seizure suppression, which provided a unique approach for demonstrating the interrogation mechanism of neural networks and treating neurological diseases involving broad neural circuits
[37]. Moreover, iLMO2 was applied in the inhibition of motor neuron activity in a noninvasive way, which played a role in addressing the mechanism of motor axon regeneration by exercise and recovery of function after peripheral nerve injury
[38].
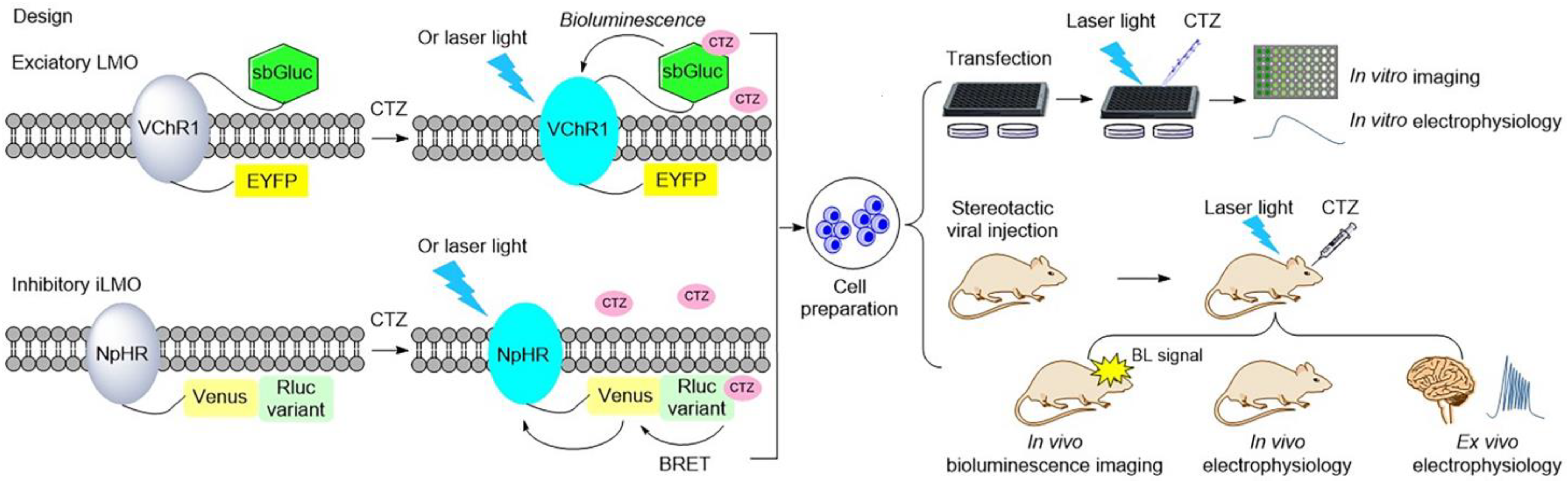
Figure 3. Illustration of typical luminopsins
[25][34][36][39][40][41][42][43] and the general experimental procedure for studying and controlling neural activity in vitro and in vivo. Luminopsins are triggered by bioluminescence or bioluminescence resonance energy transfer (BRET) with the addition of luciferin. They can also be activated by laser light.
In 2016, Berglund et al. promoted the development of luminopsins by fusing the bright Gaussia luciferase variant (sbGluc) with Volvox channelrhodopsin-1 (VChR1) to create the luminopsin LMO3 with improved light emission, which resulted in a higher photocurrent for the modulation of neuronal activity in vitro and in vivo
[39]. Another inhibitory luminopsin named iLMO was formed by use of photosensitive proton pumps from the fungus Leptosphaeria maculans (Mac) and the Gaussia luciferase variant slGluc, described in the same publication. Their study indicated that both of them were able to act as promising probes for the control of neuronal behavior in vitro and in vivo
[39].
A few years later, the luminopsin LMO3 was applied in cell transplantation therapy to enhance neuronal repair, neural network connections, and functional recovery after ischemic stroke in a mouse model
[40]. Thus, these transplanted cells containing a bioluminescent optogenetic tool provide a novel and promising treatment for neuronal functional recovery. Subsequently, Zenchak et al. demonstrated that the activation of LMO3 by CTZ led to stimulation of transplanted neural precursor cells, which improved motor skills in a mouse model of Parkinson’s disease
[44]. Moreover, LMO3 and iLMO were employed as a pair of probes to manipulate neuronal activity in hippocampal CA3 and were found to remarkably affect spatial working memory and spatial and episodic short-term memory, but rarely affected either spatial or episodic long-term memory
[41]. As the dysfunction of working memory and short-term memory is correlated to mild Alzheimer’s disease (AD), this application of excitatory and inhibitory luminopsins in studying the function of CA3 in working memory may be helpful for treatment in the early stages of AD. Furthermore, LMO3 was utilized as a tool to rehabilitate spinal cord injury in a mouse model, which provides a foundation for the use of bioluminescent optogenetic treatment for functional recovery after severe spinal cord injury
[42]. Additionally, LMO3 was also employed in noninvasive manipulation and imaging of neurons for postnatal brain development in mice
[43].
Since LMO3 is a promising tool involved in neurological disorders, scientists are focusing on investigating the optogenetic mechanism driven by bioluminescent photon emission. Gomez-Ramirez et al. characterized the relationship between bioluminescent optogenetic effects and the bioluminescent photon emission in the neocortex in vivo
[45]. Further, Zhang et al. engineered an enhanced iteration of LMO3, named eLMO3, with improved membrane trafficking, which resulted in a significant increase in photocurrents and more efficient control of neuronal activity for functional interrogation such as the examination of whisker-response effects
[46]. Subsequently, the bioluminescent optogenetics tool eLMO3 has been used to enhance axon regeneration after peripheral nerve injury
[47][48]. Recently, LMO3.2 was designed by replacing the Volvox channel used in LMO3 with a more sensitive blue-shifted opsin from Scherffelia, which resulted in a four-fold higher response after being induced by CTZ. With the increased light sensitivity and accelerated response time, LMO3.2 has the capacity to efficiently control neuronal activity by noninvasive modulation and improve locomotor function after thoracic spinal cord injury
[49]. Moreover, luminopsin 4 (LMO4) was created by fusing the brighter Gaussia luciferase mutant GlucM23 with the optogenetic element Volvox channelrhodopsin-1 (VChR1). Additionally, the combination of GlucM23 with anion channelrhodopsin iChloC produced inhibitory luminopsin 4 (iLMO4). The authors demonstrated that both LMO4 and iLMO4 were efficient probes for bimodal opto- and chemogenetic control of neural activity in a mouse model
[50]. In 2020, Berglund et al. incorporated channelrhodopsin-2 with step-function mutations as the light-sensing opsin moiety and sbGluc as the light-emitting luciferase in a new luminopsin fusion protein, named step-function luminopsin (SFLMO), which has diversified the toolbox of bioluminescence optogenetic probes for bimodal control of neuromodulation for functional interrogation or therapeutic purposes
[51]. Furthermore, scientists have also employed both the optogenetics approach and bioluminescence reporters to investigate mammalian circadian rhythms
[52][53][54].
Currently, the luminopsin toolbox contains excitatory, inhibitory, and step-function luminopsins. They are triggered both optically by light and chemically by administration of substrates and have been successfully applied in the bimodal modulation of neural activities in vitro and in vivo, especially in the interrogation of neurons in freely moving animals. Several studies of the past few years have indicated that luminopsins and the derived platforms allow noninvasive and remote control of transplanted stem cells’ activities in neurological disorders, which can potentially provide an important approach to the therapy of neurodegenerative diseases, including Parkinson’s disease, Huntington’s disease, and Alzheimer’s disease.
Apart from luminopsins, Hochgeschwender and co-workers designed an ingenious bioluminescence-driven optogenetic system that couples luciferase with photoreceptors without fusion. They took advantage of light-emitting luciferase sbGluc that is released from presynaptic vesicles and produces interluminescence to activate postsynaptic excitatory or inhibitory opsin, thus providing a platform by which to interrogate specific neural circuits with temporal and spatial control in vivo
[55].
Almost all luminopsins have been developed for the study and modulation of neuronal activity in the intact living brains of freely behaving animals. Luminopsins and luminopsin-type proteins share similar molecular designs and general experimental procedures for their application in cells and organisms.
All luminopsins and inhibitory luminopsins are the fusion proteins of blue light-emitting luciferases or luciferase-fluorescent proteins paired with the blue light-sensing photoreceptors. Specifically, LMO1 (Gluc–ChR2–EYFP), LMO2 (Gluc–VChR1–EYFP), LMO3 (sbGluc–VChR1–EYFP), and LMO4 (GlucM23–VChR1–EYFP) share the same fusion design as that of the Gluc-type luciferase, which is fused to the N-terminus of the ChR domain, while the fluorescent protein EYFP is fused to the C-terminus of the ChR domain. eLMO3 was created by inserting the Golgi trafficking signal from a neuronal potassium channel into the position between VChR1 and EYFP in the LMO3 fusion protein. The step-function series luminopsin fusion proteins were generated by coupling the luciferase mutant sbGluc to the N-terminal of the ChR2 mutation. With regard to the design and construction of inhibitory luminopsins, TagRFP–Rluc is coupled to the C-terminus of NpHR to create iLMO1. The TagRFP–Rluc in iLMO1 is replaced by a nanolantern to create iLMO2. The design of iLMO4 (GlucM23–iChloC–EYFP) is similar to that of LMO4; the GlucM23 luciferase mutant is fused to the N-terminus of iChloC, and the fluorescent protein EYFP is fused to the C-terminus of iChloC. Likewise, iLMO is a fusion protein of slGluc and Mac-EYFP.
Regarding the general approaches of these tools, the basic procedures include cell culture and transfection, luciferin solution preparation, bioluminescence imaging and activation in vitro, animal preparation, bioluminescence activation in vivo, and surgery protocols. When these luminopsin-type fusion proteins are expressed in neurons, bioluminescence from the reaction of luciferase and luciferin can excite or silence neuronal activity in cultured neurons and in brain slices, and can elicit certain behaviors in freely moving mice. In addition to general approaches for the application of luminopsins, the detailed experimental protocol and tips for LMO3 application in the study of developing neural circuits in postnatal mice can be found in Crsepo’s protocol
[43]. Tips regarding methodological procedures for LMO and iLMO (iLMO2, LMO3, or eLMO3) when addressing neuronal repair and functional regeneration after peripheral nerve injury
[38][47], spinal cord injury
[42], and ischemic stroke
[40] can be found in related articles. Furthermore, tips for implementing the light-sensitive opsins ChR2 and NpHR in studies of circadian behavior and physiology including surgical procedures can be found in Jones’s protocol
[52].