Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Phillip Myer | -- | 2602 | 2023-03-22 15:50:22 | | | |
| 2 | Sirius Huang | Meta information modification | 2602 | 2023-03-23 02:08:59 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Soltis, M.P.; Moorey, S.E.; Egert-Mclean, A.M.; Voy, B.H.; Shepherd, E.A.; Myer, P.R. Anatomy of the Rumen Affecting the Microbiome. Encyclopedia. Available online: https://encyclopedia.pub/entry/42439 (accessed on 02 March 2026).
Soltis MP, Moorey SE, Egert-Mclean AM, Voy BH, Shepherd EA, Myer PR. Anatomy of the Rumen Affecting the Microbiome. Encyclopedia. Available at: https://encyclopedia.pub/entry/42439. Accessed March 02, 2026.
Soltis, Macey P., Sarah E. Moorey, Amanda M. Egert-Mclean, Brynn H. Voy, Elizabeth A. Shepherd, Phillip R. Myer. "Anatomy of the Rumen Affecting the Microbiome" Encyclopedia, https://encyclopedia.pub/entry/42439 (accessed March 02, 2026).
Soltis, M.P., Moorey, S.E., Egert-Mclean, A.M., Voy, B.H., Shepherd, E.A., & Myer, P.R. (2023, March 22). Anatomy of the Rumen Affecting the Microbiome. In Encyclopedia. https://encyclopedia.pub/entry/42439
Soltis, Macey P., et al. "Anatomy of the Rumen Affecting the Microbiome." Encyclopedia. Web. 22 March, 2023.
Copy Citation
The rumen is a complex organ that is critical for its host to convert low-quality feedstuffs into energy. The conversion of lignocellulosic biomass to volatile fatty acids and other end products is primarily driven by the rumen microbiome and its interaction with the host. Importantly, the rumen is demarcated into five distinct rumen sacs as a result of anatomical structure, resulting in variable physiology among the sacs.
rumen
microbiome
rumen sacs
1. Introduction
Cattle are essential to the global protein supply due to their unique ability to consume plant material that is indigestible to mammals and convert it into usable energy [1]. The fermentation of plant material is performed within cattle’s four-chambered stomach, which consists of the reticulum, rumen, omasum, and abomasum [1], where the rumen provides a suitable environment for microbes to reside. Importantly, due to pillars, grooves, and folds demarcating the rumen, there are five distinct rumen sacs, creating variable sub-ruminal environments (Figure 1). Therefore, these distinct sacs may contain microbial communities that have acclimated to the varying biogeographical regions within the rumen.
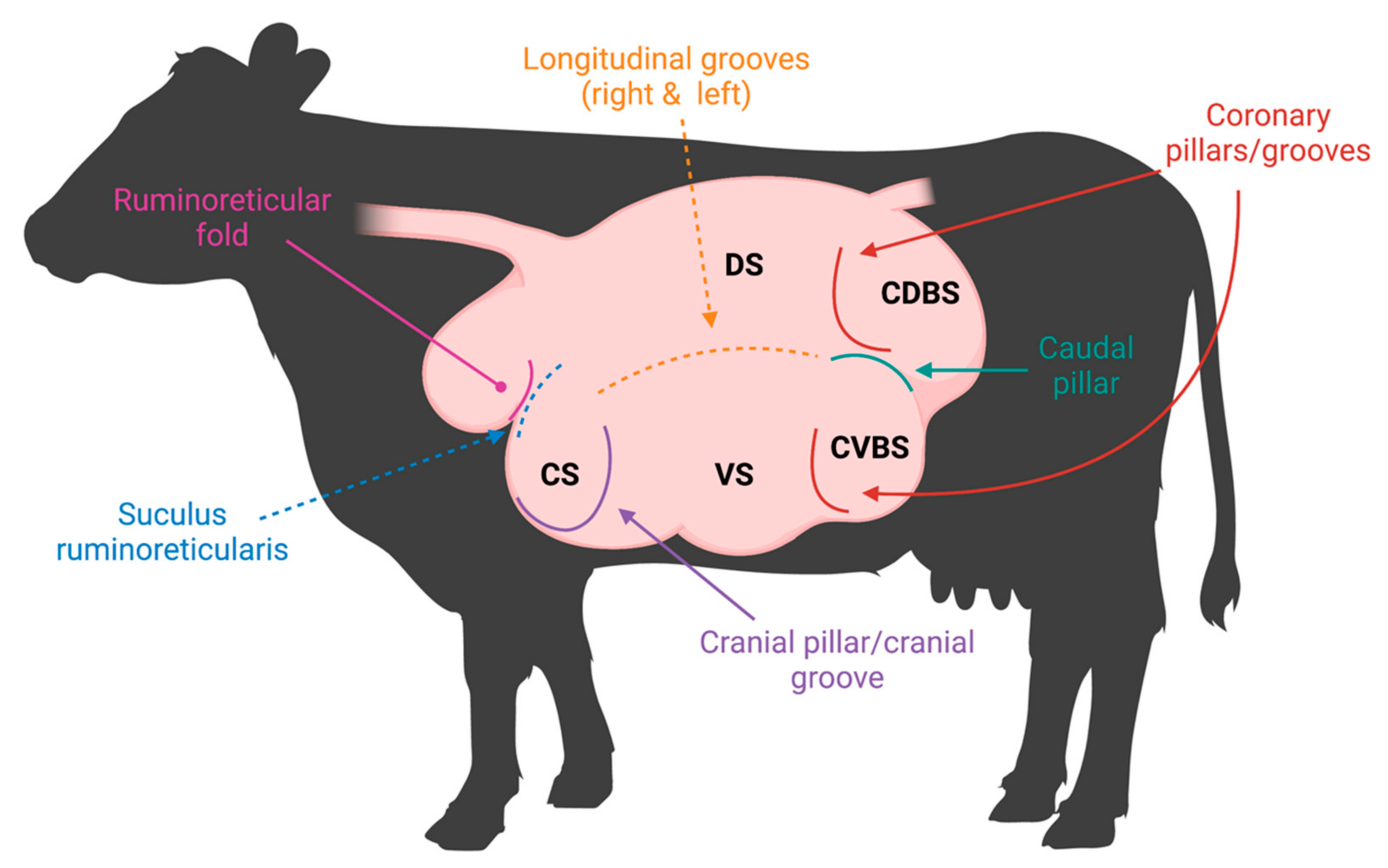
Figure 1. Rumen pillars and grooves visualized in a labeled rumen. Visual representation of musculature demarcating the rumen both internally (pillars) and externally (grooves). The rumen sacs are labeled as follows: cranial sac (CS), caudodorsal blind sac (CDBS), caudoventral blind sac (CVBS), dorsal sac (DS), and ventral sac (VS).
The anatomical configuration of the rumen creates functional variation within each rumen sac due to assorted accumulation of feed particles, saliva, and volatile fatty acids (VFAs). Investigation of different ruminal fermentation parameters, such as VFA concentration, pH, ammonia nitrogen (N), and other minerals (sodium, potassium, calcium, chloride, and phosphorus), were observed from different rumen sac sampling [2]. Cranial, dorsal, caudodorsal, caudoventral, and two different areas within the ventral sac were sampled to obtain a complete assessment of the ruminal environment. Differences were observed in fermentation parameters between the cranial sac and the other four sacs [2]. Notably, the cranial sac had greater pH and sodium concentrations, while also having lower total VFA concentrations [2], due to the increased amount of saliva and feed particles within the cranial sac. Although microbial communities were not investigated, microbial variation may occur, as favorable conditions for certain microbial species can be present [3]. Amylolytic bacteria favor and thrive in lower pH environments caused by high grain diets, while this environment will inhibit the growth of cellulolytic bacterial species [3]. Differences observed in rumen fermentation parameters are due to sample site [2], which can be attributed to anatomical distinctions made among the ruminal cavity. Pillars, grooves, and folds form the anatomical distinctions and thus, five separate rumen sacs. Environmental conditions of the rumen sacs continuously fluctuate due to ruminal contractions, stratification, and digestibility of feedstuffs occurring throughout beef cattle production. Fluctuations in each ruminal sac environment may instigate ruminal dysbiosis causing adjustments to microbial communities. Improved understanding of the five ruminal sacs, their distinct microbial communities, and the interactions between host and microbiome will reduce undesired consequences caused by unavoidable environmental, diet, and management changes occurring during production.
2. Ruminal Environment Variations and Their Impact on Microbial Community Variation
The five rumen sacs establish unique conditions that favor specific microbial species, and therefore, these microbes may occupy specific sacs with less competition [4][5] For example, in lambs, epimural bacteria appear to be less numerous in the caudal sac than other sacs of the rumen; yet, in the adult animal, the microbial colonization was greater in the dorsal sac than the ventral [5]. These differences in epimural microbial populations highlight the variation attributed to age and age × diet interactions that may occur due to these ruminal compartments or even colonization potential. Indeed, it has been shown that some epithelium-associated microbes adhere directly to the epithelium or directly to the glycocalyx of the first colonizing strains [5][6][7]. These interactions vary based on the location within the rumen and available substrates. Ruminal chemistry can also vary, as significant differences in VFA concentrations were observed when five different positions—dorsal, middle, ventral, reticulum, and dorsal posterior regions—were sampled within the reticulo-rumen [8]. However, pH was only different between ventral and dorsal regions, with an average of 0.43 pH less in the dorsal region compared to the ventral [8]. Sampling time was possibly a confounding factor among these parameters, as the dorsal region consistently contained greater VFA concentrations and was 38% greater than the ventral region [8]. This interaction between the host environmental conditions and the microbial communities is critical to appreciating the ruminal niches and how they establish.
Beyond differences in ruminal sac chemistry, stratification of rumen digesta occurs based on specific gravity differentiating rumen liquid from rumen solid digesta. The liquid layer is topped by an accumulation of low-density forage particles that form the fiber mat [9]. Ruminal stratification is best demonstrated when cattle are fed high forage diets [10] due to slower rate of fermentation and greater abundance of low specific gravity feedstuffs that rise above the rumen liquid and create the fiber mat. Vertical stratification of rumen contents can further be explained by the primary contraction cycle that facilitates the flow of denser feedstuffs to the ventral sac, where liquid digesta accumulates (Figure 2) [11]. Less dense plant materials are not cycled during primary contractions, and thus, gather atop the liquid layer forming the fiber mat in the dorsal sac [12]. Due to the primary contraction cycle enabling the accumulation of low-density plant feedstuffs in the dorsal sac, dorsal locations are more likely to be affected by stratified rumen contents [13]. When examining whether ruminal bacterial structure, population, and fermentation parameters differed between ruminal locations, rumen digesta samples were obtained from the cranial ventral, cranial dorsal, central rumen, caudal dorsal, and caudal ventral sacs [14]. No differences were observed between bacterial structure and sampling location; however, sampling was limited to the ventral and dorsal sacs. Greater pH was detected in the cranial regions [14], which influences microbial growth and is an important fermentation variable, although differences were lost due to the analysis technique [14][15]. Polymerase chain reaction–denaturing gradient gel electrophoresis (PCR-DGGE) was used to discern bacterial communities but is unable to assess bacterial species as in-depth as high-throughput sequencing (HTS). Therefore, the use of HTS is ideal to detect minor bacterial species variation between rumen sacs. Although no significant differences were observed between rumen sacs, eight specific bacterial species increased in caudal regions. Ruminal contractions impact the movement and stratification of feedstuffs and therefore microbial species which may have affected the greater populations seen among caudal regions and more basic pH among the cranial regions.
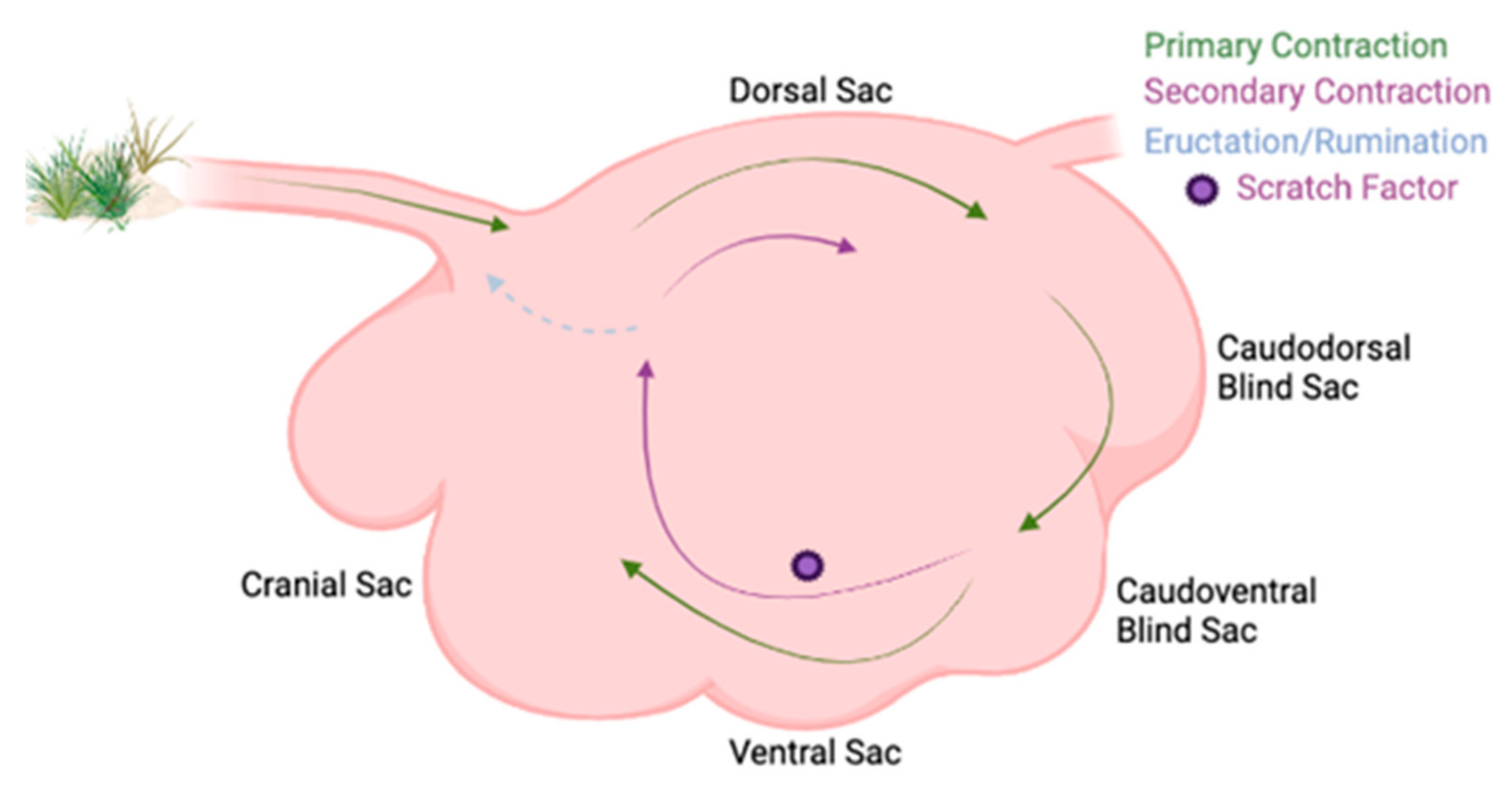
Figure 2. Ruminal contraction cycles in a labeled rumen. Labeled rumen sacs with ruminal contractions shown. The primary contraction cycle is shown with the green arrows in the clockwise cycle and it follows within the rumen. The secondary contraction cycle is shown with the purple arrows and is not explicitly explained in the text, but is where eructation, rumination, and continual fermentation occur, and the feedstuffs are not passed onto the omasum.
Considering the primary, secondary, and extraruminal contractions, rumen motility is an important factor contributing to microbial variation in the rumen [16]. Rumen motility permits the continuous colonization and interactions with ingesta, resulting in efficient fermentation processes [17][18]. Arowolo et al. examined the effect of motility in vitro using the RUSITEC (rumen simulation technique) system, hypothesizing that changes in rotation speeds would impact rumen fermentation, saturation of dissolved gases, hydrogen, and methane emissions, microbial protein synthesis, and selected microbial populations. A moderate in vitro rotation speed (15 rpm) resulted in the greatest total VFA and microbial protein concentrations, while also increasing fungi and protozoa counts in the solid contents and bacteria and fungi in the liquid contents [16]. This work was important in providing insights into the importance of motility on rumen function, fermentation, and microbial activity. In vivo, motility and the frequency of primary contractions are important in the mixing and passage of ruminal content to the omasum, which has been shown to be reduced with episodes of ruminal dysfunction, such as ruminal acidosis [19]. The change in microbiome and microbial fermentation as a result of acidosis and motility may affect VFA flow to the ventral sac, subsequently reducing the absorption of VFA from the rumen [20]. Yet, rumen motility is affected by numerous factors, such as blood flow, diet, nervous system, and rumen dysfunction [1]. In the southeastern US, where fescue toxicosis is a primary concern for beef producers, ergovaline has been shown to impact rumen motility through a decrease in frequency and amplitude of ruminal contractions, while also impacting rumen fill and intake [21]. As an example of rumen dysfunction and dietary impacts on rumen motility, these fescue ergovaline effects on rumen function are of concern, as bacterial and fungal communities are important towards the animal’s response to fescue toxicosis [22][23][24]. As continued research advances the understanding of motility and digesta passage, their effects on the rumen microbiome and the potential to interact with the host via the ecological niches established by the rumen sacs will help to define further contributions to microbiome variation.
3. Ruminal Papillae Variation among the Different Sacs
Ruminal papillae are organs of absorption, extending from the rumen epithelium to optimize surface area for short-chain fatty acid (SCFA) absorption. Short-chain fatty acids account for over 70% of the metabolizable energy supply [10][25][26]. Papillae distribution, size, width, and volume are closely related to host feeding habits, forage availability, forage quality, particle size, and digestibility [10][25][26][27]. Papillae continuously adapt to changes in host diet and stratification of digesta that occur throughout cattle production, which enable its host to better absorb VFAs. In regions of the rumen with greater amounts of digesta, dense lawns of papillae exist, improving the ability to absorb VFAs. In regions such as the dorsal sac, there is a lower density of papillae, as VFA concentrations are reduced due to limited digestion and fermentation activity. As a result of these differences in biochemistry and physiology, papillae from various sacs differ in both size and shape [28]. Variations in papillae density have been observed because of increased proportions of propionate and butyrate produced by the fermentative ruminal bacteria [29]. These VFAs can increase ruminal blood flow, stimulating mucosal mitosis and epithelial proliferation, which increases the size and number of papillae, ultimately enhancing the reticulo-rumen’s ability to absorb VFAs [10][27][30]. Papillae VFA absorption contributes to maintaining rumen VFA concentrations and pH, which supports microbial fermentation of forage feedstuffs [31][32][33][34].
Ruminal papillae morphology and count are related to ruminant type. Ruminants can be classified based on the source of feed, which has evolutionarily impacted their anatomy and physiology. These categories include concentrate selectors, intermediate, and grass/roughage eaters [35]. Importantly, each ruminant type has varying distributions of papillae based on their diets. Concentrate selectors have a more even distribution of papillae seen throughout their rumen due to the increased passage rate and digestibility of their diet. Some concentrate selectors have papillae on the pillars within their rumen. Grass/roughage eaters or browsers, including cattle, have a characteristically uneven distribution of papillae throughout the rumen, reflecting the stratification of feed particles and thus, regional differences of microbial activity within their rumen [10]. Generally, within cattle, the rumen is devoid of papillae on the pillars and in the dorsal sac, depending on diet [10]. With significant differences in papillae configurations among ruminant types, it is consistently noted that the most numerous and extended papillae are located on the cranial and caudodorsal blind sac floor. The caudodorsal blind and cranial sac papillae retain greater surface enlargement factors (SEF = 2 × papillary surface + basal surface over the basal surface) due to increased VFA concentration [10].
The ventral sac papillae structure has been well-defined, due to ease of access and its dense lawn of papillae. However, the ventral sac is unlikely to be a complete representation of rumen papillae morphology and structure. Due to factors, such as digesta stratification, pH, and feedstuff digestibility, there may be distinct differences among the papillae in different rumen sacs. Micro- and macroscopic fluctuations in papillae morphology were observed through dry and lactation periods as an impact of increased dietary concentrates [32]. Twelve rumen-cannulated Holstein cows were sampled from the ventral, caudoventral blind, and caudodorsal blind sacs. There was an overall greater surface area of papillae with increased concentrates, indicating papillae respond to enhanced microbial breakdown of the easily fermentable organic matter through the increase in papillary growth rate [32]. Further differences were identified among the three different rumen sacs, with the ventral sac containing the greatest surface area of papillae. The caudodorsal blind sac also increased in papillae density, but the caudoventral blind sac did not have significant differences after concentrates were added [32]. Increases in papillae density provide greater surface area for absorption of VFA in the rumen. The caudoventral blind sac typically has a greater amount of liquid digesta due to the stratification of rumen contents. During acclimation to high-grain diets, the passage rate increases and therefore, more liquid digesta is cycled. However, the greater liquid accumulation does not deviate far from the average environment of the caudoventral blind sac, which may explain why no significance was determined.
Papillae express a variety of genes facilitating barrier defense, pH, receptors, and transporters for absorption of VFAs. Core genes expressed in papillae remain constant throughout a ruminant’s life due to their importance in host health. However, as mentioned previously, diet, location, and stress may impact ruminal epithelial tissue. In heat-stressed cattle (28 °C, temperature–humidity index = 76), lactating Holstein dairy cows had an increased expression of proteins involved in the AMP-activated protein kinase (AMPK) and insulin signaling pathways contrasted to thermoneutral cattle (15 °C, temperature–humidity index = 60). Expression of proteins was upregulated for those involved in the chaperone-mediated folding of proteins, whereas those involved in the antioxidant defense system were downregulated [36]. These changes in expression are important for host health, describing the gross physiological adaptations of ruminal papillae to heat stress. Yet, when diet is examined as a factor, no significant differences in epithelial gene expression were observed in cannulated Holstein cows, fed varying amounts of high-quality hay and decreasing percentages of concentrates [37]. However, evidence suggests that epithelial receptor expression may differ when different diets are fed, such as in Toll-like receptor genes. Increasing dietary starch increased expression of Toll-like receptor genes in goats which may be due to the genes’ ability to bind lipopolysaccharide and initiate a host immune response [37][38][39]. In a study conducted by Kern et al., steers with either high or low intake and high or low gain were examined for differentially expressed genes. There were differentially expressed genes determined in steers with high gain and low intake (HL), against the other efficiency type steers [40]. One gene upregulated in HH steers was lipopolysaccharide-binding protein (LBP), which aids in host defense and contributes to innate immune responses [40]. It was hypothesized that more efficient steers had less immune response which may drive improved production without wasting energy on inflammation and immune responses [40]. The gene Regulator of G Protein Signaling 5 (RGS5) was also differentially expressed in HL steers, where it was upregulated when measured against the HH steers [40]. In murine studies, RGS5 knockout mice were observed to have greater fat mass regardless of diet [41], which may also influence HL steers’ residual feed intake. RGS5 was also shown to be involved with arterial wall development in mice [40][42], which may facilitate the development of a more efficient rumen vasculature arrangement. Papillae are fixed to the rumen epithelium where they aid in absorption of VFAs, metabolic activity, and barrier protection, but they also support the epithelial microbial community. An epimural microbial community establishes throughout the entirety of the rumen given that they are attached to the epithelium or within the papillae that comprise every sac. These epimural microbes are distinct from the other microbial communities in that they inhabit the rumen epithelium, scavenge oxygen, recycle rumen epithelial tissue, and serve as an interface between the host and microbiome.
References
- Krehbiel, C. Invited review: Applied nutrition of ruminants: Fermentation and digestive physiology. Prof. Anim. Sci. 2014, 30, 129–139.
- Shen, J.S.; Chai, Z.; Song, L.J.; Liu, J.X.; Wu, Y.M. Insertion depth of oral stomach tubes may affect the fermentation parameters of ruminal fluid collected in dairy cows. J. Dairy Sci. 2012, 95, 5978–5984.
- Harlow, B.E.; Flythe, M.D.; Aiken, G.E. Effect of biochanin A on corn grain (Zea mays) fermentation by bovine rumen amylolytic bacteria. J. Appl. Microbiol. 2017, 122, 870–880.
- Sbardellati, D.L.; Fischer, A.; Cox, M.S.; Li, W.; Kalscheur, K.F.; Suen, G. The bovine epimural microbiota displays compositional and structural heterogeneity across different ruminal locations. J. Dairy Sci. 2020, 103, 3636–3647.
- Rieu, F.; Fonty, G.; Gaillard, B.; Gouet, P. Electron microscopy study of the bacteria adherent to the rumen wall in young conventional lambs. Can. J. Microbiol. 1990, 36, 140–144.
- McCowan, R.; Cheng, K.; Bailey, C.; Costerton, J. Adhesion of bacteria to epithelial cell surfaces within the reticulo-rumen of cattle. Appl. Environ. Microbiol. 1978, 35, 149–155.
- Cheng, K.J.; McCowan, R.P.; Costerton, J.W. Adherent epithelial bacteria in ruminants and their roles in digestive tract function. Am. J. Clin. Nutr. 1979, 32, 139–148.
- Bryant, A. Variations in the pH and volatile fatty acid concentration within the bovine reticulo-rumen. N. Z. J. Agric. Res. 1964, 7, 694–706.
- Schmitz-Esser, S. The Rumen Epithelial Microbiota: Possible Gatekeepers of the Rumen Epithelium and Its Potential Contributions to Epithelial Barrier Function and Animal Health and Performance. Meat Muscle Biol. 2021, 4, 1–11.
- Church, D.C. The Ruminant Animal: Digestive Physiology and Nutrition; Waveland Press: Long Grove, IL, USA, 1993.
- Tafaj, M.; Junck, B.; Maulbetsch, A.; Steingass, H.; Piepho, H.P.; Drochner, W. Digesta characteristics of dorsal, middle and ventral rumen of cows fed with different hay qualities and concentrate levels. Arch. Anim. Nutr. 2004, 58, 325–342.
- Grünberg, W.; Constable, P.D. CHAPTER 6—Function and Dysfunction of the Ruminant Forestomach. In Food Animal Practice, 5th ed.; Anderson, D.E., Rings, D.M., Eds.; W.B. Saunders: Saint Louis, MO, USA, 2009; pp. 12–19.
- Millen, D.D.; De Beni Arrigoni, M.; Lauritano Pacheco, R.D. Rumenology, 1st ed.; Springer: Cham, Switzerland, 2016.
- Li, M.; Penner, G.B.; Hernandez-Sanabria, E.; Oba, M.; Guan, L.L. Effects of sampling location and time, and host animal on assessment of bacterial diversity and fermentation parameters in the bovine rumen. J. Appl. Microbiol. 2009, 107, 1924–1934.
- Rumsey, T.S.; Putnam, P.A.; Bond, J.; Oltjen, R.R. Influence of level and type of diet on ruminal pH and VFA, respiratory rate and EKG patterns of steers. J. Anim. Sci. 1970, 31, 608–616.
- Adebayo Arowolo, M.; Zhang, X.M.; Wang, M.; Wang, R.; Wen, J.N.; Hao, L.Z.; He, J.H.; Shen, W.J.; Ma, Z.Y.; Tan, Z.L. Proper motility enhances rumen fermentation and microbial protein synthesis with decreased saturation of dissolved gases in rumen simulation technique. J. Dairy Sci. 2022, 105, 231–241.
- Waghorn, G.; Reid, C. Rumen motility in sheep and cattle given different diets. N. Z. J. Agric. Res. 1983, 26, 289–295.
- Beauchemin, K. Invited review: Current perspectives on eating and rumination activity in dairy cows. J. Dairy Sci. 2018, 101, 4762–4784.
- Crichlow, E. Ruminal lactic acidosis: Forestomach epithelial receptor activation by undissociated volatile fatty acids and rumen fluids collected during loss of reticuloruminal motility. Res. Vet. Sci. 1988, 45, 364–368.
- Storm, A.C.; Kristensen, N.B. Effects of particle size and dry matter content of a total mixed ration on intraruminal equilibration and net portal flux of volatile fatty acids in lactating dairy cows. J. Dairy Sci. 2010, 93, 4223–4238.
- Ahn, G.; Ricconi, K.; Avila, S.; Klotz, J.L.; Harmon, D.L. Ruminal motility, reticuloruminal fill, and eating patterns in steers exposed to ergovaline. J. Anim. Sci. 2019, 98, skz374.
- Melchior, E.A.; Smith, J.K.; Schneider, L.G.; Mulliniks, J.T.; Bates, G.E.; Flythe, M.D.; Klotz, J.L.; Ji, H.; Goodman, J.P.; Lee, A.R.; et al. Effects of endophyte-infected tall fescue seed and red clover isoflavones on rumen microbial populations and physiological parameters of beef cattle. Transl. Anim. Sci. 2019, 3, 315–328.
- Melchior, E.A.; Myer, P.R. Fescue toxicosis and its influence on the rumen microbiome: Mitigation of production losses through clover isoflavones. J. Appl. Anim. Res. 2018, 46, 1280–1288.
- Koester, L.R.; Poole, D.H.; Serão, N.V.; Schmitz-Esser, S. Beef cattle that respond differently to fescue toxicosis have distinct gastrointestinal tract microbiota. PLoS ONE 2020, 15, e0229192.
- Bergman, E.N. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 1990, 70, 567–590.
- Steele, M.A.; Croom, J.; Kahler, M.; AlZahal, O.; Hook, S.E.; Plaizier, K.; McBride, B.W. Bovine rumen epithelium undergoes rapid structural adaptations during grain-induced subacute ruminal acidosis. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011, 300, R1515–R1523.
- Wang, B.; Wang, D.; Wu, X.; Cai, J.; Liu, M.; Huang, X.; Wu, J.; Liu, J.; Guan, L. Effects of dietary physical or nutritional factors on morphology of rumen papillae and transcriptome changes in lactating dairy cows based on three different forage-based diets. BMC Genom. 2017, 18, 353.
- Dehority, B.A. Gastrointestinal tracts of herbivores, particularly the ruminant: Anatomy, physiology and microbial digestion of plants. J. Appl. Anim. Res. 2002, 21, 145–160.
- Niwińska, B.; Hanczakowska, E.; Arciszewski, M.; Klebaniuk, R. Exogenous butyrate: Implications for the functional development of ruminal epithelium and calf performance. Animal 2017, 11, 1522–1530.
- Mach, N.; Devant, M.; Bach, A. Rumen fermentation parameters and rumen papillae characteristics in finishing bulls as affected by nonfibrous carbohydrate level and lipid source of the diet. J. Anim. Vet. Adv. 2006, 5, 220–225.
- Aschenbach, J.R.; Penner, G.B.; Stumpff, F.; Gabel, G. Ruminant Nutrition Symposium: Role of fermentation acid absorption in the regulation of ruminal pH. J. Anim. Sci. 2011, 89, 1092–1107.
- Dieho, K.; Bannink, A.; Geurts, I.A.L.; Schonewille, J.T.; Gort, G.; Dijkstra, J. Morphological adaptation of rumen papillae during the dry period and early lactation as affected by rate of increase of concentrate allowance. J. Dairy Sci. 2016, 99, 2339–2352.
- Dijkstra, J.; Ellis, J.; Kebreab, E.; Strathe, A.; López, S.; France, J.; Bannink, A. Ruminal pH regulation and nutritional consequences of low pH. Anim. Feed Sci. Technol. 2012, 172, 22–33.
- Penner, G.B.; Aschenbach, J.R.; Gabel, G.; Rackwitz, R.; Oba, M. Epithelial capacity for apical uptake of short chain fatty acids is a key determinant for intraruminal pH and the susceptibility to subacute ruminal acidosis in sheep. J. Nutr. 2009, 139, 1714–1720.
- Hofmann, R.R. Evolutionary steps of ecophysiological adaptation and diversification of ruminants: A comparative view of their digestive system. Oecologia 1989, 78, 443–457.
- Eslamizad, M.; Albrecht, D.; Kuhla, B. The effect of chronic, mild heat stress on metabolic changes of nutrition and adaptations in rumen papillae of lactating dairy cows. J. Dairy Sci. 2020, 103, 8601–8614.
- Petri, R.M.; Kleefisch, M.T.; Metzler-Zebeli, B.U.; Zebeli, Q.; Klevenhusen, F. Changes in the Rumen Epithelial Microbiota of Cattle and Host Gene Expression in Response to Alterations in Dietary Carbohydrate Composition. Appl. Environ. Microbiol. 2018, 84, e00384-18.
- Chen, Y.; Oba, M.; Guan, L.L. Variation of bacterial communities and expression of Toll-like receptor genes in the rumen of steers differing in susceptibility to subacute ruminal acidosis. Vet. Microbiol. 2012, 159, 451–459.
- Liu, J.H.; Bian, G.R.; Zhu, W.Y.; Mao, S.Y. High-grain feeding causes strong shifts in ruminal epithelial bacterial community and expression of Toll-like receptor genes in goats. Front. Microbiol. 2015, 6, 167.
- Kern, R.J.; Lindholm-Perry, A.K.; Freetly, H.C.; Snelling, W.M.; Kern, J.W.; Keele, J.W.; Miles, J.R.; Foote, A.P.; Oliver, W.T.; Kuehn, L.A.; et al. Transcriptome differences in the rumen of beef steers with variation in feed intake and gain. Gene 2016, 586, 12–26.
- Deng, W.; Wang, X.; Xiao, J.; Chen, K.; Zhou, H.; Shen, D.; Li, H.; Tang, Q. Loss of regulator of G protein signaling 5 exacerbates obesity, hepatic steatosis, inflammation and insulin resistance. PLoS ONE 2012, 7, e30256.
- Cho, H.; Kozasa, T.; Bondjers, C.; Betsholtz, C.; Kehrl, J.H. Pericyte-specific expression of Rgs5: Implications for PDGF and EDG receptor signaling during vascular maturation. FASEB J. 2003, 17, 440–442.
More
Information
Subjects:
Agriculture, Dairy & Animal Science
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
4.2K
Revisions:
2 times
(View History)
Update Date:
23 Mar 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





