
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ivan Chan-Zapata | -- | 3909 | 2023-03-18 04:50:25 | | | |
| 2 | Ivan Chan-Zapata | -18 word(s) | 3891 | 2023-03-18 05:01:16 | | | | |
| 3 | Lindsay Dong | -1 word(s) | 3890 | 2023-03-20 07:57:59 | | |
Video Upload Options
Respiratory viruses represent a world public health problem, giving rise to annual seasonal epidemics and several pandemics caused by some of these viruses, including the COVID-19 pandemic. Some antiviral drugs have been licensed for the treatment of influenza and respiratory syncytial virus, but they cause side effects, lead to resistant viral strains, or possess various limitations. On the other hand, no specific drugs are licensed to treat other viral respiratory diseases. In this sense, natural products have appeared as promising alternatives in searching for new compounds with antiviral activity. Quinones have demonstrated activity against respiratory viruses, so the activity of the different types of natural and synthetic quinones against these pathogens and their molecular targets are summarized.
1. Introduction
2. Respiratory Viruses
2.1. Parainfluenza Virus (PIV), Human Metapneumovirus (HMPV), and Respiratory Syncytial Virus (RSV)
2.2. Adenovirus (AdV)
2.3. Rhinovirus (RV)
2.4. Bocavirus (BoV)
2.5. Influenza Virus
2.6. Coronavirus (CoV)
3. Pharmacological Treatments for Respiratory Virus Infections
4. Quinones and Respiratory Viruses
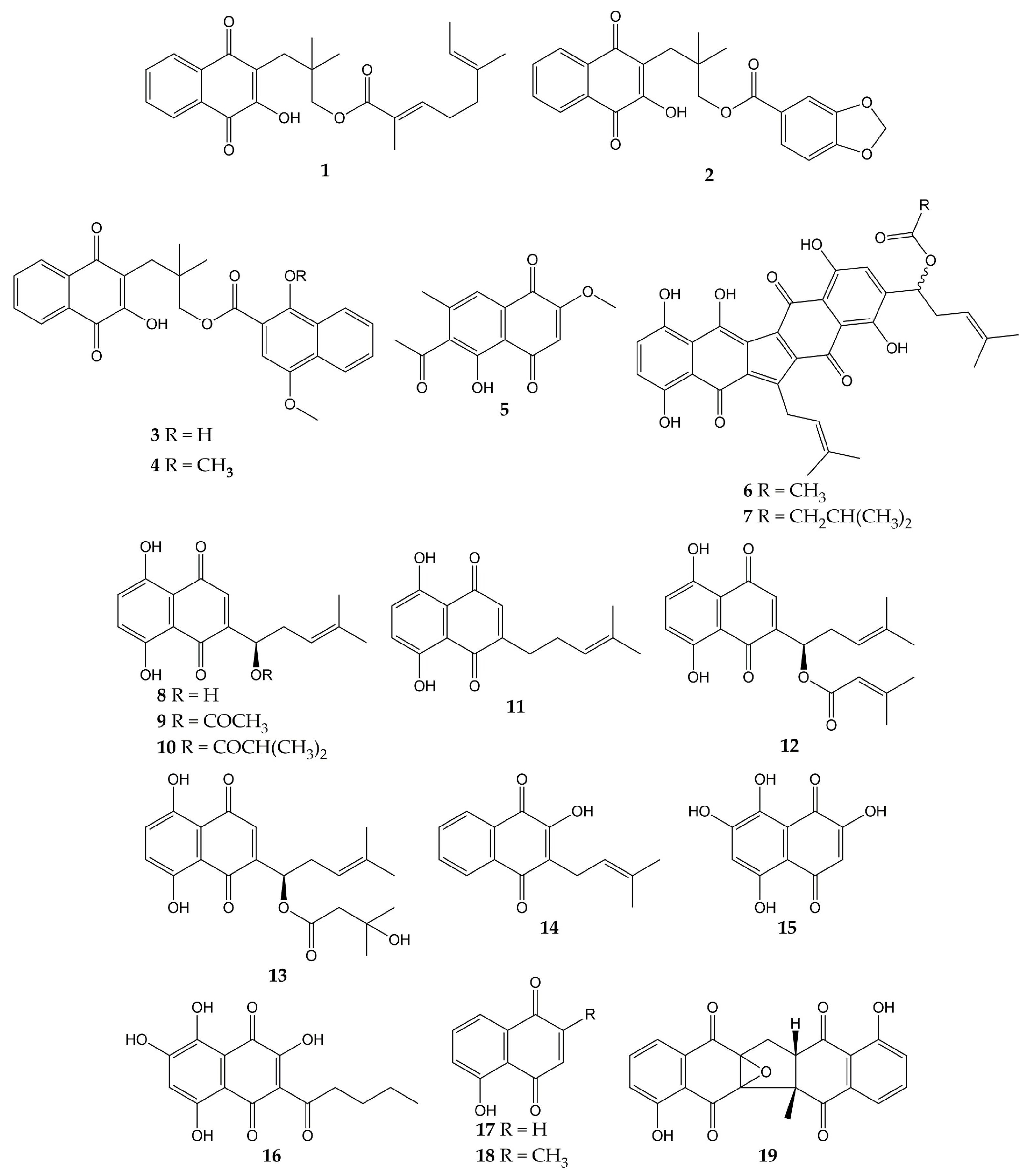
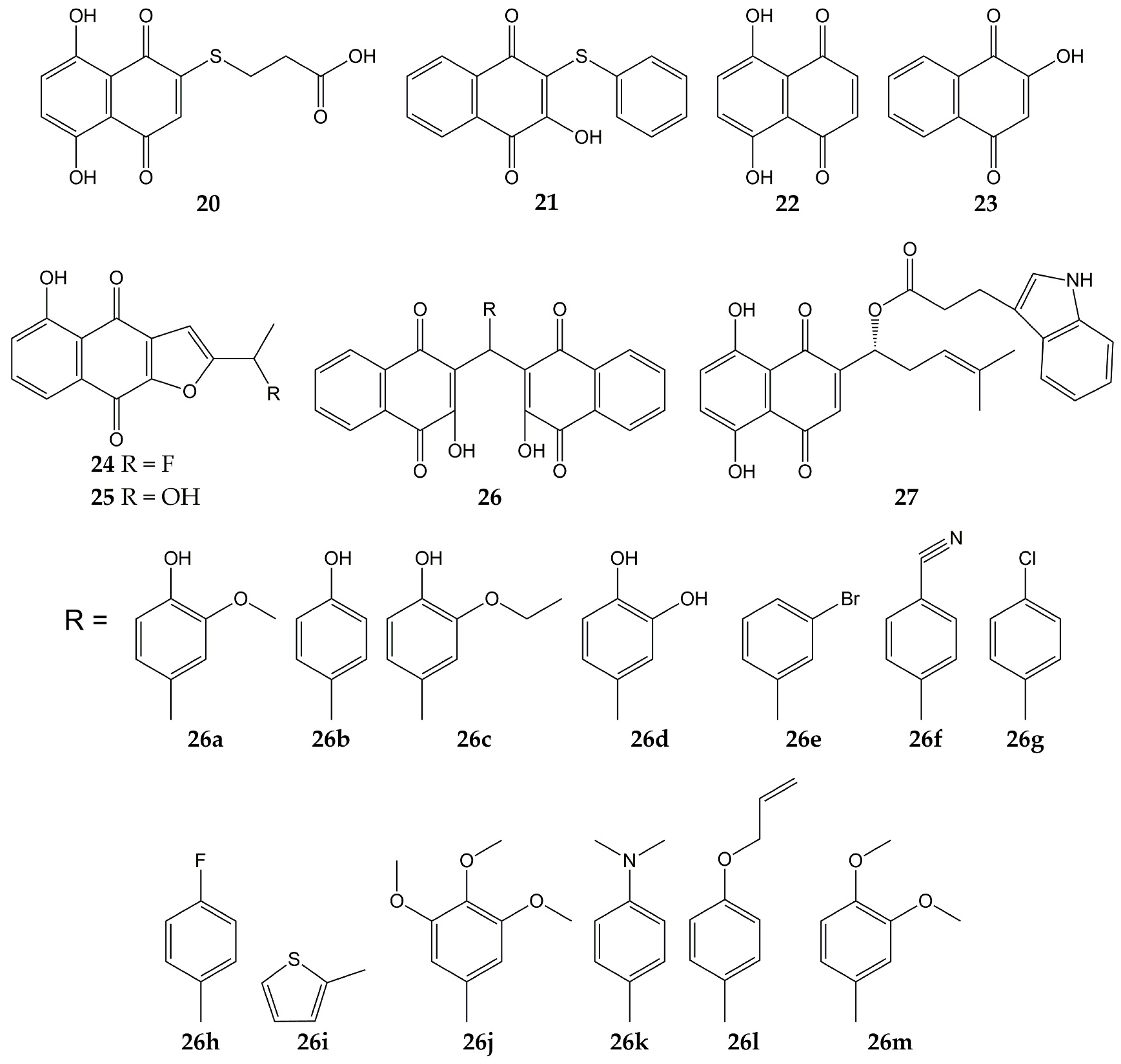
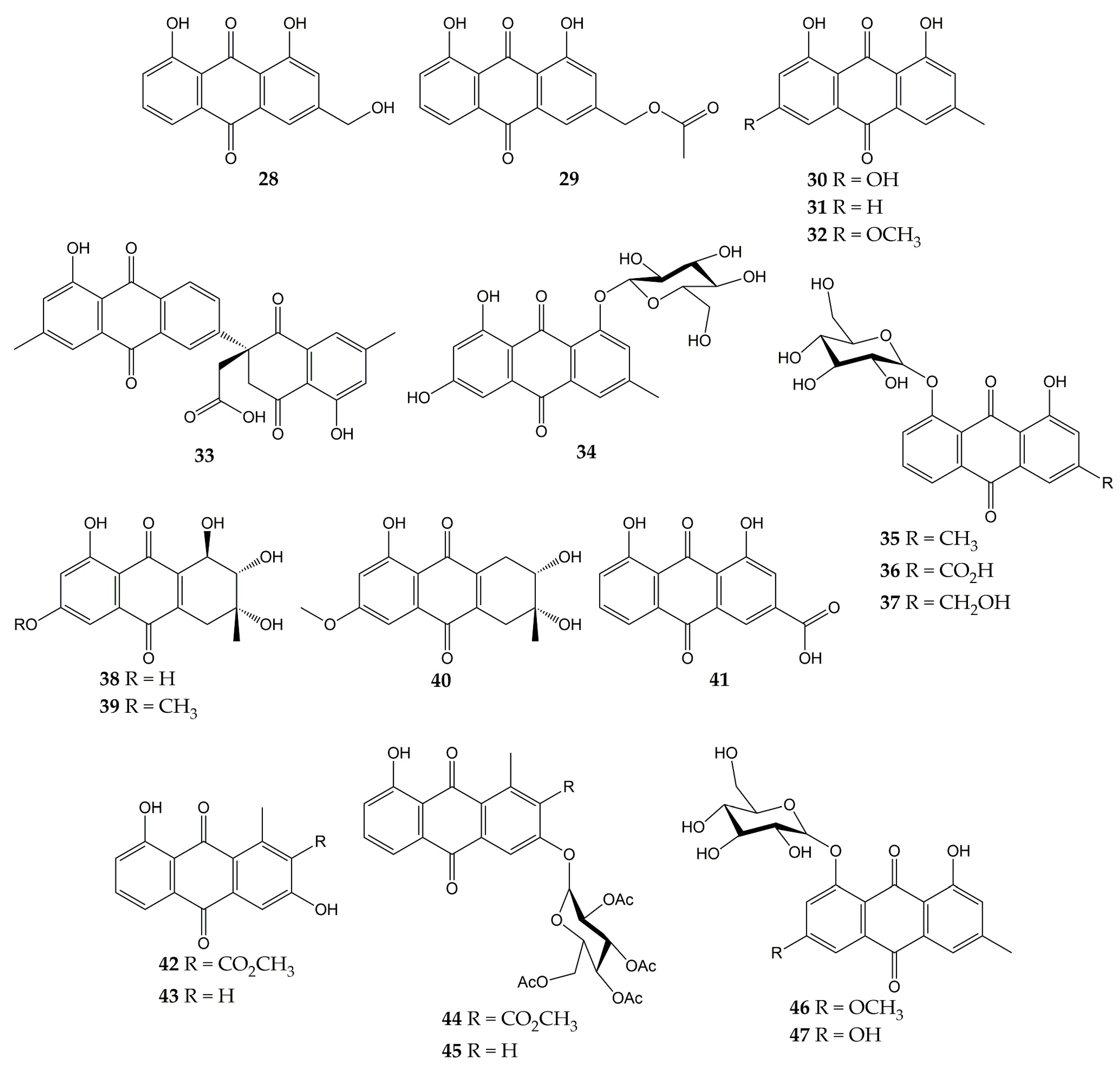
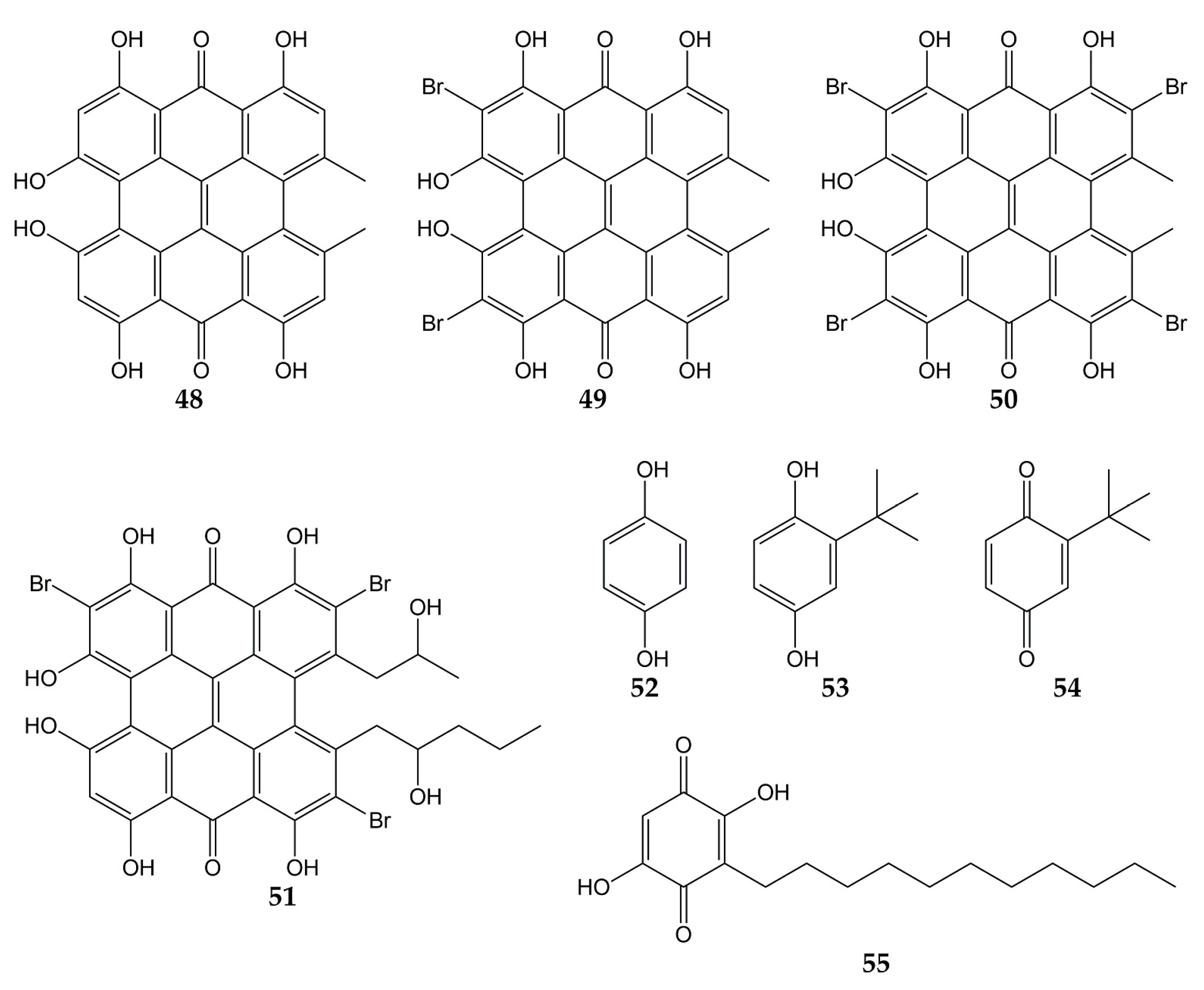
5. Conclusions
References
- Khomich, O.A.; Kochetkov, S.N.; Bartosch, B.; Ivanov, A.V. Redox Biology of Respiratory Viral Infections. Viruses 2018, 10, 392.
- Rodgers, L.; Sheppard, M.; Smith, A.; Dietz, S.; Jayanthi, P.; Yuan, Y.; Bull, L.; Wotiz, S.; Schwarze, T.; Azondekon, R.; et al. Changes in Seasonal Respiratory Illnesses in the United States During the Coronavirus Disease 2019 (COVID-19) Pandemic. Clin. Infect. Dis. 2021, 73, S110–S117.
- Leung, N.H.L. Transmissibility and transmission of respiratory viruses. Nat. Rev. Microbiol. 2021, 19, 528–545.
- Kutter, J.S.; Spronken, M.I.; Fraaij, P.L.; Fouchier, R.A.; Herfst, S. Transmission routes of respiratory viruses among humans. Curr. Opin. Virol. 2018, 28, 142–151.
- Desforges, M.; Le Coupanec, A.; Dubeau, P.; Bourgouin, A.; Lajoie, L.; Dubé, M.; Talbot, P.J. Human Coronaviruses and Other Respiratory Viruses: Underestimated Opportunistic Pathogens of the Central Nervous System? Viruses 2019, 12, 14.
- World Health Organization. Global Health Estimates: Life Expectancy and Leading Causes of Death and Disability. Available online: https://www.who.int/data/gho/data/themes/mortality-and-global-health-estimates (accessed on 11 February 2023).
- Zhang, D.; He, Z.; Xu, L.; Zhu, X.; Wu, J.; Wen, W.; Zheng, Y.; Deng, Y.; Chen, J.; Hu, Y.; et al. Epidemiology characteristics of respiratory viruses found in children and adults with respiratory tract infections in southern China. Int. J. Infect. Dis. 2014, 25, 159–164.
- Berry, M.; Gamieldien, J.; Fielding, B.C. Identification of new respiratory viruses in the new millennium. Viruses 2015, 7, 996–1019.
- Zhang, N.; Wang, L.; Deng, X.; Liang, R.; Su, M.; He, C.; Hu, L.; Su, Y.; Ren, J.; Yu, F.; et al. Recent advances in the detection of respiratory virus infection in humans. J. Med. Virol. 2020, 92, 408–417.
- Liu, W.K.; Chen, D.H.; Tan, W.P.; Qiu, S.Y.; Xu, D.; Zhang, L.; Gu, S.J.; Zhou, R.; Liu, Q. Paramyxoviruses respiratory syncytial virus, parainfluenza virus, and human metapneumovirus infection in pediatric hospitalized patients and climate correlation in a subtropical region of southern China: A 7-year survey. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 2355–2364.
- Borchers, A.T.; Chang, C.; Gershwin, M.E.; Gershwin, L.J. Respiratory syncytial virus—A comprehensive review. Clin. Rev. Allergy Immunol. 2013, 45, 331–379.
- Chatterjee, A.; Mavunda, K.; Krilov, L.R. Current State of Respiratory Syncytial Virus Disease and Management. Infect. Dis. Ther. 2021, 10, 5–16.
- Shao, N.; Liu, B.; Xiao, Y.; Wang, X.; Ren, L.; Dong, J.; Sun, L.; Zhu, Y.; Zhang, T.; Yang, F. Genetic Characteristics of Human Parainfluenza Virus Types 1-4 From Patients With Clinical Respiratory Tract Infection in China. Front. Microbiol. 2021, 12, 679246.
- Pinky, L.; Burke, C.W.; Russell, C.J.; Smith, A.M. Quantifying dose-, strain-, and tissue-specific kinetics of parainfluenza virus infection. PLoS Comput. Biol. 2021, 17, e1009299.
- Gallardo, J.; Pérez-Illana, M.; Martín-González, N.; San Martín, C. Adenovirus Structure: What Is New? Int. J. Mol. Sci. 2021, 22, 5240.
- Javanian, M.; Barary, M.; Ghebrehewet, S.; Koppolu, V.; Vasigala, V.; Ebrahimpour, S. A brief review of influenza virus infection. J. Med. Virol. 2021, 93, 4638–4646.
- Seah, A.; Loo, L.H.; Jamali, N.; Maiwald, M.; Aik, J. The influence of air quality and meteorological variations on influenza A and B virus infections in a paediatric population in Singapore. Environ. Res. 2023, 216, 114453.
- Synowiec, A.; Szczepański, A.; Barreto-Duran, E.; Lie, L.K.; Pyrc, K. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): A Systemic Infection. Clin. Microbiol. Rev. 2021, 34, e0013320.
- Flerlage, T.; Boyd, D.F.; Meliopoulos, V.; Thomas, P.G.; Schultz-Cherry, S. Influenza virus and SARS-CoV-2: Pathogenesis and host responses in the respiratory tract. Nat. Rev. Microbiol. 2021, 19, 425–441.
- Jiang, C.; Yao, X.; Zhao, Y.; Wu, J.; Huang, P.; Pan, C.; Liu, S. Comparative review of respiratory diseases caused by coronaviruses and influenza A viruses during epidemic season. Microbes Infect. 2020, 22, 236–244.
- Abed, Y.; Boivin, G. Treatment of respiratory virus infections. Antivir. Res. 2006, 70, 1–16.
- Mahajan, S.; Choudhary, S.; Kumar, P.; Tomar, S. Antiviral strategies targeting host factors and mechanisms obliging +ssRNA viral pathogens. Bioorg. Med. Chem. 2021, 46, 116356.
- Tompa, D.R.; Immanuel, A.; Srikanth, S.; Kadhirvel, S. Trends and strategies to combat viral infections: A review on FDA approved antiviral drugs. Int. J. Biol. Macromol. 2021, 172, 524–541.
- Toots, M.; Plemper, R.K. Next-generation direct-acting influenza therapeutics. Transl. Res. 2020, 220, 33–42.
- Farrukee, R.; Hurt, A.C. Antiviral Drugs for the Treatment and Prevention of Influenza. Curr. Treat. Options Infect. Dis. 2017, 9, 318–332.
- Lackenby, A.; Besselaar, T.G.; Daniels, R.S.; Fry, A.; Gregory, V.; Gubareva, L.V.; Huang, W.; Hurt, A.C.; Leang, S.K.; Lee, R.T.C.; et al. Global update on the susceptibility of human influenza viruses to neuraminidase inhibitors and status of novel antivirals, 2016–2017. Antivir. Res. 2018, 157, 38–46.
- Lina, B.; Boucher, C.; Osterhaus, A.; Monto, A.S.; Schutten, M.; Whitley, R.J.; Nguyen-Van-Tam, J.S. Five years of monitoring for the emergence of oseltamivir resistance in patients with influenza A infections in the Influenza Resistance Information Study. Influenza Other Respir. Viruses 2018, 12, 267–278.
- Domachowske, J.B.; Anderson, E.J.; Goldstein, M. The Future of Respiratory Syncytial Virus Disease Prevention and Treatment. Infect. Dis. Ther. 2021, 10, 47–60.
- Chibanga, V.P.; Dirr, L.; Guillon, P.; El-Deeb, I.M.; Bailly, B.; Thomson, R.J.; von Itzstein, M. New antiviral approaches for human parainfluenza: Inhibiting the haemagglutinin-neuraminidase. Antivir. Res. 2019, 167, 89–97.
- Van Den Bergh, A.; Bailly, B.; Guillon, P.; von Itzstein, M.; Dirr, L. Antiviral strategies against human metapneumovirus: Targeting the fusion protein. Antivir. Res. 2022, 207, 105405.
- Dodge, M.J.; MacNeil, K.M.; Tessier, T.M.; Weinberg, J.B.; Mymryk, J.S. Emerging antiviral therapeutics for human adenovirus infection: Recent developments and novel strategies. Antivir. Res. 2021, 188, 105034.
- Bizot, E.; Bousquet, A.; Charpié, M.; Coquelin, F.; Lefevre, S.; Le Lorier, J.; Patin, M.; Sée, P.; Sarfati, E.; Walle, S.; et al. Rhinovirus: A Narrative Review on Its Genetic Characteristics, Pediatric Clinical Presentations, and Pathogenesis. Front. Pediatr. 2021, 9, 643219.
- Alkhalf, H.; Almutairi, A.R.; Almutairi, A.; Almutairi, R.K.; AlGhnam, S.; Aljohani, S.; Alqanatish, J.T.; Babiker, A. Prevalence and Clinical Characterization of Bocavirus Infection in a Specialized Children’s Hospital in Saudi Arabia. Cureus 2022, 14, e22127.
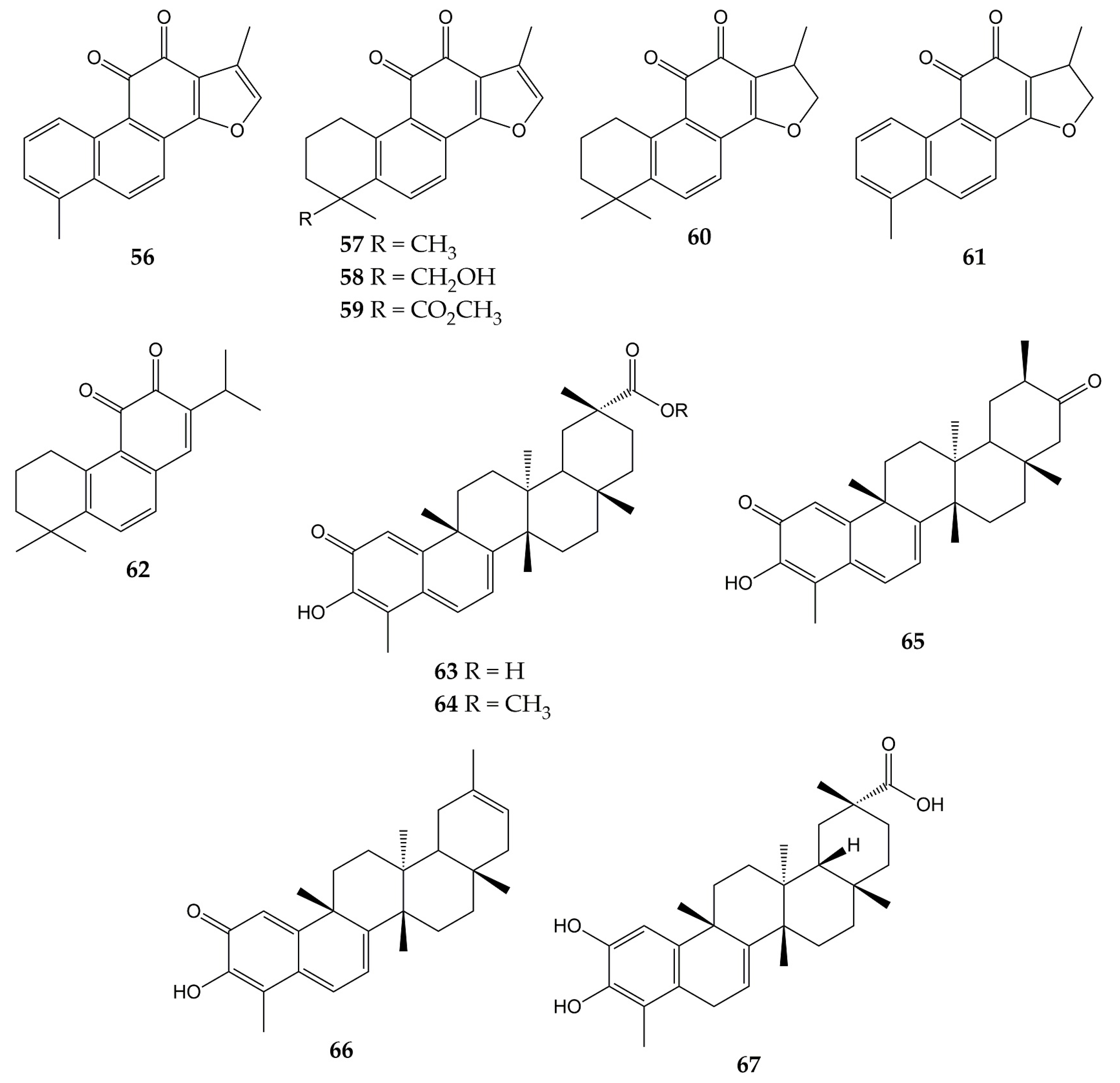
- Ngoc, T.M.; Phuong, N.T.T.; Khoi, N.M.; Park, S.; Kwak, H.J.; Nhiem, N.X.; Trang, B.T.T.; Tai, B.H.; Song, J.H.; Ko, H.J.; et al. A new naphthoquinone analogue and antiviral constituents from the root of Rhinacanthus nasutus. Nat. Prod. Res. 2019, 33, 360–366.
- Uddin, Z.; Song, Y.H.; Curtis-Long, M.J.; Kim, J.Y.; Yuk, H.J.; Park, K.H. Potent bacterial neuraminidase inhibitors, anthraquinone glucosides from Polygonum cuspidatum and their inhibitory mechanism. J. Ethnopharmacol. 2016, 193, 283–292.
- Yang, Y.; Zhao, D.; Yuan, K.; Zhou, G.; Wang, Y.; Xiao, Y.; Wang, C.; Xu, J.; Yang, W. Two new dimeric naphthoquinones with neuraminidase inhibitory activity from Lithospermum erythrorhizon. Nat. Prod. Res. 2015, 29, 908–913.
- Kim, J.Y.; Jeong, H.J.; Park, J.Y.; Kim, Y.M.; Park, S.J.; Cho, J.K.; Park, K.H.; Ryu, Y.B.; Lee, W.S. Selective and slow-binding inhibition of shikonin derivatives isolated from Lithospermum erythrorhizon on glycosyl hydrolase 33 and 34 sialidases. Bioorg. Med. Chem. 2012, 20, 1740–1748.
- Meng, X.; Wang, Y. Drug Repurposing for Influenza Virus Polymerase Acidic (PA) Endonuclease Inhibitor. Molecules 2021, 26, 7326.
- Zima, V.; Radilová, K.; Kožíšek, M.; Albiñana, C.B.; Karlukova, E.; Brynda, J.; Fanfrlík, J.; Flieger, M.; Hodek, J.; Weber, J.; et al. Unraveling the anti-influenza effect of flavonoids: Experimental validation of luteolin and its congeners as potent influenza endonuclease inhibitors. Eur. J. Med. Chem. 2020, 208, 112754.
- Yang, Z.; Yang, Y.; Wu, F.; Feng, X. Computational investigation of interaction mechanisms between juglone and influenza virus surface glycoproteins. Mol. Simul. 2013, 39, 788–795.
- Kadam, R.U.; Juraszek, J.; Brandenburg, B.; Buyck, C.; Schepens, W.B.G.; Kesteleyn, B.; Stoops, B.; Vreeken, R.J.; Vermond, J.; Goutier, W.; et al. Potent peptidic fusion inhibitors of influenza virus. Science 2017, 358, 496–502.
- Gamblin, S.J.; Vachieri, S.G.; Xiong, X.; Zhang, J.; Martin, S.R.; Skehel, J.J. Hemagglutinin Structure and Activities. Cold Spring Harb. Perspect. Med. 2021, 11, a038638.
- Chavan, R.; Shinde, P.; Girkar, K.; Mandage, R.; Chowdhary, A. Identification of Potent Natural Inhibitors against H1N1/A/2009 Virus using In Silico Subtractive Genomics Approach and Docking Technology. Int. J. Pharm. Res. 2014, 6, 105–113.
- Wohlgemuth, N.; Lane, A.P.; Pekosz, A. Influenza A Virus M2 Protein Apical Targeting Is Required for Efficient Virus Replication. J. Virol. 2018, 92, e0142518.
- Tandon, V.K.; Singh, R.V.; Yadav, D.B. Synthesis and evaluation of novel 1,4-naphthoquinone derivatives as antiviral, antifungal and anticancer agents. Bioorg. Med. Chem. Lett. 2004, 14, 2901–2904.
- Rudnicka, M.; Ludynia, M.; Karcz, W. Effects of Naphthazarin (DHNQ) Combined with Lawsone (NQ-2-OH) or 1,4-Naphthoquinone (NQ) on the Auxin-Induced Growth of Zea mays L. Coleoptile Segments. Int. J. Mol. Sci. 2019, 20, 1788.
- Sharma, G.; Vasanth Kumar, S.; Wahab, H.A. Molecular docking, synthesis, and biological evaluation of naphthoquinone as potential novel scaffold for H5N1 neuraminidase inhibition. J. Biomol. Struct. Dyn. 2018, 36, 233–242.
- Zhang, Y.; Han, H.; Qiu, H.; Lin, H.; Yu, L.; Zhu, W.; Qi, J.; Yang, R.; Pang, Y.; Wang, X.; et al. Antiviral activity of a synthesized shikonin ester against influenza A (H1N1) virus and insights into its mechanism. Biomed. Pharmacother. 2017, 93, 636–645.
- Wang, X.M.; Lin, H.Y.; Kong, W.Y.; Guo, J.; Shi, J.; Huang, S.C.; Qi, J.L.; Yang, R.W.; Gu, H.W.; Yang, Y.H. Synthesis and biological evaluation of heterocyclic carboxylic acyl shikonin derivatives. Chem. Biol. Drug Des. 2014, 83, 334–343.
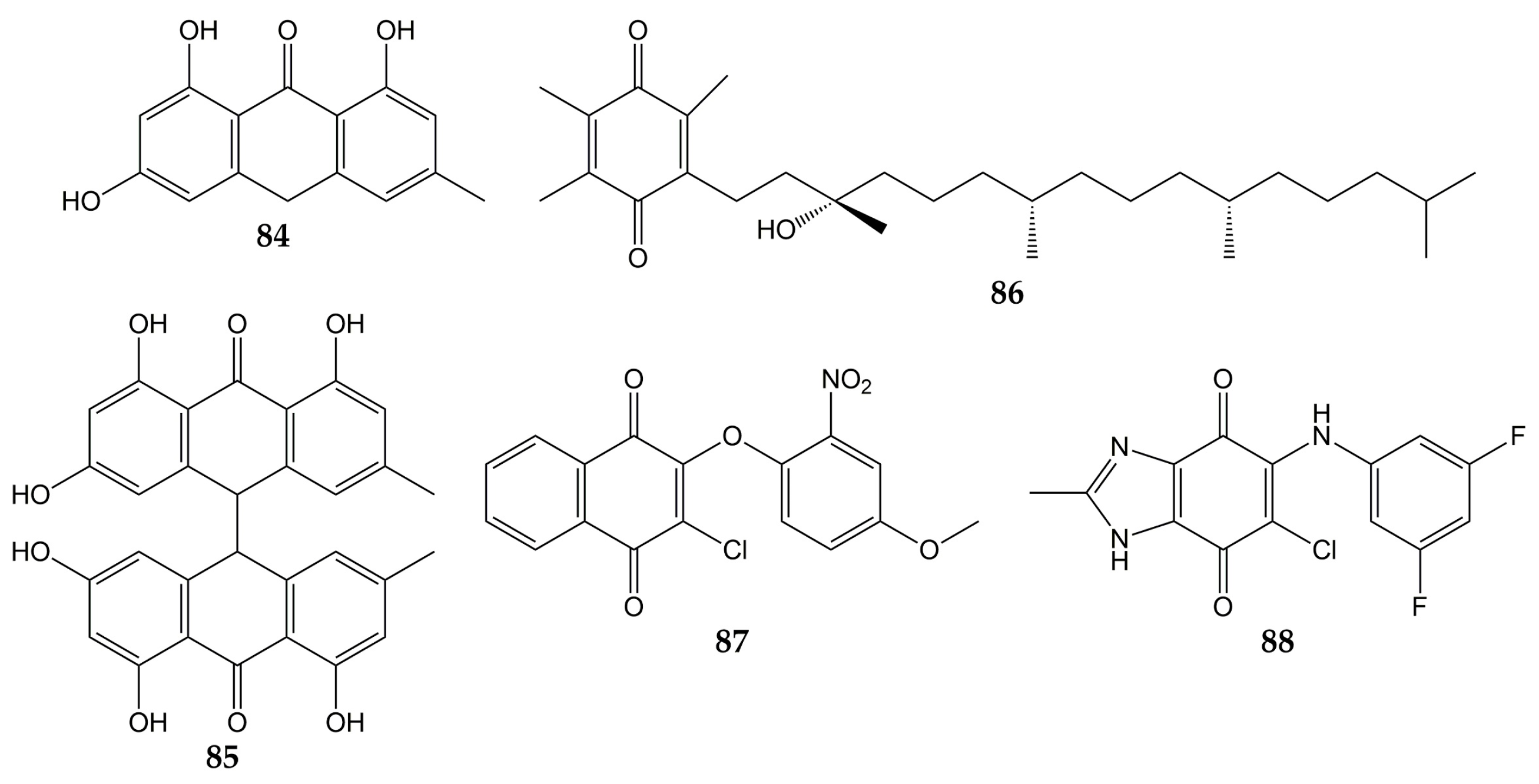
- Mohammed, M.M.D.; El-Souda, S.S.; El-Hallouty, S.M.; Kobayashi, N. Antiviral and cytotoxic activities of anthraquinones isolated from Cassia roxburghii Linn. leaves. Herba Pol. 2014, 59, 33–44.
- Pang, X.; Zhao, J.Y.; Liu, N.; Chen, M.H.; Zheng, W.; Zhang, J.; Chen, X.J.; Cen, S.; Yu, L.Y.; Ma, B.P. Anthraquinone analogues with inhibitory activities against influenza a virus from Polygonatum odoratum. J. Asian Nat. Prod. Res. 2021, 23, 717–723.
- Bei, Y.; Tia, B.; Li, Y.; Guo, Y.; Deng, S.; Huang, R.; Zeng, H.; Li, R.; Wang, G.F.; Dai, J. Anti-influenza A Virus Effects and Mechanisms of Emodin and Its Analogs via Regulating PPARα/γ-AMPK-SITR1 Pathway and Fatty Acid Metabolism. BioMed Res. Int. 2021, 2021, 9066938.
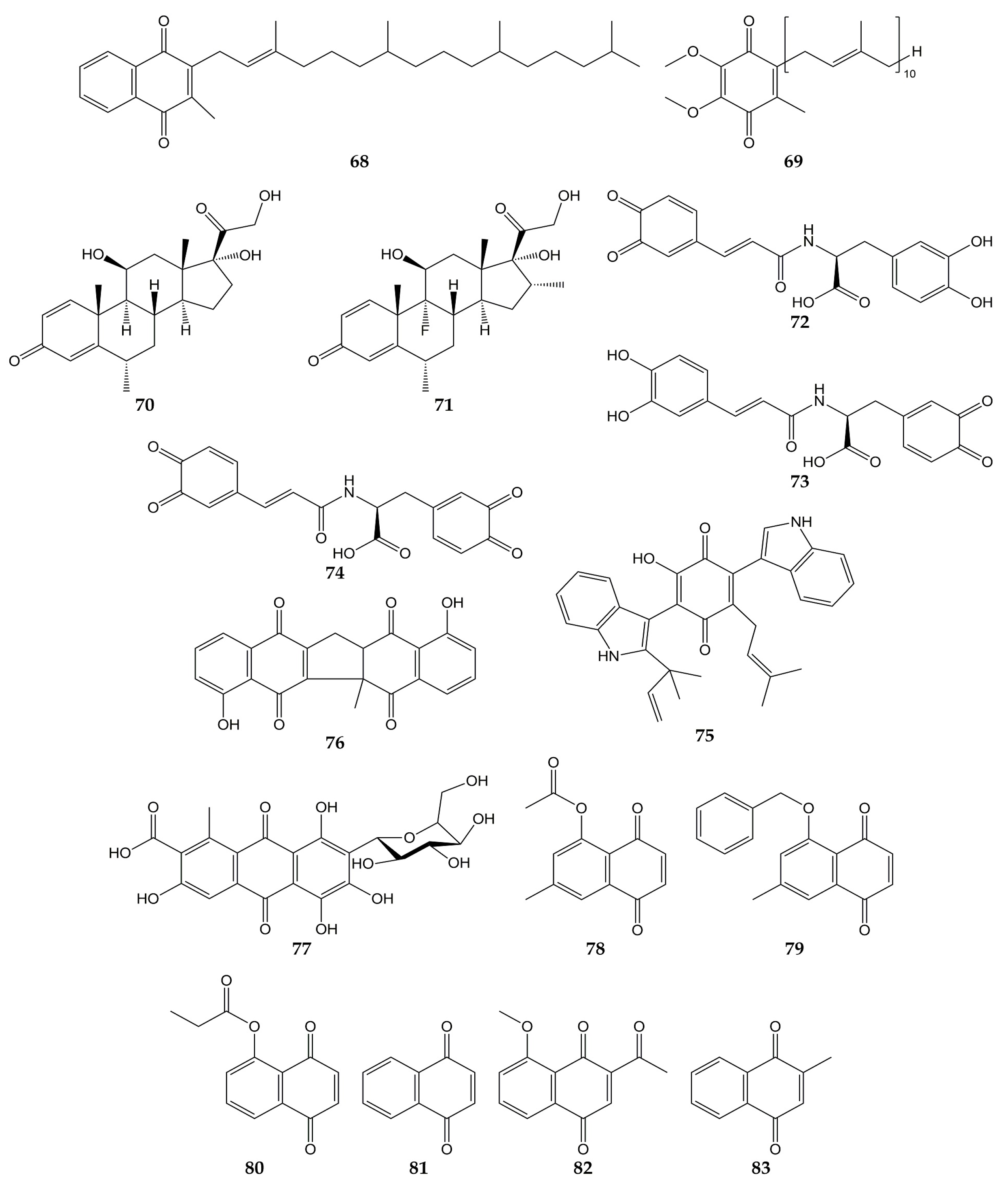
- Tang, J.; Colacino, J.M.; Larsen, S.H.; Spitzer, W. Virucidal activity of hypericin against enveloped and non-enveloped DNA and RNA viruses. Antivir. Res. 1990, 13, 313–325.
- Hudson, J.B.; Delaey, E.; de Witte, P.A. Bromohypericins are potent photoactive antiviral agents. Photochem. Photobiol. 1999, 70, 820–822.
- Dao, N.; Jang, Y.; Kim, M.; Nguyễn, H.; Pham, D.; Dang, Q.; Nguyen, J.; Yun, B.-S.; Pham, Q.; Kim, J.-C.; et al. Chemical Constituents and Anti-influenza Viral Activity of the Leaves of Vietnamese Plant Elaeocarpus tonkinensis. Rec. Nat. Prod. 2018, 13, 71–80.
- Xiong, M.; Su, H.; Zhao, W.; Xie, H.; Shao, Q.; Xu, Y. What coronavirus 3C-like protease tells us: From structure, substrate selectivity, to inhibitor design. Med. Res. Rev. 2021, 41, 1965–1998.
- Lin, C.W.; Tsai, F.J.; Tsai, C.H.; Lai, C.C.; Wan, L.; Ho, T.Y.; Hsieh, C.C.; Chao, P.D. Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds. Antivir. Res. 2005, 68, 36–42.
- Sternberg, A.; Naujokat, C. Structural features of coronavirus SARS-CoV-2 spike protein: Targets for vaccination. Life Sci. 2020, 257, 118056.
- Ho, T.Y.; Wu, S.L.; Chen, J.C.; Li, C.C.; Hsiang, C.Y. Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction. Antivir. Res. 2007, 74, 92–101.
- Caruso, F.; Rossi, M.; Pedersen, J.Z.; Incerpi, S. Computational studies reveal mechanism by which quinone derivatives can inhibit SARS-CoV-2. Study of embelin and two therapeutic compounds of interest, methyl prednisolone and dexamethasone. J. Infect. Public Health 2020, 13, 1868–1877.
- Wood, S.; Huffman, J.; Weber, N.; Andersen, D.; North, J.; Murray, B.; Sidwell, R.; Hughes, B. Antiviral activity of naturally occurring anthraquinones and anthraquinone derivatives. Planta Med. 1990, 56, 651–652.
- Liu, Z.; Ma, N.; Zhong, Y.; Yang, Z.Q. Antiviral Effect of Emodin from Rheum palmatum against Coxsakievirus B5 and Human Respiratory Syncytial Virus In Vitro. J. Huazhong Univ. Sci. Technol. Med. Sci. 2015, 35, 916–922.
- Luo, D.; Xiong, S.; Li, Q.G.; Jiang, L.; Niu, Q.W.; He, L.J.; Li, Y.L.; Zhang, Y.B.; Wang, G.C. Terpenoids from the stems of Celastrus hindsii and their anti-RSV activities. Fitoterapia 2018, 130, 118–124.
- Jung, E.; Lee, J.Y.; Kim, H.J.; Ryu, C.K.; Lee, K.I.; Kim, M.; Lee, C.K.; Go, Y.Y. Identification of quinone analogues as potential inhibitors of picornavirus 3C protease in vitro. Bioorg. Med. Chem. Lett. 2018, 28, 2533–2538.




