Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Mudrika Tripathi | -- | 1436 | 2023-03-16 13:02:15 | | | |
| 2 | Conner Chen | + 5 word(s) | 1441 | 2023-03-20 02:39:22 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Tripathi, M.; Colige, A.; Deroanne, C.F. Similarities and Differences of RhoGDI1 and RhoGDI2. Encyclopedia. Available online: https://encyclopedia.pub/entry/42267 (accessed on 07 February 2026).
Tripathi M, Colige A, Deroanne CF. Similarities and Differences of RhoGDI1 and RhoGDI2. Encyclopedia. Available at: https://encyclopedia.pub/entry/42267. Accessed February 07, 2026.
Tripathi, Mudrika, Alain Colige, Christophe F. Deroanne. "Similarities and Differences of RhoGDI1 and RhoGDI2" Encyclopedia, https://encyclopedia.pub/entry/42267 (accessed February 07, 2026).
Tripathi, M., Colige, A., & Deroanne, C.F. (2023, March 16). Similarities and Differences of RhoGDI1 and RhoGDI2. In Encyclopedia. https://encyclopedia.pub/entry/42267
Tripathi, Mudrika, et al. "Similarities and Differences of RhoGDI1 and RhoGDI2." Encyclopedia. Web. 16 March, 2023.
Copy Citation
RhoGDI1 and RhoGDI2 (guanine nucleotide dissociation inhibitor (GDI)) have been implicated in multiple human cancers through their involvement in cancer cell migration, invasion and metastasis and, thus, are regarded as attractive targets for cancer biology. RhoGDI2 has largely remained in RhoGDI1′s shadow because of its lower abundancy and more restrained distribution.
RhoGDI2 1
RhoGDI1 2
Rho GTPases 3
cancer 4
1. Introduction
Rho GTPases are highly conserved members of the Ras superfamily, which are best known to organize the actin and microtubule cytoskeleton thereby defining the cell shape and migration. They also control a wide variety of signaling pathways that regulate crucial biological processes such as vesicle transport, cell division and gene transcription [1][2][3]. Rho GTPases cycle between an active GTP-bound form and an inactive GDP-bound form. This activity is regulated by three classes of proteins: guanine nucleotide exchange factors (GEFs) catalyze the exchange of GDP for GTP to activate the GTPase; whereas GTPase-activating proteins (GAPs) increase the intrinsic GTP hydrolysis rate of the GTPase and inactivate it; and guanine nucleotide dissociation inhibitors (GDIs) sequester the GDP-bound form of GTPases in the cytosol to prevent their activation by GEFs or ubiquitin-mediated degradation (Figure 1) [4]. Aberrant signaling of Rho GTPases and their regulators is commonly found in many human cancers and has been attributed to several mechanisms [5][6][7][8][9][10].
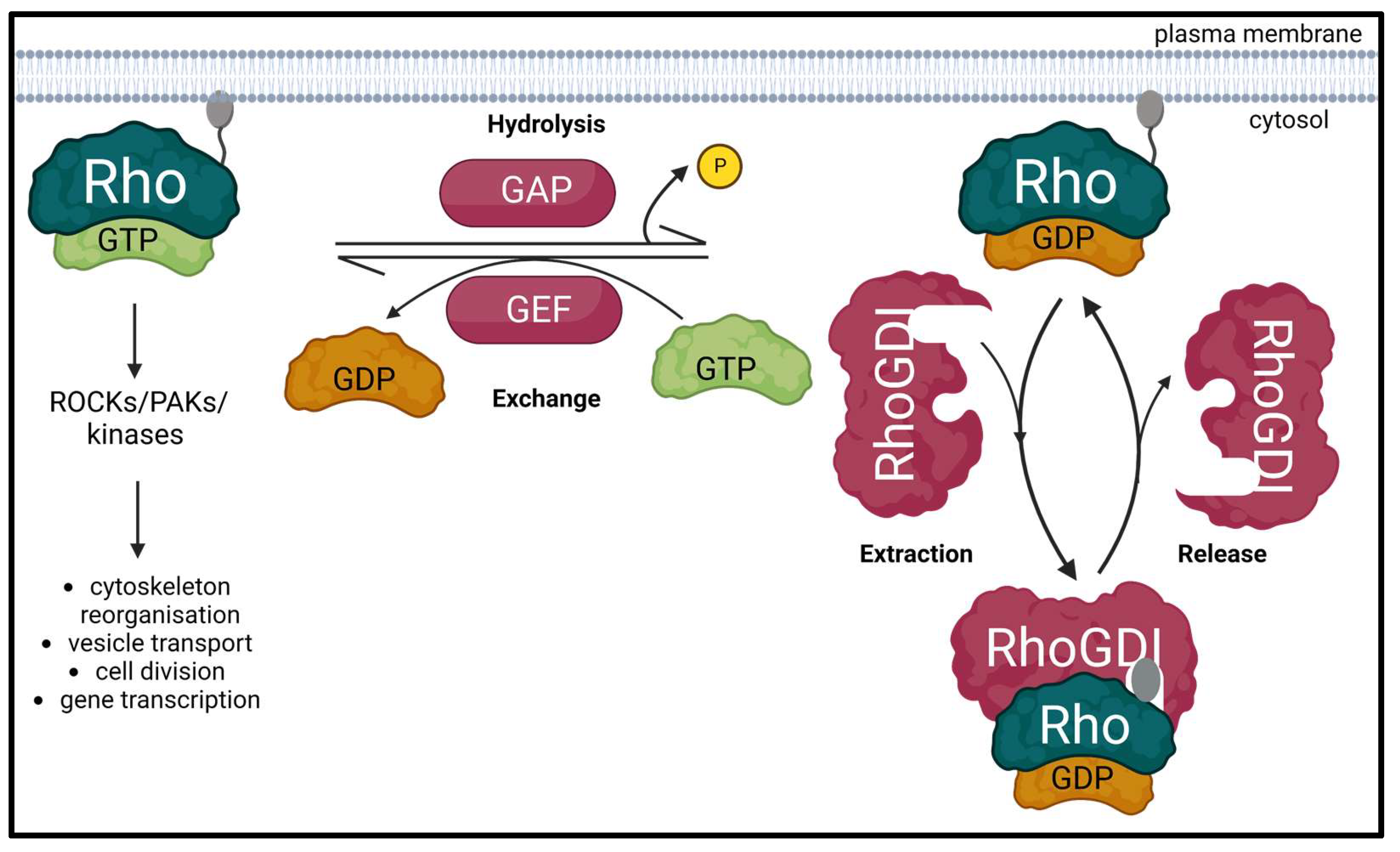
Figure 1. Schematic diagram of the Rho GTPase regulatory cycle. Inactive Rho GTPase is dissociated from its GDP and uptakes GTP to get activated through a process promoted by the Rho guanine exchange factors (RhoGEFs). Active GTP-bound Rho GTPase can then interact with its effectors such as Rho-associated coiled-coil containing kinases (ROCKs), PAK family of serine/threonine kinases (PAKs) and other kinases to participate in various biological processes. This interaction ceases when Rho GTPase-activating proteins (RhoGAPs) stimulate the hydrolysis of the bound GTP to GDP, thereby inactivating the Rho GTPase. The inactive GDP-bound form of Rho GTPase is free to bind to and be sequestered by Rho guanine nucleotide dissociation inhibitors (RhoGDIs). This induces their relocation to the cytoplasm, prevents their ubiquitin-mediated degradation and regulates the activation of Rho GTPases by GEFs. Created with BioRender.com (accessed on 28 January 2023).
To this date, nearly 85 RhoGEFs and 66 RhoGAPs have been identified for nearly 20 Rho GTPase family members, wherein, in stark contrast, only three human RhoGDIs have been identified so far: RhoGDI1 (or RhoGDIα), RhoGDI2 (or RhoGDIβ or D4-GDI or Ly-GDI) and RhoGDI3 (or RhoGDIγ) [8]. All three reside exclusively in the cytoplasm wherein RhoGDI1 is ubiquitously expressed [11][12]. RhoGDI2 was initially believed to be expressed specifically in hematopoietic cells [13][14] but subsequently has also been found in various other cell types and tissues, including cancer cells [8]. RhoGDI3 is primarily expressed in the brain, lung, kidney, testis and pancreas where it targets the Golgi, and shows specificity towards RhoB and RhoG [15].
RhoGDI1 and RhoGDI2 have been implicated in multiple human cancers through their involvement in cancer cell migration, invasion and metastasis and, thus, are regarded as attractive targets for cancer biology [8]. RhoGDI2 has largely remained in RhoGDI1′s shadow because of its lower abundancy and more restrained distribution. It is, however, starting to garner more attention due to discoveries hinting that RhoGDI2 may play more complex roles in multiple human cancers and many key cellular processes.
2. RhoGDI1 and RhoGDI2: Similarities and Differences
RhoGDI1 was the first RhoGDI to be discovered in rabbit intestine and bovine brain cytosol in 1989 and is widely considered to be the prototype of RhoGDIs. Subsequently, corresponding human cDNA was isolated and a RhoGDI protein was also identified in yeast [11]. Leffers et al. characterized RhoGDI2 and found that it was largely expressed in hematopoietic cells [16]. RhoGDIs interact with the GDP-bound Rho GTPases and extract Rho GTPases from the membrane to regulate them from undergoing the GDP/GTP exchange cycle. [17]. The N-terminal domain of the RhoGDIs interacts with the switch 1 and switch 2 regions of GDP-bound Rho GTPases which prevents the exchange of GDP for GTP and therefore keeps them in their inactive form [18][19], whereas the C-terminal domain also contributes towards their inhibition by extracting Rho GTPases from the membrane [17][20].
RhoGDIs may also shuttle inactive Rho GTPases towards membranes leading to their activation [17][21]. Moreover, RhoGDIs can protect its interacting Rho GTPases from proteasomal degradation [22], demonstrating that RhoGDIs are not merely inhibitors for Rho GTPases but also have a key role in their regulation and signaling. Quite expectedly in view of these functions, both the RhoGDIs are involved in the regulation of multiple biological processes such as actin cytoskeletal organization, cell migration and immune response [23][24][25][26]. As mentioned previously, they are also implicated in many human cancers where they can either be upregulated or downregulated (Table 1).
Table 1. Expression of RhoGDI1 and RhoGDI2 in human cancers. Biphasic, up and then down.
| Cancer | RhoGDI | Regulation | Reference(s) |
|---|---|---|---|
| Colorectal cancer | RhoGDI1 | Up | [27] |
| RhoGDI2 | Up | [28] | |
| Breast cancer | RhoGDI1 | Up | [29] |
| RhoGDI2 | Up | [30][31] | |
| Biphasic | [32][33] | ||
| Hepatocellular carcinoma | RhoGDI1 | Down | [34] |
| RhoGDI2 | Up | [35] | |
| Bladder cancer | RhoGDI1 | Down | [36] |
| RhoGDI2 | Down | [37] | |
| Ovarian cancer | RhoGDI2 | Down | [38][39] |
| Hodgkin’s lymphoma | RhoGDI2 | Down | [40][41] |
| Gastric cancer | RhoGDI2 | Up | [42][43] |
| Pancreatic cancer | RhoGDI2 | Up | [44][45][46] |
| Melanoma | RhoGDI2 | Up | [47] |
| Lung cancer | RhoGDI2 | Down | [48][49] |
| Osteosarcoma | RhoGDI2 | Down | [50][51] |
| Leukemias | RhoGDI2 | Down | [52][53] |
Although their extreme N-terminal domain (25 and 22 amino acids for RhoGDI 1 and 2, respectively) are completely divergent, RhoGDI 1 and 2 show 73.6% identity for the remaining C-terminal sequence (Figure 2). RhoGDI1 and RhoGDI2 interact with and form complexes with the classical Rho GTPases, i.e., RhoA, RhoC, Rac1, Rac2, Rac3, RhoG and Cdc42 [19][54][55][56]. However, the interaction potency of RhoGDI2 with Cdc42 is 10–20 folds lower than that of RhoGDI1. Platko et al. observed that a single residue (Ile 177 in RhoGDI1/Asn 174 in RhoGDI2) is responsible for this difference in their affinity for Cdc42 [57].
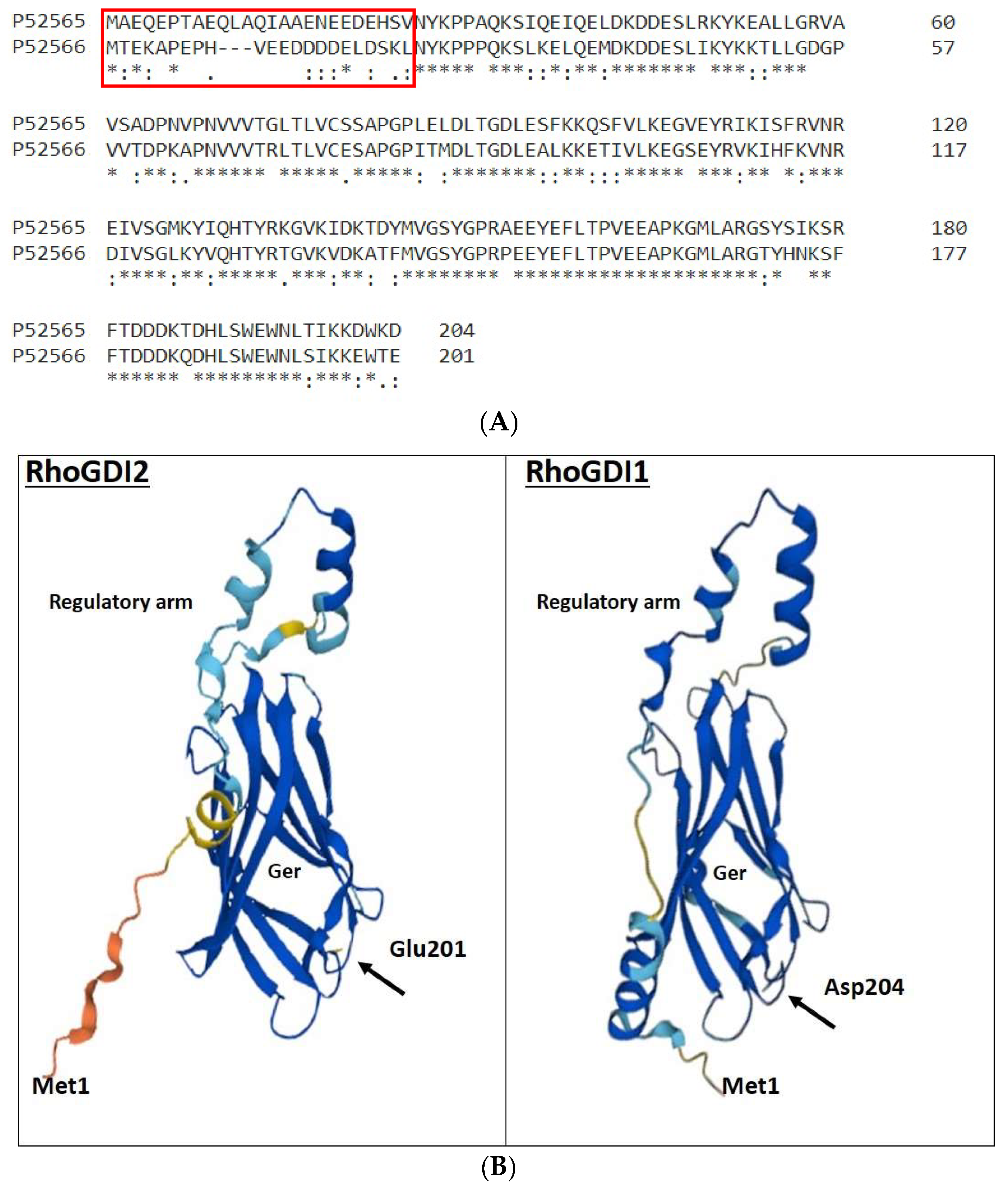
Figure 2. Comparison of primary and tertiary structures of human RhoGDI1 and RhoGDI2. (A) Protein sequences were compared using EMBL-EBI’s Clustal Omega tool. The accession numbers are as follows: human RhoGDI1, P52565 and human RhoGDI2, P52566. Identical residues are indicated by asterisks; substitutions for amino acids possessing highly similar or somehow similar characteristics are indicated by double and single dots, respectively. The highly divergent extreme N-terminal domains are enclosed in the red box. RhoGDI2 is phosphorylated at Y24, S31 and Y153 by β2 integrin-related kinases Src, c-Abl and Syk in response to PSGL-1 antibody ligation. On the contrary, RhoGDI1 is phosphorylated at S45, S48 and T52 by calcium-dependent protein kinase CPK3. (B) Predictions of the 3D structure of RhoGDI1 and RhoGDI2 are from the AlphaFold project (AF-P52565-F1 and AF-P52566-F1, respectively). Confidence regarding the 3D structure corresponding to different parts of the proteins is provided by color code, with dark blue representing the highest confidence and orange the lowest confidence. Ger: pocket accommodating the geranylgeranyl moiety of the Rho GTPases.
Several other proteins that are not part of the Rho GTPase family have been found to interact with RhoGDI 1 or 2 or both, mainly through high throughput experiments. Upon examining Uniprot (https://www.uniprot.org (accessed on 23 January 2023)) and Biogrid (https://thebiogrid.org (accessed on 23 January 2023)) databases, the interactors of both RhoGDIs can be extrapolated. Both RhoGDI 1 and 2 have been found to interact with ubiquitin-fold modifier 1 (UFM1), small ubiquitin-like modifier 4 (SUMO4), U2 small nuclear RNA auxiliary factor 2 (U2AF2) and DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 (DDX58). However, RhoGDI1 interacts with Cullin3, whereas RhoGDI2 does not. RhoGDI1 also interacts with EWS RNA-binding protein 1, ezrin, moesin and radixin. On the other hand, RhoGDI2 interacts with RhoGEF Vav1, whereas RhoGDI1 is unable to do so. RhoGDI2 also interacts with acyl-CoA thioesterase 7, B cell CLL/lymphoma 6 and cadherin1. Perhaps the differences in functions of both RhoGDIs may be attributed to their interactions with different proteins that do not belong to the Rho GTPase family.
Mouse models were used to identify the respective functions of RhoGDI 1 and 2. Yin et al. generated RhoGDI2-null mice to explore its functions in lymphocytes. They observed that there were no abnormalities in lymphoid development and immune responses. However, in vitro cultivation of B and T cells from these mice showed de-regulated interactions and other impaired phenotypes. They inferred that RhoGDI2 regulates Rho GTPases in lymphocyte survival and responsiveness, wherein the absence of RhoGDI2 can be compensated in vivo by other Rho GTPase regulatory proteins [26]. It was later shown that RhoGDI1-null mice display abnormalities in the kidneys and reproductive system in adulthood and that levels of RhoGDI2 expression did not change in WT and RhoGDI1-null mice [58]. Double knockouts of RhoGDI 1 and 2 were then generated in order to get a better insight into their specific and shared functions. These mice are characterized by aberrant homeostasis of lymphocytes and an increased eosinophil population. T cells derived from the mice display defective in vitro proliferation and development and lower levels of CD3 expression. These results show that RhoGDI 1 and 2 share similar functions and can partly substitute for each other in lymphocytic migration and development [59].
References
- Vega, F.M.; Ridley, A.J. Rho GTPases in cancer cell biology. FEBS Lett. 2008, 582, 2093–2101.
- Rodenburg, W.S.; van Buul, J.D. Rho GTPase signalling networks in cancer cell transendothelial migration. Vasc. Biol. 2021, 3, R77–R95.
- Magalhaes, Y.T.; Farias, J.O.; Silva, L.E.; Forti, F.L. GTPases, genome, actin: A hidden story in DNA damage response and repair mechanisms. DNA Repair 2021, 100, 103070.
- Mosaddeghzadeh, N.; Ahmadian, M. The RHO Family GTPases: Mechanisms of Regulation and Signaling. Cells 2021, 10, 1831.
- Svensmark, J.H.; Brakebusch, C. Rho GTPases in cancer: Friend or foe? Oncogene 2019, 38, 7447–7456.
- Kreider-Letterman, G.; Carr, N.M.; Garcia-Mata, R. Fixing the GAP: The role of RhoGAPs in cancer. Eur. J. Cell Biol. 2022, 101, 151209.
- Maldonado, M.D.M.; Medina, J.I.; Velazquez, L.; Dharmawardhane, S. Targeting Rac and Cdc42 GEFs in Metastatic Cancer. Front. Cell Dev. Biol. 2020, 8, 201.
- Cho, H.J.; Kim, J.-T.; Baek, K.E.; Kim, B.-Y.; Lee, H.G. Regulation of Rho GTPases by RhoGDIs in Human Cancers. Cells 2019, 8, 1037.
- Crosas-Molist, E.; Samain, R.; Kohlhammer, L.; Orgaz, J.L.; George, S.L.; Maiques, O.; Barcelo, J.; Sanz-Moreno, V. Rho GTPase signaling in cancer progression and dissemination. Physiol. Rev. 2022, 102, 455–510.
- Kowluru, A. Roles of GTP and Rho GTPases in pancreatic islet beta cell function and dysfunction. Small GTPases 2020, 12, 323–335.
- Fukumoto, Y.; Kaibuchi, K.; Hori, Y.; Fujioka, H.; Araki, S.; Ueda, T.; Kikuchi, A.; Takai, Y. Molecular cloning and characterization of a novel type of regulatory protein (GDI) for the rho proteins, ras p21-like small GTP-binding proteins. Oncogene 1990, 5, 1321–1328.
- Kowluru, A.; Gleason, N.F. Underappreciated roles for Rho GDP dissociation inhibitors (RhoGDIs) in cell function: Lessons learned from the pancreatic islet β-cell. Biochem. Pharmacol. 2021, 197, 114886.
- Lelias, J.M.; Adra, C.N.; Wulf, G.M.; Guillemot, J.C.; Khagad, M.; Caput, D.; Lim, B. cDNA cloning of a human mRNA preferentially expressed in hematopoietic cells and with homology to a GDP-dissociation inhibitor for the rho GTP-binding proteins. Proc. Natl. Acad. Sci. USA 1993, 90, 1479–1483.
- Scherle, P.; Behrens, T.; Staudt, L.M. Ly-GDI, a GDP-dissociation inhibitor of the RhoA GTP-binding protein, is expressed preferentially in lymphocytes. Proc. Natl. Acad. Sci. USA 1993, 90, 7568–7572.
- De León-Bautista, M.P.; Cardenas-Aguayo, M.D.C.; Casique-Aguirre, D.; Almaraz-Salinas, M.; Parraguirre-Martinez, S.; Olivo-Diaz, A.; Thompson-Bonilla, M.D.R.; Vargas, M. Immunological and Functional Characterization of RhoGDI3 and Its Molecular Targets RhoG and RhoB in Human Pancreatic Cancerous and Normal Cells. PLoS ONE 2016, 11, e0166370.
- Leffers, H.; Nielsen, M.S.; Andersen, A.H.; Honoré, B.; Madsen, P.; Vandekerckhove, J.; Celis, J.E. Identification of Two Human Rho GDP Dissociation Inhibitor Proteins Whose Overexpression Leads to Disruption of the Actin Cytoskeleton. Exp. Cell Res. 1993, 209, 165–174.
- Garcia-Mata, R.; Boulter, E.; Burridge, K. The ‘invisible hand’: Regulation of RHO GTPases by RHOGDIs. Nat. Rev. Mol. Cell Biol. 2011, 12, 493–504.
- Keep, N.; Barnes, M.; Barsukov, I.; Badii, R.; Lian, L.-Y.; Segal, A.W.; Moody, P.C.; Roberts, G.C. A modulator of rho family G proteins, rhoGDI, binds these G proteins via an immunoglobulin-like domain and a flexible N-terminal arm. Structure 1997, 5, 623–633.
- Mosaddeghzadeh, N.; Jasemi, N.S.K.; Majolée, J.; Zhang, S.-C.; Hordijk, P.L.; Dvorsky, R.; Ahmadian, M.R. Electrostatic Forces Mediate the Specificity of RHO GTPase-GDI Interactions. Int. J. Mol. Sci. 2021, 22, 12493.
- Kardol-Hoefnagel, T.; van Logtestijn, S.A.; Otten, H.G. A Review on the Function and Regulation of ARHGDIB/RhoGDI2 Expression Including the Hypothetical Role of ARHGDIB/RhoGDI2 Autoantibodies in Kidney Transplantation. Transplant. Direct 2020, 6, e548.
- Choi, E.-K.; Kim, J.-G.; Kim, H.-J.; Cho, J.-Y.; Jeong, H.; Park, Y.; Islam, R.; Cap, C.K.; Park, J.-B. Regulation of RhoA GTPase and novel target proteins for ROCK. Small GTPases 2017, 11, 95–102.
- Majolée, J.; Podieh, F.; Hordijk, P.L.; Kovačević, I. The interplay of Rac1 activity, ubiquitination and GDI binding and its consequences for endothelial cell spreading. PLoS ONE 2021, 16, e0254386.
- Chinchole, A.N.; Lone, K.A.; Tyagi, S. MLL regulates actin cytoskeleton and cell migration by stabilizing Rho GTPases via the transcription of RhoGDI1. J. Cell Sci. 2022, 135, jcs260042.
- Liu, W.; Wang, X.; Wang, S.; Ba, X.; Xu, T.; Wang, X.; Zeng, X. RhoGDI2 positively regulates the Rho GTPases activation in response to the β2 outside-in signaling in T cells adhesion and migration on ICAM-1. J. Leukoc. Biol. 2019, 106, 431–446.
- Nagar, H.; Kim, S.; Lee, I.; Choi, S.-J.; Piao, S.; Jeon, B.H.; Shong, M.; Kim, C.-S. CRIF1 deficiency suppresses endothelial cell migration via upregulation of RhoGDI2. PLoS ONE 2021, 16, e0256646.
- Yin, L.; Schwartzberg, P.; Scharton-Kerstenj, T.M.; Staudt, L.; Lenardo, M. Immune responses in mice deficient in Ly-GDI, a lymphoid-specific regulator of Rho GTPases. Mol. Immunol. 1997, 34, 481–491.
- Zhu, G.-F.; Xu, Y.-W.; Li, J.; Niu, H.-L.; Ma, W.-X.; Xu, J.; Zhou, P.-R.; Liu, X.; Ye, D.-L.; Liu, X.-R.; et al. Mir20a/106a-WTX axis regulates RhoGDIa/CDC42 signaling and colon cancer progression. Nat. Commun. 2019, 10, 112.
- Li, X.; Wang, J.; Zhang, X.; Zeng, Y.; Liang, L.; Ding, Y. Overexpression of RhoGDI2 Correlates with Tumor Progression and Poor Prognosis in Colorectal Carcinoma. Ann. Surg. Oncol. 2011, 19, 145–153.
- Cho, H.J.; Kim, J.-T.; Lee, S.-J.; Hwang, Y.S.; Park, S.Y.; Kim, B.-Y.; Yoo, J.; Hong, K.S.; Min, J.-K.; Lee, C.-H.; et al. Protein phosphatase 1B dephosphorylates Rho guanine nucleotide dissociation inhibitor 1 and suppresses cancer cell migration and invasion. Cancer Lett. 2018, 417, 141–151.
- Zhang, Y.; Zhang, B. D4-GDI, a Rho GTPase Regulator, Promotes Breast Cancer Cell Invasiveness. Cancer Res 2006, 66, 5592–5598.
- Wang, X.; Bi, X.; Huang, X.; Wang, B.; Guo, Q.; Wu, Z. Systematic investigation of biomarker-like role of ARHGDIB in breast cancer. Cancer Biomark. 2020, 28, 101–110.
- Hu, L.; Zou, H.; Zhan, S.; Cao, K. Biphasic expression of RhoGDI2 in the progression of breast cancer and its negative relation with lymph node metastasis. Oncol. Rep. 2007, 17, 1383–1389.
- Joly, M.M.; Williams, M.M.; Hicks, D.J.; Jones, B.; Sanchez, V.; Young, C.D.; Sarbassov, D.D.; Muller, W.J.; Brantley-Sieders, D.; Cook, R.S. Two distinct mTORC2-dependent pathways converge on Rac1 to drive breast cancer metastasis. Breast Cancer Res. 2017, 19, 74.
- Liang, L.; Li, Q.; Huang, L.Y.; Li, D.W.; Wang, Y.W.; Li, X.X.; Cai, S.J. Loss of ARHGDIA expression is associated with poor prognosis in HCC and promotes invasion and metastasis of HCC cells. Int. J. Oncol. 2014, 45, 659–666.
- Fang, Y.; Yi, J.; Lizhi, L.; Qiucheng, C. Rho GDP Dissociation Inhibitor Beta Promotes Cell Proliferation and Invasion by Modulating the AKT Pathway in Hepatocellular Carcinoma. DNA Cell Biol. 2014, 33, 781–786.
- Liu, L.; Cui, J.; Zhao, Y.; Liu, X.; Chen, L.; Xia, Y.; Wang, Y.; Chen, S.; Sun, S.; Shi, B.; et al. KDM6A-ARHGDIB axis blocks metastasis of bladder cancer by inhibiting Rac1. Mol. Cancer 2021, 20, 77.
- Ahmed, M.; Sottnik, J.L.; Dancik, G.M.; Sahu, D.; Hansel, D.E.; Theodorescu, D.; Schwartz, M.A. An Osteopontin/CD44 Axis in RhoGDI2-Mediated Metastasis Suppression. Cancer Cell 2016, 30, 432–443.
- Stevens, E.V.; Banet, N.; Onesto, C.; Plachco, A.; Alan, J.K.; Nikolaishvili-Feinberg, N.; Midkiff, B.R.; Kuan, P.F.; Liu, J.; Miller, C.R.; et al. RhoGDI2 antagonizes ovarian carcinoma growth, invasion and metastasis. Small GTPases 2011, 2, 202–210.
- Xia, B.; Wang, J. Adenosine Inhibits Ovarian Cancer Growth Through Regulating RhoGDI2 Protein Expression. Drug Des. Dev. Ther. 2019, 13, 3837–3844.
- Küppers, R.; Klein, U.; Schwering, I.; Distler, V.; Bräuninger, A.; Cattoretti, G.; Tu, Y.; Stolovitzky, G.A.; Califano, A.; Hansmann, M.-L.; et al. Identification of Hodgkin and Reed-Sternberg cell-specific genes by gene expression profiling. J. Clin. Investig. 2003, 111, 529–537.
- Ma, L.; Xu, G.; Sotnikova, A.; Szczepanowski, M.; Giefing, M.; Krause, K.; Krams, M.; Siebert, R.; Jin, J.; Klapper, W. Loss of expression of LyGDI (ARHGDIB), a rho GDP-dissociation inhibitor, in Hodgkin lymphoma. Br. J. Haematol. 2007, 139, 217–223.
- Zeng, Y.; Ren, M.; Li, Y.; Liu, Y.; Chen, C.; Su, J.; Su, B.; Xia, H.; Liu, F.; Jiang, H.; et al. Knockdown of RhoGDI2 represses human gastric cancer cell proliferation, invasion and drug resistance via the Rac1/Pak1/LIMK1 pathway. Cancer Lett. 2020, 492, 136–146.
- Kim, H.-J.; Ryu, K.-J.; Kim, M.; Kim, T.; Kim, S.-H.; Han, H.; Kim, H.; Hong, K.-S.; Song, C.Y.; Choi, Y.; et al. RhoGDI2-Mediated Rac1 Recruitment to Filamin A Enhances Rac1 Activity and Promotes Invasive Abilities of Gastric Cancer Cells. Cancers 2022, 14, 255.
- Yi, B.; Hu, Y.; Qin, G.; Gu, W.; Zhu, X.; He, S.; Zhou, J.; Li, D. Depletion of RhoGDI2 expression inhibits the ability of invasion and migration in pancreatic carcinoma. Int. J. Mol. Med. 2014, 34, 205–212.
- Zhang, M.; Ding, G.; Zhou, L.; Shen, T.; Xu, X.; Zhao, T.; Jia, S.; Cao, L. Interferon Gamma Inhibits CXCL8-Induced Proliferation and Migration of Pancreatic Cancer BxPC-3 Cell Line via a RhoGDI2/Rac1/NF-κB Signaling Pathway. J. Interferon Cytokine Res. 2018, 38, 413–422.
- Yi, B.; Hu, Y.; Zhu, D.; Yao, J.; Zhou, J.; Zhang, Y.; He, Z.; Zhang, L.; Zhang, Z.; Yang, J.; et al. RhoGDI2 induced malignant phenotypes of pancreatic cancer cells via regulating Snail expression. Genes Genom. 2022, 44, 561–569.
- Liu, Y.; Wei, X.; Guan, L.; Xu, S.; Yuan, Y.; Lv, D.; He, X.; Zhan, J.; Kong, Y.; Guo, J.; et al. Unconventional myosin VIIA promotes melanoma progression. J. Cell Sci. 2018, 131, jcs209924.
- Niu, H.; Wu, B.; Jiang, H.; Li, H.; Zhang, Y.; Peng, Y.; He, P. Mechanisms of RhoGDI2 Mediated Lung Cancer Epithelial-Mesenchymal Transition Suppression. Cell. Physiol. Biochem. 2014, 34, 2007–2016.
- Niu, H.; Wu, B.; Peng, Y.; Jiang, H.; Zhang, Y.; Wang, J.; Zhang, Y.; He, P. RNA interference-mediated knockdown of RhoGDI2 induces the migration and invasion of human lung cancer A549 cells via activating the PI3K/Akt pathway. Tumor Biol. 2014, 36, 409–419.
- Cao, J.; Wang, Y.; Dong, R.; Lin, G.; Zhang, N.; Wang, J.; Lin, N.; Gu, Y.; Ding, L.; Ying, M.; et al. Hypoxia-Induced WSB1 Promotes the Metastatic Potential of Osteosarcoma Cells. Cancer Res 2015, 75, 4839–4851.
- Che, J.; Jin, Z.; Yan, F.; You, J.; Xie, J.; Chen, B.; Cheng, G.; Zhu, H.; He, Q.; Hu, Y.; et al. Discovery of 5,6-Bis(4-methoxy-3-methylphenyl)pyridin-2-amine as a WSB1 Degrader to Inhibit Cancer Cell Metastasis. J. Med. Chem. 2021, 64, 8621–8643.
- Nakata, Y.; Kondoh, K.; Fukushima, S.; Hashiguchi, A.; Du, W.; Hayashi, M.; Fujimoto, J.-I.; Hata, J.-I.; Yamada, T. Mutated D4-guanine diphosphate–dissociation inhibitor is found in human leukemic cells and promotes leukemic cell invasion. Exp. Hematol. 2008, 36, 37–50.
- Luo, J.; Wang, J.; Zheng, H.; Wang, L. Rho GDP-Dissociation Inhibitor 2 Inhibits C-X-C Chemokine Receptor Type 4-Mediated Acute Lymphoblastic Leukemia Cell Migration. Front. Oncol. 2020, 10, 1512.
- Mokhtar, A.M.B.A.; Ahmed, S.B.M.; Darling, N.J.; Harris, M.; Mott, H.R.; Owen, D. A Complete Survey of RhoGDI Targets Reveals Novel Interactions with Atypical Small GTPases. Biochemistry 2021, 60, 1533–1551.
- Kasper, B.; Tidow, N.; Grothues, D.; Welte, K. Differential expression and regulation of GTPases (RhoA and Rac2) and GDIs (LyGDI and RhoGDI) in neutrophils from patients with severe congenital neutropenia. Blood 2000, 95, 2947–2953.
- Scheffzek, K.; Stephan, I.; Jensen, O.N.; Illenberger, D.; Gierschik, P. The Rac-RhoGDI complex and the structural basis for the regulation of Rho proteins by RhoGDI. Nat. Struct. Biol. 2000, 7, 122–126.
- Platko, J.V.; Leonard, A.D.; Adra, C.N.; Shaw, R.J.; Cerione, A.R.; Lim, B. A single residue can modify target-binding affinity and activity of the functional domain of the Rho-subfamily GDP dissociation inhibitors. Proc. Natl. Acad. Sci. USA 1995, 92, 2974–2978.
- Togawa, A.; Miyoshi, J.; Ishizaki, H.; Tanaka, M.; Takakura, A.; Nishioka, H.; Yoshida, H.; Doi, T.; Mizoguchi, A.; Matsuura, N.; et al. Progressive impairment of kidneys and reproductive organs in mice lacking Rho GDIα. Oncogene 1999, 18, 5373–5380.
- Ishizaki, H.; Togawa, A.; Tanaka-Okamoto, M.; Hori, K.; Nishimura, M.; Hamaguchi, A.; Imai, T.; Takai, Y.; Miyoshi, J. Defective Chemokine-Directed Lymphocyte Migration and Development in the Absence of Rho Guanosine Diphosphate-Dissociation Inhibitors α and β. J. Immunol. 2006, 177, 8512–8521.
More
Information
Subjects:
Oncology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
862
Revisions:
2 times
(View History)
Update Date:
20 Mar 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




