Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Balaji Ramachandran | -- | 1357 | 2023-03-14 07:29:11 | | | |
| 2 | Conner Chen | Meta information modification | 1357 | 2023-03-16 06:37:32 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Chandrasekar, N.; Balaji, R.; Perala, R.S.; Nik Humaidi, N.Z.; Shanmugam, K.; Liao, Y.; Hwang, M.T.; Govindaraju, S. Significance of Synthetic DNA Constructs. Encyclopedia. Available online: https://encyclopedia.pub/entry/42150 (accessed on 08 February 2026).
Chandrasekar N, Balaji R, Perala RS, Nik Humaidi NZ, Shanmugam K, Liao Y, et al. Significance of Synthetic DNA Constructs. Encyclopedia. Available at: https://encyclopedia.pub/entry/42150. Accessed February 08, 2026.
Chandrasekar, Narendhar, Ramachandran Balaji, Ramaswamy Sandeep Perala, Nik Zulkarnine Nik Humaidi, Kirubanandan Shanmugam, Ying-Chih Liao, Michael Taeyoung Hwang, Saravanan Govindaraju. "Significance of Synthetic DNA Constructs" Encyclopedia, https://encyclopedia.pub/entry/42150 (accessed February 08, 2026).
Chandrasekar, N., Balaji, R., Perala, R.S., Nik Humaidi, N.Z., Shanmugam, K., Liao, Y., Hwang, M.T., & Govindaraju, S. (2023, March 14). Significance of Synthetic DNA Constructs. In Encyclopedia. https://encyclopedia.pub/entry/42150
Chandrasekar, Narendhar, et al. "Significance of Synthetic DNA Constructs." Encyclopedia. Web. 14 March, 2023.
Copy Citation
The prevalence of mutated species of COVID-19 antigens has provided a strong impetus for identifying a cost-effective, rapid and facile strategy for identifying the viral loads in public places. The ever-changing genetic make-up of SARS-CoV-2 posts a significant challenfge for the research community to identify a robust mechanism to target, bind and confirm the presence of a viral load before it spreads. Synthetic DNA constructs are a novel strategy to design complementary DNA sequences specific for antigens of interest as SARS-CoV-2 antigens.
SARS
complementary DNA
protein–DNA
1. Introduction
In the pandemic era, the uncontrollable spread of COVID-19 has necessitated the development of a novel sensor which can efficiently detect the virus in its current form as well as in future mutated forms. Early detection can be performed by the rapid analytical techniques using graphene-based materials. The COVID-19 antigens are among the best targeting sites for early detection of viral infection in the human system. This can be performed via the reliable and rapid process of a label-free diagnostic method coupled with synthetic DNA constructs. Generally, the synthesis of DNA constructs and fabrication into synthons, genes, biological circuits and construction of genomes maps via chemical synthesis methods are emerging research area in the field of molecular diagnostic technology. Alternatively, these DNA constructs can be used in the low-cost rapid diagnostic methods for various viral infections [1]. Since December 2019, COVID-19 was declared as a pandemic by the WHO, urging the development of highly sensitive biomarker-based detection methods capable of operating in robust conditions for detection of COVID-19 and their variants in biofluids [2].
The rapid sensing technique for detecting COVID-19 biomarkers in all phases of infection stages is a challenging task in molecular virology and provides a platform for controlling the infection in human. The diagnostic method for COVID-19 can be broadly divided into: (1) detection and investigation of the nucleic acid genome of the SARS-CoV-2 via qualitative reverse transcriptase polymerase chain reaction (qRT-PCR) and loop-mediated isothermal amplification (LAMP), (2) detection of viral antigens/proteins by the enzyme-linked immunosorbent assay (ELISA) or lateral flow assay (LFA), (3) studying antibodies produced by the human immune system due to viral infections. The US-FDA (United States–Food and Drug Administration) authenticated the above methods for commercialization and they are currently in practice. However, the Abbott-based COVID-19 diagnostic method gave false responses even at the early stage of coronavirus infection due to poor sensitivity and specificity problems. This occurred due to the low availability of biomarkers such as viral RNA, viral proteins and antibodies in the serum of the human blood samples collected from the coronavirus patients.
RT-PCR is now the most desired standard technique for detection of COVID-19. RT-PCR is an accurate test for diagnosis of COVID-19 in humans. However, it takes longer, up to days, to obtain results since skilled personnel are required to operate the instrument. In the design and engineering of a diagnostic method for detection of COVID-19, the bottlenecks in analytical techniques to assay COVID-19-associated biomarkers in saliva and blood/plasma/serum and/or nasal swabs are: (1) the inability to assay different types of biomolecules in one instrument—for example, the RT-PCR and LAMP techniques are capable of assaying viral nucleic acids but not proteins; (2) ELISA and LFA only detect proteins but not RNAs; and (3) general inadequacy in assay sensitivity/limit of detections to quantify a low abundance of biomarkers while retaining high specificity and accuracy [3].
The most commonly relied upon COVID-19 sensing techniques ELISA, PCR and RT-PCR are reliable but require expansive instrumentation, training and time for diagnosis. PCR is predominantly used to detect specific organisms such as viruses. PCR is a very powerful technique as it can identify the residual fragments of a virus even after the disease disappears and so it has enabled rapid analysis to diagnose diseases in humans. RT-PCR is a laboratory-based investigation process which produces copies of specific DNA/RNA sequences for investigation. Enzymes belonging to RT are manipulated and used to vary a specific sequence of RNA to a matching piece of DNA. After producing numerous pieces of DNA in large quantities through DNA polymerase, it can identify a specific mRNA sequence which makes up a gene. RT-PCR can help to efficiently diagnose a disease and monitor an infection. So, in standard PCR analysis, a DNA template is amplified and a thermocycler is used; but in RT-PCR, RNA is utilized as a template and is reverse transcribed into complementary DNA. PCR is typically employed for viruses that have DNA for amplification [1]. Similarly, SARS-CoV-2 has only RNA and it can be detected through RT-PCR. Figure 1 shows the structure of and components present in SARS-CoV-2.
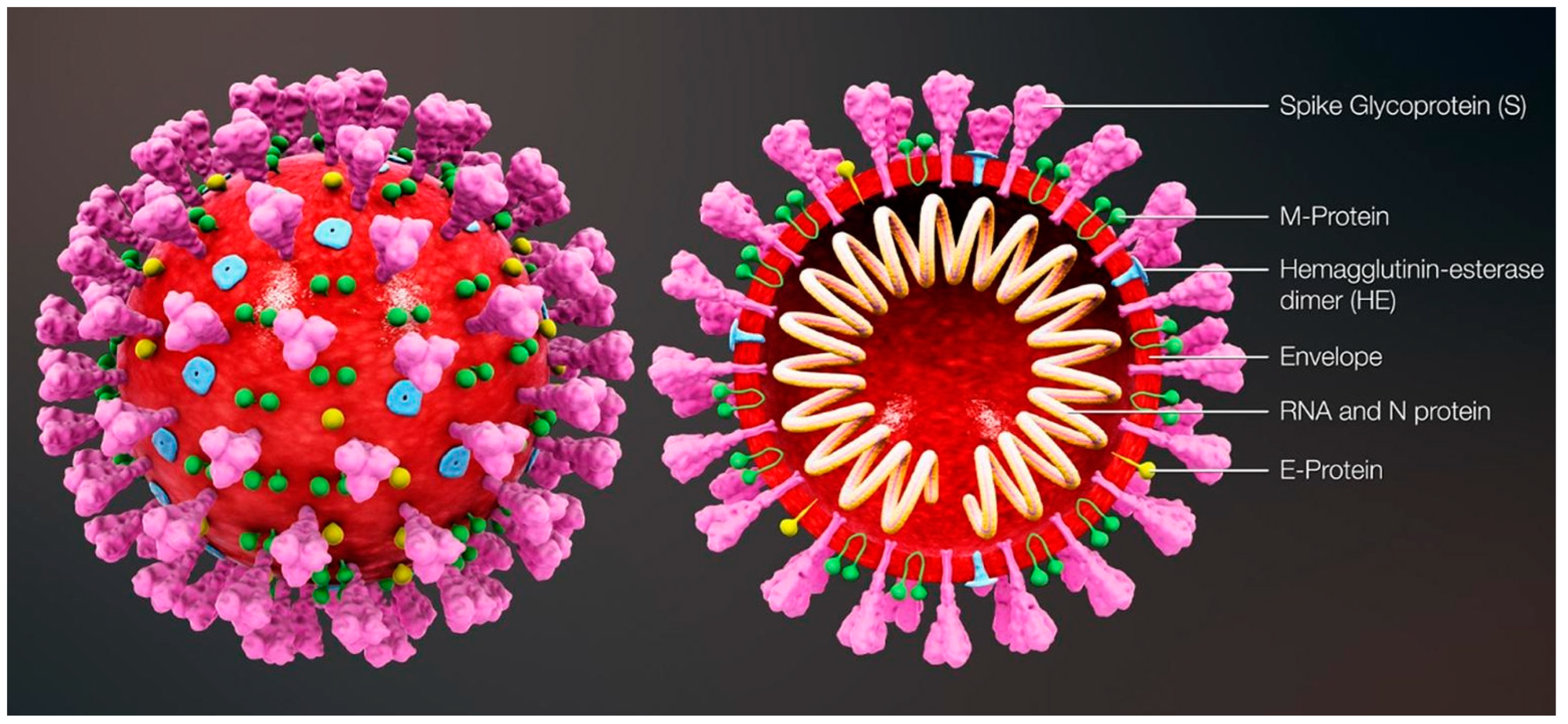
Figure 1. Structure and components of SARS-CoV-2. Reproduced with permission from Ref. [1]. Copyright 2022 Elsevier.
The aforementioned detection techniques [1][2] suffer from certain demerits, such as the need for trained and qualified personnel and also more demand for expensive chemical reagents. Importantly, traditional techniques are not suited for large-scale diagnosis in quick time frames. Although these techniques exhibit great sensitivity, sometimes they provide false positive or negative test results. It is noteworthy to mention that the sensitivity and the limit of detection (LOD) are respective to the COVID-19 viral dose and infective virus dose, which becomes an issue for sensing platforms. For RT-PCR testing of nasal samples, standard COVID-19 detection techniques provide an average LOD of 100 copies of viral RNA per mL [4]. Having said that, more than 10,000 fold the LOD can be achieved through electrochemical-based detection techniques (Figure 2).
Figure 2. Schematic of an electrochemical COVID-19 detection technique using voltametric, impedimetric and potentiometric sensors. Reproduced with permission from Ref. [1]. Copyright 2022 Elsevier.
Electrochemical detection is among the significant, low-cost and high-performance sensing techniques [2][3] which can effectively detect changes in the current, voltage or charge generated from surface interactions with a working electrode and analyte. The salient features of the electrochemical sensing platform are their user-friendly operation, low-cost fabrication, rapid detection capability and a superior LOD. These advantages can achieve the global need for diagnosing COVID-19 at larger scales. The overall performance of the electrochemical technique is improved through integration of nanomaterials into the system. Nanomaterials such as graphene family materials (GFM) can help in the successful interaction between analytes and sensors largely due to their superior specific surface area that aids in precise, rapid detection of virus biomarkers.
2. Significance of Synthetic DNA Constructs
The ease of access through which oligonucleotides are being synthesized by chemical methods has resulted in the construction of custom-made synthetic DNA strands or synthons being used as recognition units for biosensors. The ever-adapting SARS-CoV-2 is tough to track for sudden mutations which are specific to certain locations. However, identification of the commonly occurring nucleotide sequences in the SARS-CoV-2 antigens can be targeted by the synthetic constructs.
Batches of such synthetic DNA constructs can be fabricated by using controlled porous glass beads, wherein the oligonucleotides are grown over the glass bead surface as the feed chemicals are made to flow over the surface. Microarray-based synthesis facilitates the growth of several unique strands of DNA at the same time as enabling the methodology to produce novel and customized nucleotide pools which can be used as probes or complementary strands for target analytes such as COVID-19 antigens [1][2][3].
A single microchip can be fabricated for a variety of such oligonucleotide pools, which can then be segmented into specific sub pools so that they can be utilized to generate assembly-specific harvests. Such high-throughput fabrication of oligonucleotides can be employed to target several mutant strains or different species of microbes altogether [3][4][5].
Kosuri et al. [6] describes the scalable gene synthesis on a DNA chip from the OLS pool without distinguishing dsDNA and ssDNA, cleaved to form an oligonucleotide pool. Yellow or brown indexed primers are used to amplify separate plate sub pools with DNA to assemble different genes. Blue-colored sequences are assembly specific and are used to amplify assembly sub pools that have the DNA required to make one unique gene. The primer sequences are cleaved, forming dsDNA by using type IIS restriction enzymes; or forming ssDNA by DpnII/USER/λ exonuclease. Construction primers are employed to proceed with assembly PCR reaction to build one unique gene from each assembly sub pool. The assembled products are cloned and validated by enzyme-mediated correction.
References
- Hamidi-Asl, E.; Heidari-Khoshkelat, L.; Raoof, J.B.; Richard, T.P.; Farhad, S.; Ghani, M. A review on the recent achievements on coronaviruses recognition using electrochemical detection methods. Microchem. J. 2022, 178, 107322.
- Yakoh, A.; Pimpitak, U.; Rengpipat, S.; Hirankarn, N.; Chailapakul, O.; Chaiyo, S. Paper-based electrochemical biosensor for diagnosing COVID-19: Detection of SARS-CoV-2 antibodies and antigen. Biosens. Bioelectron. 2021, 176, 112912.
- Alireza Hashemi, S.; Bahrani, S.; Mojtaba Mousavi, S.; Omidifar, N.; Ghaleh Golab Behbahan, N.; Arjmand, M.; Ramakrishna, S.; Bagheri Lankarani, K.; Moghadami, M.; Shokripour, M.; et al. Ultra-precise label-free nanosensor based on integrated graphene with Au nanostars toward direct detection of IgG antibodies of SARS-CoV-2 in blood. J. Electroanal. Chem. 2021, 894, 115341.
- Arnaout, R.; Lee, R.A.; Lee, G.R.; Callahan, C.; Yen, C.F.; Smith, K.P.; Arora, R.; Kirby, J.E. SARS-CoV2 Testing: The Limit of Detection Matters. bioRxiv 2020.
- Hughes, R.A.; Ellington, A.D. Synthetic DNA Synthesis and Assembly: Putting the Synthetic in Synthetic Biology. Cold Spring Harb. Perspect. Biol. 2017, 9, a023812.
- Kosuri, S.; Eroshenko, N.; LeProust, E.M.; Super, M.; Way, J.; Li, J.B.; Church, G. Scalable gene synthesis by selective amplification of DNA pools from high-fidelity microchips. Nat. Biotechnol. 2010, 28, 1295–1299.
More
Information
Subjects:
Engineering, Chemical
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
597
Revisions:
2 times
(View History)
Update Date:
16 Mar 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




