Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Sajid Ali | -- | 2763 | 2023-03-14 06:08:09 | | | |
| 2 | Conner Chen | + 2 word(s) | 2765 | 2023-03-15 03:31:49 | | | | |
| 3 | Conner Chen | + 3 word(s) | 2768 | 2023-03-16 02:06:38 | | | | |
| 4 | Conner Chen | Meta information modification | 2768 | 2023-03-16 07:16:12 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Ali, S.; Khan, M.; Al Azzawi, T.N.I.; Yun, B. Nitric Oxide Biosynthesis. Encyclopedia. Available online: https://encyclopedia.pub/entry/42148 (accessed on 08 February 2026).
Ali S, Khan M, Al Azzawi TNI, Yun B. Nitric Oxide Biosynthesis. Encyclopedia. Available at: https://encyclopedia.pub/entry/42148. Accessed February 08, 2026.
Ali, Sajid, Murtaza Khan, Tiba Nazar Ibrahim Al Azzawi, Byung-Wook Yun. "Nitric Oxide Biosynthesis" Encyclopedia, https://encyclopedia.pub/entry/42148 (accessed February 08, 2026).
Ali, S., Khan, M., Al Azzawi, T.N.I., & Yun, B. (2023, March 14). Nitric Oxide Biosynthesis. In Encyclopedia. https://encyclopedia.pub/entry/42148
Ali, Sajid, et al. "Nitric Oxide Biosynthesis." Encyclopedia. Web. 14 March, 2023.
Copy Citation
Nitric oxide (NO), a colorless gaseous molecule, is a lipophilic free radical that easily diffuses through the plasma membrane. These characteristics make NO an ideal autocrine (i.e., within a single cell) and paracrine (i.e., between adjacent cells) signalling molecule. As a chemical messenger, NO plays a crucial role in plant growth, development, and responses to biotic and abiotic stresses. Furthermore, NO interacts with reactive oxygen species, antioxidants, melatonin, and hydrogen sulfide. It regulates gene expression, modulates phytohormones, and contributes to plant growth and defense mechanisms. In plants, NO is mainly produced via redox pathways.
nitric oxide
antioxidants
phytohormones
1. Introduction
Nitric oxide (NO) was first discovered in plants by Klepper [1]. However, the discovery of the critical functions of NO in mammals prompted its research in plants. In animals, Palmer et al. [2] investigated the role of NO as an endothelium-derived relaxing factor. This discovery grabbed the attention of scientists to further explore the role of NO as a chemical messenger in other living organisms under different environmental conditions. Early studies on NO focused on its environmental impact, since it is a significant pollutant. NO is abundantly produced in the atmosphere due to industrial combustion and automobile emissions, and it damages the ozone layer and causes acid rain [3]. Therefore, in the beginning, NO was considered a toxic gaseous molecule. However, later on, NO was explored as a significant signalling molecule in both plants and animals to regulate their physiological, molecular, and biochemical aspects under both normal and stressful conditions [4]. Further studies explored that NO is produced in living organisms via different enzymatic and nonenzymatic reactions and that NO can also be applied to plants in the form of different NO donors such as sodium nitroprusside (SNP) [5]. As a multifunctional molecule, in 1992, NO was regarded as a molecule of the year [6].
NO is a small, colorless, lipophilic free radical and a diatomic gas molecule that consists of nitrogen and oxygen. Therefore, NO can easily diffuse through cell membranes and can work as an excellent intracellular and intercellular chemical messenger in both plants and animals under normal and stressful conditions [4]. Under normal conditions, NO significantly contributes to the breaking of seed dormancy and induction of seed germination, root development, lateral root formation, primary root growth, adventitious root formation, root hair development, chlorophyll biosynthesis, vegetative growth, vascular differentiation, symbiosis nodule formation, stomatal moment, iron homeostasis, leaf senescence, flowering control, and fruit ripening [7]. Furthermore, NO also plays a pivotal role to protect plants from various environmental stresses, including wounding, pathogen invasion, hypoxia, ultra violet (UV) radiation, drought stress, salt stress, temperature extremes, and heavy metals stresses [5][8][9].
Compared with animals, NO production in plants is still a subject of controversy. In mammals, the main pathway responsible for NO production involves the conversion of L-arginine to citrulline by nitric oxide synthase (NOS). Although NOS-like activities in plants are susceptible to mammalian NOS inhibitors, no typical NOS sequence was discovered in the more than 1000 sequenced transcriptomes of terrestrial plants [10]. However, Foresi, et al. [11] reported the presence of NOS-like enzyme in green algae. In addition, in plants, NO is produced from different enzymatic and nonenzymatic routs, which are in continuous examination [12]. Cytosolic nitrate reductase (NR) has been considered as one of the main enzymatic sources of NO production in plants under aerobic conditions [13]. NR-dependent NO production pathway is involved in the production of NO during pathogen-induced defense responses, drought stress, cold stress, regulation of stomatal opening and closing, mitigation of symptoms caused by iron deficiency, and root-growth-associated responses [14]. Furthermore, NO can also be synthesized non-enzymatically from NO2- in the presence of a reductant, such as ascorbate [15]. NO is produced in different plant organelles, including chloroplasts, mitochondria, peroxisomes, cytoplasm, cell wall, and cell membrane [16].
Although reactive oxygen species (ROS) are produced in the plant as a part of normal cellular metabolism, and at optimum concentrations can act as signaling molecules, if overproduced they can cause oxidative stress damages [4]. Plants primarily deal with the oxidative stress via an antioxidant defense system [4]. During abiotic stress, ROS are overproduced, resulting in an increased NO production which indicates the interaction between ROS and reactive nitrogen species (RNS). For instance, the application of hydrogen peroxide (H2O2) significantly enhanced the production of NO in Arabidopsis, which leads to the induction of antioxidants activities and reestablishment of redox balance [17]. On the other hand, NO production was reduced with the removal of H2O2 [18]. Furthermore, calcium ion channel’s inhibitors also inhibit H2O2-induced NO synthesis [19]). Similarly, H2O2 is crucial for the production of NO mediated by abscisic acid [20]. In contrast, the production of H2O2 mediated by ABA does not depend on NO [21]). Importantly, both H2O2 and NO are essential for signal transduction as well as phytotoxicity [22]. The NO and ROS signaling pathways in the defense system of plants are closely connected. During stress conditions, NO and ROS can alter gene transcription profiles and the production of plant hormones and metabolites. For instance, NO reacts with superoxide anion (O2−) to form peroxynitrite (ONOO−), which leads to post-translational modification (PTM). Several studies over the last two decades have demonstrated the involvement of NO and ROS in plant defense systems, where they show a symbiotic relationship [23]. Furthermore, in plants, melatonin also acts as a multipurpose molecule and regulates plant growth and development, and defense responses against biotic and abiotic stress conditions [24][25]. NO and melatonin (MEL) interact to induce plant growth and development under biotic and abiotic stress conditions [24]. Along with NO, MEL promotes plant growth and modulates the ability of plants to withstand abiotic stress by strengthening the antioxidant system [26]. MEL can induce NO synthesis by boosting the activities of NOS and scavenging NO if overproduced [27][28]. NO and MEL combine to form NOmela, a newly discovered chemical that may exert a more potent signaling effect than either MEL or NO alone [29]. In addition, melatonin also acts as a chemical scavenger for ROS and reactive nitrogen species (RNS) to improve the antioxidant system of the plants and protect the plants from oxidative damages [30].
Interactions between NO and antioxidants and plant hormones further broaden the signaling role of NO in plant growth and development under normal and stressful conditions. It has been reported that NO affects the activities of antioxidant enzymes, such as catalase (CAT), superoxide dismutase (SOD), various peroxiredoxins (Prxs) and enzymes involved in the ascorbate-glutathione (Asa-GSH) cycle, through PTMs [31]. In recent studies it has been reported that different NO donors such as SNP, S-nitrosocysteine (CySNO) and S-nitrosoglutathione (GSNO) are used to exogenously to supply NO to the plants to improve their defense system against different stresses [5][9]. Moreover, SNP has been widely used as a NO donor in pharmacologic studies to investigate its function in NO synthesis. While in plants, SNP has been used as a NO donor to elicit the synthesis of important bioactive secondary metabolites [32]. Application of SNP (a NO donor) on Panax ginseng and rice plants, significantly enhanced the activities of antioxidant enzymes such as CAT, SOD, peroxidase (POD), polyphenol peroxidase (PPO) and APX to protect the plants from the toxic effects of salt and lead stress [5][33]. NO also acts synergistically with auxin (AUX) to control a variety of plant responses, including root organogenesis, gravitropic responses, formation of root nodules, responses to iron deficiency, activation of cell division, formation of embryogenic cells, and stimulation of nitrate reductase (NR) activity [34]. Application of SNP and AUX significantly enhanced photosynthetic efficiency and antioxidant system of Brassica juncea plants under salt stress. Moreover, the combined application of SNP and AUX was more effective than their individual treatment [35]. However, AUX has been found to have little to no impact on the activation of NO production under specific experimental conditions or in specific cell types [36]. On the other hand, NO has been found to exert both synergistic and antagonistic effects on cytokinin (CK) metabolism. NO has been reported to promote the activation of CK to correct the meristematic abnormalities of the noa1 mutant in root tissues [37]. Furthermore, high concentrations of NO are known to inhibit CK signaling [38]. Under stress conditions, NO interacts with ABA to induce the plant defense system [39]. The interaction between NO and ABA regulates crassulacean acid metabolism (CAM) expression [40], thereby aiding plant survival under drought stress and nutrient deficiency. NO-induced AtAO3 and AtNCED3 significantly improved drought stress tolerance in Arabidopsis thaliana [39]. NO interacts both synergistically and antagonistically with gibberellins (GAs) and has been reported to function upstream of GA to regulate its production, perception, and transduction [41]. In contrast, in one study, endogenous GA concentration was significantly reduced in Arabidopsis plants treated with SNP [41]. Most of the recent studies have shown a negative relationship between NO and ethylene (ETH). For instance, exogenous application of NO delays the senescence of both vegetative and reproductive organs by inhibiting factors involved in ETH generation [42].
The role of NO as a chemical messenger has been studied extensively with respect to plant growth and development under normal and stressful conditions. Pandey et al. [43] reported that during seed germination, NO suppresses cytochrome oxidase (COX) to increase respiration and thereby provide more energy for germination. Furthermore, NO has also been reported to significantly contribute to plant growth and development, symbiotic associations, defense responses against biotic and abiotic stressors, and vegetative and reproductive growth [4][7]. NO-induced AtILL6 gene positively regulated plant’s basal defense, defense mediated by resistance (R) gene and systemic acquired resistance (SAR) against virulent and avirulent pathogenic bacteria in Arabidopsis thaliana [9]. Similar findings were also observed by Shahid, et al. [44], and they stated that NO-induced AtCLV1 and AtCLV2 positively regulated the plant’s immunity against pathogenic bacteria. In contrast, Nabi et al. [45] reported that NO-induced AtBZIP62 negatively regulated plants’ basal defense and SAR in A. thaliana. However, the NO-induced AtBZIP62 significantly enhanced the resistance of Arabidopsis thaliana against drought stress [8]. Furthermore, NO-induced AtAO3 and AtNCED3 negatively regulated immunity of A. thaliana but positively regulated the resistance of it to drought stress [39]. Application of NO-donors also significantly improved the defense system of different plants against abiotic stress conditions. For instance, the application of SNP significantly improved the defense system of rice against salinity and lead stress [5][46]. Similarly, SNP application increased the defense system of soybean plants against flooding stress [47].
2. NO Biosynthesis
NO was first studied by Joseph Priestley in 1772, who noted that NO was a colorless, toxic gas [6]. In 1879, William Murrell first used nitroglycerin to treat angina pectoris, but it was not until 1977 that Ferid Murad discovered that the release of NO causes nitroglycerin to exert positive pharmacological effects on vascular smooth muscle [6]. Furthermore, Palmer, Ferrige and Moncada [2] identified NO as an endothelial relaxation factor. Eventually, in 1992, NO was regarded as the “molecule of the year” [6]. In plants, NO was first discovered by Klepper [1]. In animals, NO is produced in the presence of NADPH and O2 by the conversion of L-arginine to N-hydroxyarginine via NOS, which also results in the production of L-citrulline. However, in higher plants, it is unknown whether NOS is present. The production of NO via NOS, nitrate/nitrite reductase, other enzymes, and non-enzymatic reaction pathways is discussed in the following subsections.
2.1. NO Production through NOS
NOS has not yet been discovered in higher plants, but has been found in Ostreococcus tauri, a unicellular alga. This suggests that higher plants may have evolved different mechanisms of NO production than those found in animals [48]. However, some studies have shown that NO production using NOS-associated pathways also exists in plants, since experiments conducted on maize, pea, and tobacco found that the use of NOS inhibitors and antibodies (monoclonal and polyclonal) reduced the production of NO [49][50]. Furthermore, it has also been demonstrated that the NO levels of Arabidopsis were raised in response to the expression of rat neuron-type NOS [51]. However, the genes and protein sequences of plant NOS-like enzymes are substantially dissimilar from those found in animals. For example, multiple NOS-like proteins in maize were found to show limited protein sequence homology with animal-derived NOS sequences [52]. Guo, Okamoto and Crawford [36] discovered that the protein produced by the AtNOS1 gene has a similar sequence to one involved in NO generation in gastropods (snails), but that this differs from other animal NOS proteins. Later research by Moreau et al. [53] revealed that AtNOS1 is a circularly-permuted GTPase (cGTPase), which led to the renaming of this protein to AtNOA1. It may play a role in mitochondrial and ribosomal biosynthesis and translation and may act indirectly to regulate NO generation [53][54].
2.2. NO Production via the Nitrate or Nitrite Reductase and Other Enzymes
The cytoplasm within plant cells contains an enzyme called NR, which catalyzes the conversion of nitrate and nitrite to NO. This conversion is thought to be the main source of NO in plants [55][56]. A previous study reported that nitrite and endogenous NO levels were considerably lower in Arabidopsis NR deletion double mutant atnia1/atnia2 relative to wild-type (WT) plants and concluded that the NR genes AtNIA1 and AtNIA2 are involved in NO production [57]. NR activity was subsequently detected in a variety of plants, such as maize, cucumber, sunflower, and spinach [56][58]. NR activity can be controlled through PTM of proteins, including phosphorylation, and via redox pathways; both forms of regulation can impact NO generation [59].
Research has demonstrated that the nitrite reductase (NiR) reaction pathway occurs in tobacco root cells [60]. The presence of high NO concentrations in transgenic tobacco plants containing an antisense version of the NiR gene suggests that NiR is involved in the generation of endogenous NO in tobacco plants [61]. Since NiR derived from plastids reduces nitrite to ammonia under normal circumstances, the nitrite content of plant cells is significantly lower than their nitrate content [56]. This is thought to inhibit NiR activity via competitive inhibition. As a result, when NADH and nitrate levels are sufficient, NR is much less effective at catalyzing the conversion of nitrite to NO than it is at catalyzing the conversion of nitrate to NO [56]. However, further research is needed to determine the exact molecular regulatory mechanism by which NR effectively converts nitrite to NO.
In addition, NR can function as a cofactor to help other enzymes produce NO. For instance, in a unicellular algae (Chlamydomonas reinhardtii), NR was observed to help the amidoxime-reducing component (ARC) to convert nitrite to NO and replace the electron transfer functions of cytochrome b5 (cytb5) and cytochrome b5 reductase (cytb5R) [62]. The major function of plant ARC proteins may be the manufacturing of NO, which is why they are also known as NO-forming nitrite reductases. The Arabidopsis genome has two ARC genes that show a high degree of evolutionary conservation and are similar to those found in C. rhinoceros [63].
Numerous enzymes, such as horseradish peroxidase (HPOX) [64], xanthine oxidoreductase (XOR), xanthine dehydrogenase (XDH) [65], cyt P450 [66], polyamine oxidase (PAO), polyamine oxidoreductase (POR) [48], and copperamine oxidase (CuAO) [41], also produce NO in plant cells. For instance, PAO uses polyamine (PA) and POR uses hydroxylamine (HA) as their respective substrates for the production of NO. CuAO in Arabidopsis has been found to increase arginine and NO levels by decreasing the activity of arginase [67]. However, further investigation of proteins or protein complexes in plants associated with arginine-dependent NO generation is warranted. Several molybdoenzymes, including xanthine oxidase (XO), aldehyde oxidase (AO), and sulfite oxidase (SO), also reduce nitrite to NO under low oxygen conditions [68], although the precise reduction pathways involved and their associated molecular regulatory mechanisms remain poorly understood.
2.3. NO Production via Nonenzymatic Reaction Pathways
It has been demonstrated that NO can also be formed nonenzymatically by the reduction of nitrite in the presence of antioxidants, acidic and reducing agents, or as a byproduct of direct chemical interactions between nitrogen oxides and plant metabolites [69]. Exogenous nitrite treatments used in earlier studies were found to partially restore NO levels and Pseudomonas syringae resistance in Arabidopsis nia1/nia2 mutants. This raises the possibility that plants may have a mechanism for producing NO that is totally independent of enzymatic processes [59][70]. For example, Bethke et al., found that following exogenous application of nitrite, the ectoderm of barley pasteurized cells underwent a non-enzymatic reaction to reduce nitrite to NO [71]. Furthermore, some arginine metabolites, including PAs and HAs, can be used by plants to produce NO [68]. However, further research is required to fully understand the physiological and biochemical functions of these pathways and their underlying molecular mechanisms. A summary of different sources and pathways related to NO production discussed above is shown in Figure 1.
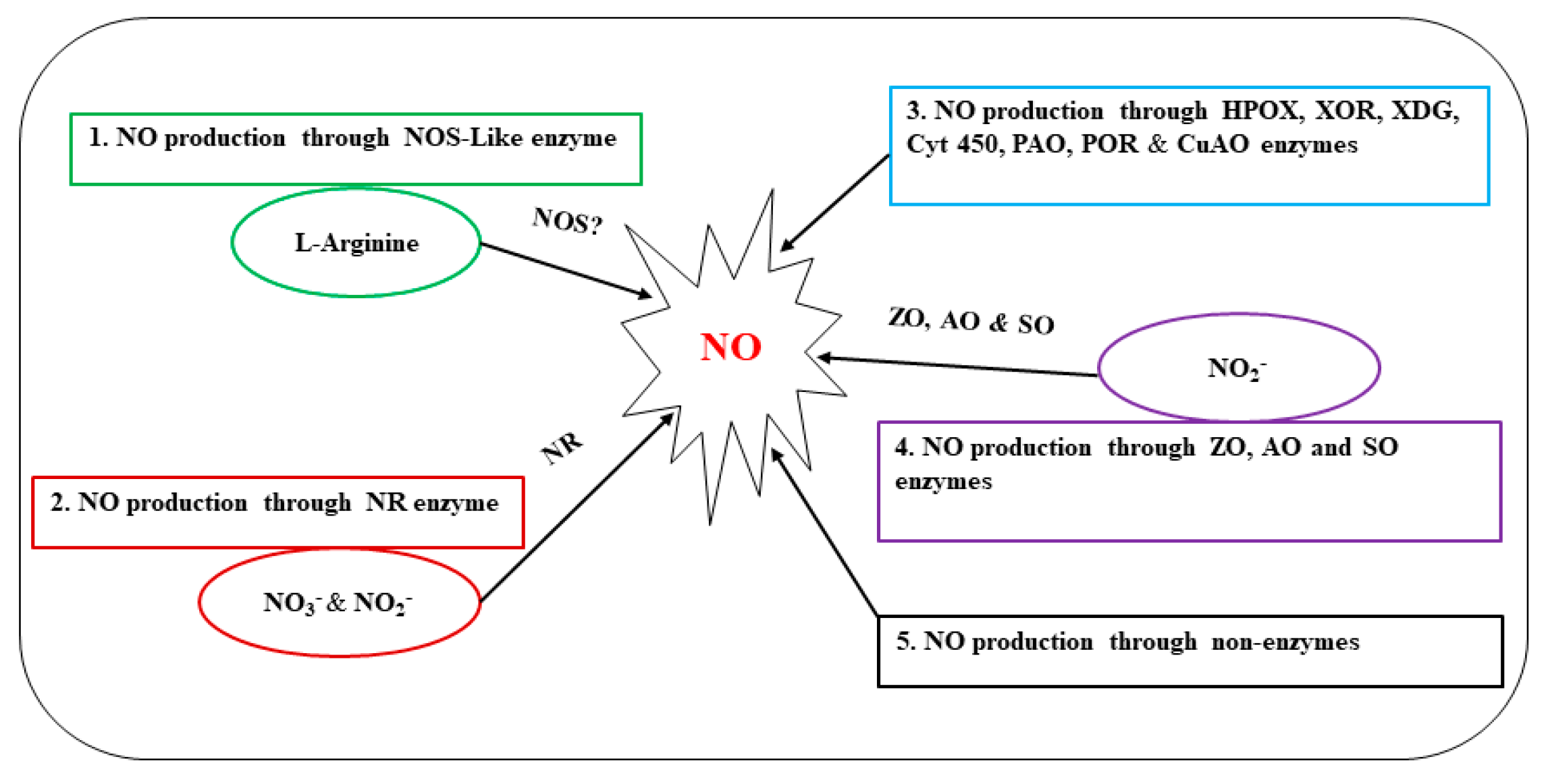
Figure 1. Production of NO through different pathways. Nitric oxide synthase (NOS), not discovered in higher plants (NOS?), Nitrogen oxide (NO), nitric oxide synthase (NOS), nitrate reductase (NR), nitrite ion (NO3−), nitrate ion (NO2−), horseradish peroxidase (HPOX), xanthine oxidoreductase (XOR), xanthine dehydrogenase (XDG), cytochrome (cyt P450), polyamine oxidase (PAO), polyamine oxidoreductase (POR), copperamine oxidase (CuAO), xanthine oxidase (ZO), aldehyde oxidase (AO), and sulfite oxidase (SO).
References
- Klepper, L. Nitric oxide (NO) and nitrogen dioxide (NO2) emissions from herbicide-treated soybean plants. Atmos. Environ. 1979, 13, 537–542.
- Palmer, R.M.J.; Ferrige, A.G.; Moncada, S. Nitric oxide release accounts for the biological activity of endothelium-derived relaxing factor. Nature 1987, 327, 524–526.
- Salgado, I.; Carmen Martínez, M.; Oliveira, H.C.; Frungillo, L. Nitric oxide signaling and homeostasis in plants: A focus on nitrate reductase and S-nitrosoglutathione reductase in stress-related responses. Braz. J. Bot. 2013, 36, 89–98.
- Khan, M.; Ali, S.; Al Azzawi, T.N.I.; Saqib, S.; Ullah, F.; Ayaz, A.; Zaman, W. The Key Roles of ROS and RNS as a Signaling Molecule in Plant–Microbe Interactions. Antioxidants 2023, 12, 268.
- Rahim, W.; Khan, M.; Al Azzawi, T.N.I.; Pande, A.; Methela, N.J.; Ali, S.; Imran, M.; Lee, D.-S.; Lee, G.-M.; Mun, B.-G. Exogenously Applied Sodium Nitroprusside Mitigates Lead Toxicity in Rice by Regulating Antioxidants and Metal Stress-Related Transcripts. Int. J. Mol. Sci. 2022, 23, 9729.
- Levine, A.B.; Punihaole, D.; Levine, T.B. Characterization of the role of nitric oxide and its clinical applications. Cardiology 2012, 122, 55–68.
- Simontacchi, M.; Galatro, A.; Ramos-Artuso, F.; Santa-María, G.E. Plant survival in a changing environment: The role of nitric oxide in plant responses to abiotic stress. Front. Plant Sci. 2015, 6, 977.
- Rolly, N.K.; Imran, Q.M.; Shahid, M.; Imran, M.; Khan, M.; Lee, S.-U.; Hussain, A.; Lee, I.-J.; Yun, B.-W. Drought-induced AtbZIP62 transcription factor regulates drought stress response in Arabidopsis. Plant Physiol. Biochem. 2020, 156, 384–395.
- Khan, M.; Nazar, T.; Pande, A.; Mun, B.-G.; Lee, D.; Hussain, A.; Lee, B.-H.; Yun, B.-W. The Role of Nitric Oxide-Induced ATILL6 in Growth and Disease Resistance in Arabidopsis thaliana. Front. Plant Sci. 2021, 12, 1314.
- Jeandroz, S.; Wipf, D.; Stuehr, D.J.; Lamattina, L.; Melkonian, M.; Tian, Z.; Zhu, Y.; Carpenter, E.J.; Wong, G.K.-S.; Wendehenne, D. Occurrence, structure, and evolution of nitric oxide synthase–like proteins in the plant kingdom. Sci. Signal. 2016, 9, re2.
- Foresi, N.; Correa-Aragunde, N.; Parisi, G.; Calo, G.; Salerno, G.; Lamattina, L. Characterization of a nitric oxide synthase from the plant kingdom: NO generation from the green alga Ostreococcus tauri is light irradiance and growth phase dependent. Plant Cell 2010, 22, 3816–3830.
- Foresi, N.; Correa-Aragunde, N.; Lamattina, L. Synthesis, actions, and perspectives of nitric oxide in photosynthetic organisms. In Nitric Oxide; Elsevier: Amsterdam, The Netherlands, 2017; pp. 125–136.
- Yamasaki, H.; Sakihama, Y.; Takahashi, S. An alternative pathway for nitric oxide production in plants: New features of an old enzyme. Trends Plant Sci. 1999, 4, 128–129.
- Del Castello, F.; Nejamkin, A.; Cassia, R.; Correa-Aragunde, N.; Fernández, B.; Foresi, N.; Lombardo, C.; Ramirez, L.; Lamattina, L. The era of nitric oxide in plant biology: Twenty years tying up loose ends. Nitric Oxide 2019, 85, 17–27.
- Baldet, P.; Devaux, C.; Chevalier, C.; Brouquisse, R.; Just, D.; Raymond, P. Contrasted responses to carbohydrate limitation in tomato fruit at two stages of development. Plant Cell Environ. 2002, 25, 1639–1649.
- Farnese, F.S.; Menezes-Silva, P.E.; Gusman, G.S.; Oliveira, J.A. When bad guys become good ones: The key role of reactive oxygen species and nitric oxide in the plant responses to abiotic stress. Front. Plant Sci. 2016, 7, 471.
- Hasanuzzaman, M.; Nahar, K.; Rahman, A.; Inafuku, M.; Oku, H.; Fujita, M. Exogenous nitric oxide donor and arginine provide protection against short-term drought stress in wheat seedlings. Physiol. Mol. Biol. Plants 2018, 24, 993–1004.
- Bright, J.; Desikan, R.; Hancock, J.T.; Weir, I.S.; Neill, S.J. ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. Plant J. 2006, 45, 113–122.
- González, A.; Cabrera, M.d.L.Á.; Henríquez, M.J.; Contreras, R.A.; Morales, B.; Moenne, A. Cross talk among calcium, hydrogen peroxide, and nitric oxide and activation of gene expression involving calmodulins and calcium-dependent protein kinases in Ulva compressa exposed to copper excess. Plant Physiol. 2012, 158, 1451–1462.
- Zhang, A.; Jiang, M.; Zhang, J.; Ding, H.; Xu, S.; Hu, X.; Tan, M. Nitric oxide induced by hydrogen peroxide mediates abscisic acid-induced activation of the mitogen-activated protein kinase cascade involved in antioxidant defense in maize leaves. New Phytol. 2007, 175, 36–50.
- Qiao, W.; Li, C.; Fan, L.-M. Cross-talk between nitric oxide and hydrogen peroxide in plant responses to abiotic stresses. Environ. Exp. Bot. 2014, 100, 84–93.
- Palma, J.M.; Gupta, D.K.; Corpas, F.J. Hydrogen peroxide and nitric oxide generation in plant cells: Overview and queries. Nitric. Oxide Hydrog. Peroxide Signal. High. Plants 2019, 2019, 1–16.
- Scheler, C.; Durner, J.; Astier, J. Nitric oxide and reactive oxygen species in plant biotic interactions. Curr. Opin. Plant Biol. 2013, 16, 534–539.
- Khan, M.; Ali, S.; Manghwar, H.; Saqib, S.; Ullah, F.; Ayaz, A.; Zaman, W. Melatonin function and crosstalk with other phytohormones under normal and stressful conditions. Genes 2022, 13, 1699.
- Imran, M.; Aaqil Khan, M.; Shahzad, R.; Bilal, S.; Khan, M.; Yun, B.-W.; Khan, A.L.; Lee, I.-J. Melatonin ameliorates thermotolerance in soybean seedling through balancing redox homeostasis and modulating antioxidant defense, phytohormones and polyamines biosynthesis. Molecules 2021, 26, 5116.
- He, H.; He, L.F. Crosstalk between melatonin and nitric oxide in plant development and stress responses. Physiol. Plant. 2020, 170, 218–226.
- Aghdam, M.S.; Luo, Z.; Jannatizadeh, A.; Sheikh-Assadi, M.; Sharafi, Y.; Farmani, B.; Fard, J.R.; Razavi, F. Employing exogenous melatonin applying confers chilling tolerance in tomato fruits by upregulating ZAT2/6/12 giving rise to promoting endogenous polyamines, proline, and nitric oxide accumulation by triggering arginine pathway activity. Food Chem. 2019, 275, 549–556.
- Liu, J.; Yang, J.; Zhang, H.; Cong, L.; Zhai, R.; Yang, C.; Wang, Z.; Ma, F.; Xu, L. Melatonin inhibits ethylene synthesis via nitric oxide regulation to delay postharvest senescence in pears. J. Agric. Food Chem. 2019, 67, 2279–2288.
- Martínez-Lorente, S.E.; Pardo-Hernández, M.; Martí-Guillén, J.M.; López-Delacalle, M.; Rivero, R.M. Interaction between melatonin and no: Action mechanisms, main targets, and putative roles of the emerging molecule nomela. Int. J. Mol. Sci. 2022, 23, 6646.
- Debnath, B.; Islam, W.; Li, M.; Sun, Y.; Lu, X.; Mitra, S.; Hussain, M.; Liu, S.; Qiu, D. Melatonin mediates enhancement of stress tolerance in plants. Int. J. Mol. Sci. 2019, 20, 1040.
- Begara-Morales, J.C.; Sánchez-Calvo, B.; Chaki, M.; Valderrama, R.; Mata-Pérez, C.; Padilla, M.N.; Corpas, F.J.; Barroso, J.B. Antioxidant systems are regulated by nitric oxide-mediated post-translational modifications (NO-PTMs). Front. Plant Sci. 2016, 7, 152.
- Ma, Y.J.; Li, X.P.; Wang, Y.; Wang, J.W. Nitric oxide donor sodium nitroprusside-induced transcriptional changes and hypocrellin biosynthesis of Shiraia sp. S9. Microb. Cell Factories 2021, 20, 92.
- Tewari, R.K.; Hahn, E.-J.; Paek, K.-Y. Function of nitric oxide and superoxide anion in the adventitious root development and antioxidant defence in Panax ginseng. Plant Cell Rep. 2008, 27, 563–573.
- Pande, A.; Mun, B.-G.; Lee, D.-S.; Khan, M.; Lee, G.-M.; Hussain, A.; Yun, B.-W. No network for plant–microbe communication underground: A review. Front. Plant Sci. 2021, 12, 658679.
- Shiraz, M.; Sami, F.; Siddiqui, H.; Yusuf, M.; Hayat, S. Interaction of auxin and nitric oxide improved photosynthetic efficiency and antioxidant system of Brassica juncea plants under salt stress. J. Plant Growth Regul. 2021, 40, 2379–2389.
- Guo, F.-Q.; Okamoto, M.; Crawford, N.M. Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 2003, 302, 100–103.
- Shen, Q.; Wang, Y.-T.; Tian, H.; Guo, F.-Q. Nitric oxide mediates cytokinin functions in cell proliferation and meristem maintenance in Arabidopsis. Mol. Plant 2013, 6, 1214–1225.
- Liu, W.-Z.; Kong, D.-D.; Gu, X.-X.; Gao, H.-B.; Wang, J.-Z.; Xia, M.; Gao, Q.; Tian, L.-L.; Xu, Z.-H.; Bao, F. Cytokinins can act as suppressors of nitric oxide in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 1548–1553.
- Khan, M.; Imran, Q.M.; Shahid, M.; Mun, B.-G.; Lee, S.-U.; Khan, M.A.; Hussain, A.; Lee, I.-J.; Yun, B.-W. Nitric oxide-induced AtAO3 differentially regulates plant defense and drought tolerance in Arabidopsis thaliana. BMC Plant Biol. 2019, 19, 602.
- Mioto, P.T.; Mercier, H. Abscisic acid and nitric oxide signaling in two different portions of detached leaves of Guzmania monostachia with CAM up-regulated by drought. J. Plant Physiol. 2013, 170, 996–1002.
- Lozano-Juste, J.; León, J. Nitric oxide regulates DELLA content and PIF expression to promote photomorphogenesis in Arabidopsis. Plant Physiol. 2011, 156, 1410–1423.
- Manjunatha, G.; Gupta, K.J.; Lokesh, V.; Mur, L.A.J.; Neelwarne, B. Nitric oxide counters ethylene effects on ripening fruits. Plant Signal. Behav. 2012, 7, 476–483.
- Pandey, S.; Kumari, A.; Shree, M.; Kumar, V.; Singh, P.; Bharadwaj, C.; Loake, G.J.; Parida, S.K.; Masakapalli, S.K.; Gupta, K.J. Nitric oxide accelerates germination via the regulation of respiration in chickpea. J. Exp. Bot. 2019, 70, 4539–4555.
- Shahid, M.; Imran, Q.M.; Hussain, A.; Khan, M.; Lee, S.U.; Mun, B.G.; Yun, B.-W. Comprehensive analyses of nitric oxide-induced plant stem cell-related genes in Arabidopsis thaliana. Genes 2019, 10, 190.
- Nabi, R.B.S.; Rolly, N.K.; Tayade, R.; Khan, M.; Shahid, M.; Yun, B.-W. Enhanced Resistance of atbzip62 against Pseudomonas syringae pv. tomato Suggests Negative Regulation of Plant Basal Defense and Systemic Acquired Resistance by AtbZIP62 Transcription Factor. Int. J. Mol. Sci. 2021, 22, 11541.
- Adamu, T.A.; Mun, B.-G.; Lee, S.-U.; Hussain, A.; Yun, B.-W. Exogenously applied nitric oxide enhances salt tolerance in rice (Oryza sativa L.) at seedling stage. Agronomy 2018, 8, 276.
- Lee, I.J. Nitric Oxide Modulates Glycine Max L. Growth and Physio-Molecular Responses during Flooding Stress; Austin Publishing Group: Middlesex County, NJ, USA, 2022.
- Santolini, J.; André, F.; Jeandroz, S.; Wendehenne, D. Nitric oxide synthase in plants: Where do we stand? Nitric Oxide 2017, 63, 30–38.
- Meng, Y.; Jing, H.; Huang, J.; Shen, R.; Zhu, X. The Role of Nitric Oxide Signaling in Plant Responses to Cadmium Stress. Int. J. Mol. Sci. 2022, 23, 6901.
- Garcia-Cardena, G.; Oh, P.; Liu, J.; Schnitzer, J.E.; Sessa, W.C. Targeting of nitric oxide synthase to endothelial cell caveolae via palmitoylation: Implications for nitric oxide signaling. Proc. Natl. Acad. Sci. USA 1996, 93, 6448–6453.
- Shi, H.-T.; Li, R.-J.; Cai, W.; Liu, W.; Wang, C.-L.; Lu, Y.-T. Increasing nitric oxide content in Arabidopsis thaliana by expressing rat neuronal nitric oxide synthase resulted in enhanced stress tolerance. Plant Cell Physiol. 2012, 53, 344–357.
- Butt, Y.; Lum, J.; Lo, S. Proteomic identification of plant proteins probed by mammalian nitric oxide synthase antibodies. Planta 2003, 216, 762–771.
- Moreau, M.; Lee, G.I.; Wang, Y.; Crane, B.R.; Klessig, D.F. AtNOS/AtNOA1 is a functional Arabidopsis thaliana cGTPase and not a nitric-oxide synthase. J. Biol. Chem. 2008, 283, 32957–32967.
- Zemojtel, T.; Fröhlich, A.; Palmieri, M.C.; Kolanczyk, M.; Mikula, I.; Wyrwicz, L.S.; Wanker, E.E.; Mundlos, S.; Vingron, M.; Martasek, P. Plant nitric oxide synthase: A never-ending story? Trends Plant Sci. 2006, 11, 524–525.
- Xu, Y.C.; Zhao, B.L. The main origin of endogenous NO in higher non-leguminous plants. Plant Physiol. Biochem. 2003, 41, 833–838.
- Rockel, P.; Strube, F.; Rockel, A.; Wildt, J.; Kaiser, W.M. Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J. Exp. Bot. 2002, 53, 103–110.
- Wilkinson, J.Q.; Crawford, N.M. Identification and characterization of a chlorate-resistant mutant of Arabidopsis thaliana with mutations in both nitrate reductase structural genes NIA1 and NIA2. Mol. Gen. Genet. MGG 1993, 239, 289–297.
- de la Haba, P.; Agüera, E.; Benítez, L.; Maldonado, J.M. Modulation of nitrate reductase activity in cucumber (Cucumis sativus) roots. Plant Sci. 2001, 161, 231–237.
- Bellin, D.; Asai, S.; Delledonne, M.; Yoshioka, H. Nitric oxide as a mediator for defense responses. Mol. Plant-Microbe. Interact. 2013, 26, 271–277.
- StoÈhr, C.; Ullrich, W.R. Generation and possible roles of NO in plant roots and their apoplastic space. J. Exp. Bot. 2002, 53, 2293–2303.
- Morot-Gaudry-Talarmain, Y.; Rockel, P.; Moureaux, T.; Quillere, I.; Leydecker, M.; Kaiser, W.; Morot-Gaudry, J. Nitrite accumulation and nitric oxide emission in relation to cellular signaling in nitrite reductase antisense tobacco. Planta 2002, 215, 708–715.
- Chamizo-Ampudia, A.; Sanz-Luque, E.; Llamas, Á.; Ocaña-Calahorro, F.; Mariscal, V.; Carreras, A.; Barroso, J.B.; Galván, A.; Fernández, E. A dual system formed by the ARC and NR molybdoenzymes mediates nitrite-dependent NO production in Chlamydomonas. Plant Cell Environ. 2016, 39, 2097–2107.
- Chamizo-Ampudia, A.; Sanz-Luque, E.; Llamas, A.; Galvan, A.; Fernandez, E. Nitrate reductase regulates plant nitric oxide homeostasis. Trends Plant Sci. 2017, 22, 163–174.
- Huang, J.; Sommers, E.M.; Kim-Shapiro, D.B.; King, S.B. Horseradish peroxidase catalyzed nitric oxide formation from hydroxyurea. J. Am. Chem. Soc. 2002, 124, 3473–3480.
- Harrison, R. Structure and function of xanthine oxidoreductase: Where are we now? Free Radic. Biol. Med. 2002, 33, 774–797.
- Mansuy, D.; Boucher, J.-L. Oxidation of N-hydroxyguanidines by cytochromes P450 and NO-synthases and formation of nitric oxide. Drug Metab. Rev. 2002, 34, 593–606.
- Groß, F.; Rudolf, E.-E.; Thiele, B.; Durner, J.; Astier, J. Copper amine oxidase 8 regulates arginine-dependent nitric oxide production in Arabidopsis thaliana. J. Exp. Bot. 2017, 68, 2149–2162.
- Maia, L.B.; Moura, J.J.G. Nitrite reduction by molybdoenzymes: A new class of nitric oxide-forming nitrite reductases. JBIC J. Biol. Inorg. Chem. 2015, 20, 403–433.
- Bethke, P.C.; Badger, M.R.; Jones, R.L. Apoplastic synthesis of nitric oxide by plant tissues. Plant Cell 2004, 16, 332–341.
- Modolo, L.V.; Augusto, O.; Almeida, I.M.G.; Magalhaes, J.R.; Salgado, I. Nitrite as the major source of nitric oxide production by Arabidopsis thaliana in response to Pseudomonas syringae. FEBS Lett. 2005, 579, 3814–3820.
- Bethke, P.C.; Libourel, I.G.L.; Aoyama, N.; Chung, Y.-Y.; Still, D.W.; Jones, R.L. The Arabidopsis aleurone layer responds to nitric oxide, gibberellin, and abscisic acid and is sufficient and necessary for seed dormancy. Plant Physiol. 2007, 143, 1173–1188.
More
Information
Subjects:
Plant Sciences
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.1K
Revisions:
4 times
(View History)
Update Date:
16 Mar 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




