
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Vladimir Simirskii | + 1101 word(s) | 1101 | 2020-03-05 04:13:38 | | | |
| 2 | Nicole Yin | + 3 word(s) | 1104 | 2020-10-27 06:53:43 | | | | |
| 3 | Conner Chen | Meta information modification | 1104 | 2021-08-04 11:15:35 | | | | |
| 4 | Conner Chen | Meta information modification | 1104 | 2021-09-22 02:47:28 | | |
Video Upload Options
The retinal arhitecture is similar across all vertebrates despite the morphological and functional peculiarities. The retina is composed of two main parts: the single-layred retinal pigment epithelium and the multilayred neuroretina which includes a number of neuronal and glial cell types. Homeobox genes from different classes are accepted as critical for eye field specification and retinal cells type differentiation by a broad array of loss- or gain-of-function models. Among these genes are some that are known to cause inherited retinal diseases (IRDs) that disturb the formation, function, and survival of rod and cone photoreceptors, ganglion cells, or retinal pigment epithelial cells [4,6–8]. The advances in the field of genetics and high-throughput next-generation sequencing and cell technologies allow for deepening of knowledge of the genetic basis of inherited retinal diseases (IRDs), as well as improve their diagnostics and therapy.
1. Introduction
The general plan of the retinal architecture is similar across all vertebrates and humans, despite the morphological and functional peculiarities[1]. The retina is composed of two parts: The single-layered retinal pigment epithelium (RPE) on the posterior side and the neuroretina on the anterior side of the eye.
2. In the Mirror of Homeobox Genes
The neuroretina is a highly organized multilayered tissue that includes interconnecting layers of specialized cells: Six main types of neurons (photoreceptors, bipolar cells, horizontal cells, amacrine cells, ganglion cells, and interplexiform neurons) and four types of radial glia cells (Muller cells, astrocytes, microglia, and oligodendrocytes) (Figure 1).
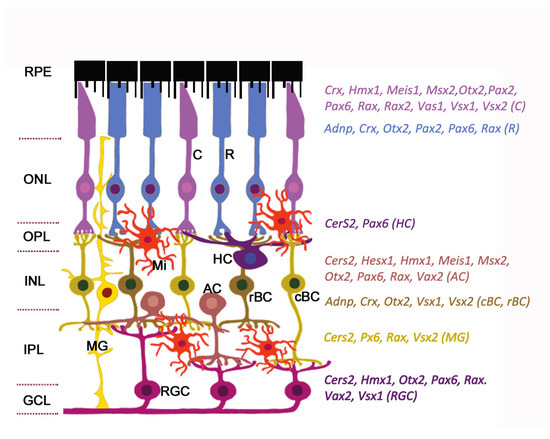
Figure 1. Expression of homeobox genes in the adult mouse retina. The retinal architecture is detailed in the review. The cell-specific expression of genes is detected by single-cell RNA sequencing (data from https://eyeintegration.nei.nih.gov). The only genes which are known to associate with eye/retinal malformations in humans are shown. On the left: Retinal layers; on the right: homeobox genes indicated in the same color as the cell types expressed them. Abbreviations in parentheses show corresponding cell types. RPE, retinal pigment epithelium; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer; C, cone; R, rod; HC, horizontal cell; AC, amacrine cell; cBC, cone bipolar cell; rBC, rod bipolar cell; MG, Muller glia; RGC, retinal ganglion cell; Mi, microglial cell. Modified from[2]. License to reproduce: https://creativecommons.org/licenses/by/4.0/.
Radial glia of the retina line the bottom and lateral surface of the optic cup, forming the radial layers[3][4]. Three nuclear layers, consisting of different types of sensory neurons, and two plexiform layers (outer and inner) representing synaptic connections between retinal neurons of the border nuclear layer, are distinguished in the retina. The outer nuclear layer (ONL) of the retina includes light-sensitive cells (rods and cones of photoreceptors). The outer segments of photoreceptors are in close interaction with the RPE (single row layer of intensely pigmented epithelial cells). RPE cells are located between the photoreceptors and the choroid and perform a number of physiological functions: Protection of photoreceptors from excess light, transduction of visual signal, retinal homeostasis (growth factor secretion, regulation of ion balance in subretinal space), and phagocytosis of exfoliated discs of outer segments of photoreceptors[5][6]. The RPE is underlain by Bruch’s membrane which consists of the components of the choroid endothelium and the fibrillar layer of the RPE basal plate[7]. The second (inner) nuclear retinal layer (INL) is formed by interneurons: bipolars and amacrine and horizontal cells. Bipolars participate in transmission of visual signal from the photoreceptors into the ganglion, and horizontal and amacrine cells connect all cells of the retina[4]. Ganglion cells are the neurons of the second order, forming the third ganglion nuclear layer. Their axons take part in the formation of the optic nerve[8]. The outer plexiform layer (OPL) is formed by synaptic contacts between rods/cones and bipolar cells, while the inner plexiform layer provides a connection between bipolar and ganglion neurons, as well as a horizontal connection between amacrine and horizontal neurons[9][10]. The INL is where the bodies of Muller glia cells are localized. These retinal macroglia permeates the entire retina from external to internal border basal membranes (formed by outgrowths of these cells) localized on the border of plexiform tissue layers. Muller glia performs structural and neurotrophic functions in the retina[11]. Functions of the other elements of retinal glia, such as astrocytes, are associated with maintaining the structure and metabolic activity of retinal neurons. Astrocytes can secrete vasoactive substances that regulate retinal vascular tone[12]. Retinal microglia present as a resident population of immunocompetent cells[13]. The retina blood supply comes from the central retinal artery feeding the inner retinal layers and choriocapillaries for the outer layers: photoreceptor layer, ONL, and OPL[14]. Retinal neurons, macroglia, and microglia, as well as the wall cells of the microvessels (endotheliocytes and pericytes) interact with each other and form a blood–retinal barrier that regulates the supply of oxygen and trophic factors to retinal neurons and is involved in recycling of metabolic products[15][16][17].
Retinal development in vertebrate embryogenesis is under strict spatial and temporal regulation by the overlapping gene networks[18][19]. Homeobox transcription factors, an evolutionarily conserved class among the transcription factors, are key regulators of developmental processes such as regional specification, cells migration, and differentiation and morphogenesis of the tissues and organs, by regulating the expression of specific sets of target genes. The developing retina is marked by distinct boundaries of homeobox gene expression at different developmental time points. During retinal development the homeobox genes play multiple roles such as regulation of patterning of the retina along the dorsoventral and nasotemporal axes. These genes are essential for the control of proliferation and the choice of cells fate, the order of differentiation of specific neuronal and glial subtypes through instruction signals from surrounding tissues, and retinal cells survival[20][21][22]. Homeobox-containing genes continue to be expressed in adult eye tissues, the retina in particular, ensuring eye homeostasis and supporting axon function[23][24].
Constantly enriching the data on the human retina genetic map will help us to understand the molecular profile of individual retinal cell types that enable cells to keep functioning and contribute to healthy vision, and will help us to study how those genes impact different kinds of cells. The molecular atlas of healthy cells with cells from retinal diseases and across different stages of human development will help identify the factors and signals that cause cells to stop functioning and lead to vision loss and blindness[25].
It is important to take into account species barriers and tissue-specific features of regulating the expression of regulatory transcription factors, among which are the homeobox genes, in attempts to transmit data from experimental animal models to the human eye[26].
References
- Kolb, H. Simple Anatomy of the Retina. In Webvision: The Organization of the Retina and Visual System [Internet]; Kolb, H., Fernandez, E., Nelson, R., Eds.; University of Utah Health Sciences Center: Salt Lake City, UT, USA, 2005.
- Fenge Li; Danye Jiang; Melanie A. Samuel; Microglia in the developing retina. Neural Development 2019, 14, 1-13, 10.1186/s13064-019-0137-x.
- Hoon, M.; Okawa, H.; Della Santina, L.; Wong, R.O. Functional architecture of the retina: Development and disease. Prog. Retin. Eye Res. 2014, 42, 44–84.
- Tsukamoto, Y. Morphological Survey from Neurons to Circuits of the Mouse Retina. Methods Mol. Biol. 2018, 1753, 3–25.
- Strauss, O. The retinal pigment epithelium in visual function. Physiol. Rev. 2005, 85, 845–881.
- Mazzolini, M.; Facchetti, G.; Andolfi, L.; Proietti Zaccaria, R.; Tuccio, S.; Treu, J.; Altafini, C.; Di Fabrizio, E.M.; Lazzarino, M.; Rapp, G.; et al. The phototransduction machinery in the rod outer segment has a strong efficacy gradient. Proc. Natl. Acad. Sci. USA 2015, 112, E2715–E2724.
- Benjamin Amram; Yamit Cohen-Tayar; Ahuvit David; Ruth Ashery-Padan; The retinal pigmented epithelium – from basic developmental biology research to translational approaches. The International Journal of Developmental Biology 2017, 61, 225-234, 10.1387/ijdb.160393ra.
- Joshua R. Sanes; Richard H. Masland; The Types of Retinal Ganglion Cells: Current Status and Implications for Neuronal Classification. Annual Review of Neuroscience 2015, 38, 221-246, 10.1146/annurev-neuro-071714-034120.
- Kolb, H. Outer Plexiform Layer. In Webvision: The Organization of the Retina and Visual System [Internet]; Kolb, H., Fernandez, E., Nelson, R., Eds.; University of Utah Health Scisence. Center: Salt Lake City, UT, USA, 2005.
- Kolb, H. Inner Plexiform Layer. In Webvision: The Organization of the Retina and Visual System [Internet]; Kolb, H., Fernandez, E., Nelson, R., Eds.; University of Utah Health Sciences Center: Salt Lake City, UT, USA, 2001.
- Paula V. Subirada; María C. Paz; Magali E. Ridano; Valeria E. Lorenc; María V. Vaglienti; Pablo F. Barcelona; José D. Luna; María C. Sánchez; A journey into the retina: Müller glia commanding survival and death. European Journal of Neuroscience 2018, 47, 1429-1443, 10.1111/ejn.13965.
- Harold K. Kimelberg; Functions of Mature Mammalian Astrocytes: A Current View. The Neuroscientist 2010, 16, 79-106, 10.1177/1073858409342593.
- Sem Genini; William A. Beltran; Veronika M. Stein; Gustavo D. Aguirre; Isolation and ex vivo characterization of the immunophenotype and function of microglia/macrophage populations in normal dog retina.. Single Molecule and Single Cell Sequencing 2014, 801, 339-345, 10.1007/978-1-4614-3209-8_43.
- Senthil Selvam; Tejas Kumar; Marcus Fruttiger; Retinal vasculature development in health and disease. Progress in Retinal and Eye Research 2018, 63, 1-19, 10.1016/j.preteyeres.2017.11.001.
- Cunha-Vaz, J.; Bernardes, R.; Lobo, C. Blood-retinal barrier. Eur. J. Ophthalmol. 2011, 21 (Suppl. 6), S3–S9.
- Díaz-Coránguez, M.; Ramos, C.; Antonetti, D.A. The inner blood-retinal barrier: Cellular basis and development. Vision Res. 2017, 139, 123–137.
- Trost, A.; Bruckner, D.; Rivera, F.J.; Reitsamer, H.A. Pericytes in the Retina. Adv. Exp. Med. Biol. 2019, 1122, 1–26.
- Sabine Fuhrmann; Eye Morphogenesis and Patterning of the Optic Vesicle. Current Topics in Developmental Biology 2010, 93, 61-84, 10.1016/b978-0-12-385044-7.00003-5.
- Cheryl Y. Gregory-Evans; Valerie A. Wallace; Kevin Gregory-Evans; Gene networks: Dissecting pathways in retinal development and disease. Progress in Retinal and Eye Research 2013, 33, 40-66, 10.1016/j.preteyeres.2012.10.003.
- Dyer, M.A.; Livesey, F.J.; Cepko, C.L.; Oliver, G. Prox1 function controls progenitor cell proliferation and horizontal cell genesis in the mammalian retina. Nat. Genet. 2003, 34, 53–58.
- Wigle, J.T.; Eisenstat, D.D. Homeobox genes in vertebrate forebrain development and disease. Clin. Genet. 2008, 73, 212–226.
- Giudetti, G.; Giannaccini, M.; Biasci, D.; Mariotti, S.; Degl’innocenti, A.; Perrotta, M.; Barsacchi, G.; Andreazzoli, M. Characterization of the Rx1-dependent transcriptome during early retinal development. Dev. Dyn. 2014, 243, 1352–1361.
- J.L. Zagozewski; Q. Zhang; D.D. Eisenstat; Genetic regulation of vertebrate eye development. Clinical Genetics 2014, 86, 453-460, 10.1111/cge.12493.
- Amanda Miles; Vincent Tropepe; Coordinating progenitor cell cycle exit and differentiation in the developing vertebrate retina.. Neurogenesis 2016, 3, e1161697, 10.1080/23262133.2016.1161697.
- Samuel W Lukowski; Camden Y Lo; Alexei A Sharov; Quan Nguyen; Lyujie Fang; Sandy Sc Hung; Ling Zhu; Ting Zhang; Ulrike Grünert; Tu Nguyen; et al.Anne SenabouthJafar S JabbariEmily WelbyJane C SowdenHayley S WaughAdrienne MackeyGraeme PollockTrevor D LambPeng‐Yuan WangAlex W HewittMark C GilliesJoseph E PowellRaymond Cb Wong A single‐cell transcriptome atlas of the adult human retina. The EMBO Journal 2019, 38, e100811, 10.15252/embj.2018100811.
- Pilar Cacheiro; the International Mouse Phenotyping Consortium and the Monarch Initiative; Melissa A. Haendel; Damian Smedley; New models for human disease from the International Mouse Phenotyping Consortium. Mammalian Genome 2019, 30, 143-150, 10.1007/s00335-019-09804-5.




