
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Delin Xu | -- | 1576 | 2023-02-24 06:47:53 | | | |
| 2 | Catherine Yang | Meta information modification | 1576 | 2023-02-24 07:04:39 | | |
Video Upload Options
Endophytes, which are widely found in host plants and have no harmful effects, are a vital biological resource. Plant endophytes promote plant growth and enhance plants’ resistance to diseases, pests, and environmental stresses. In addition, they enhance the synthesis of important secondary metabolites in plants and improve the potential applicability of plants in agriculture, medicine, food, and horticulture. Secondary metabolites (SMs) are the products formed by interactions with the environment during plant growth and development. SMs mainly include alkaloids, flavonoids, terpenoids, peptides, phenols, sterols, and additional minor molecular organic compounds. As essential substances used by plants for self-protection to cope with their environment, SMs have various physiological functions, such as regulating plant growth and biological defense, and they are also involved in the plant response abiotic stresses, such as drought, low temperature, salinity, and metals
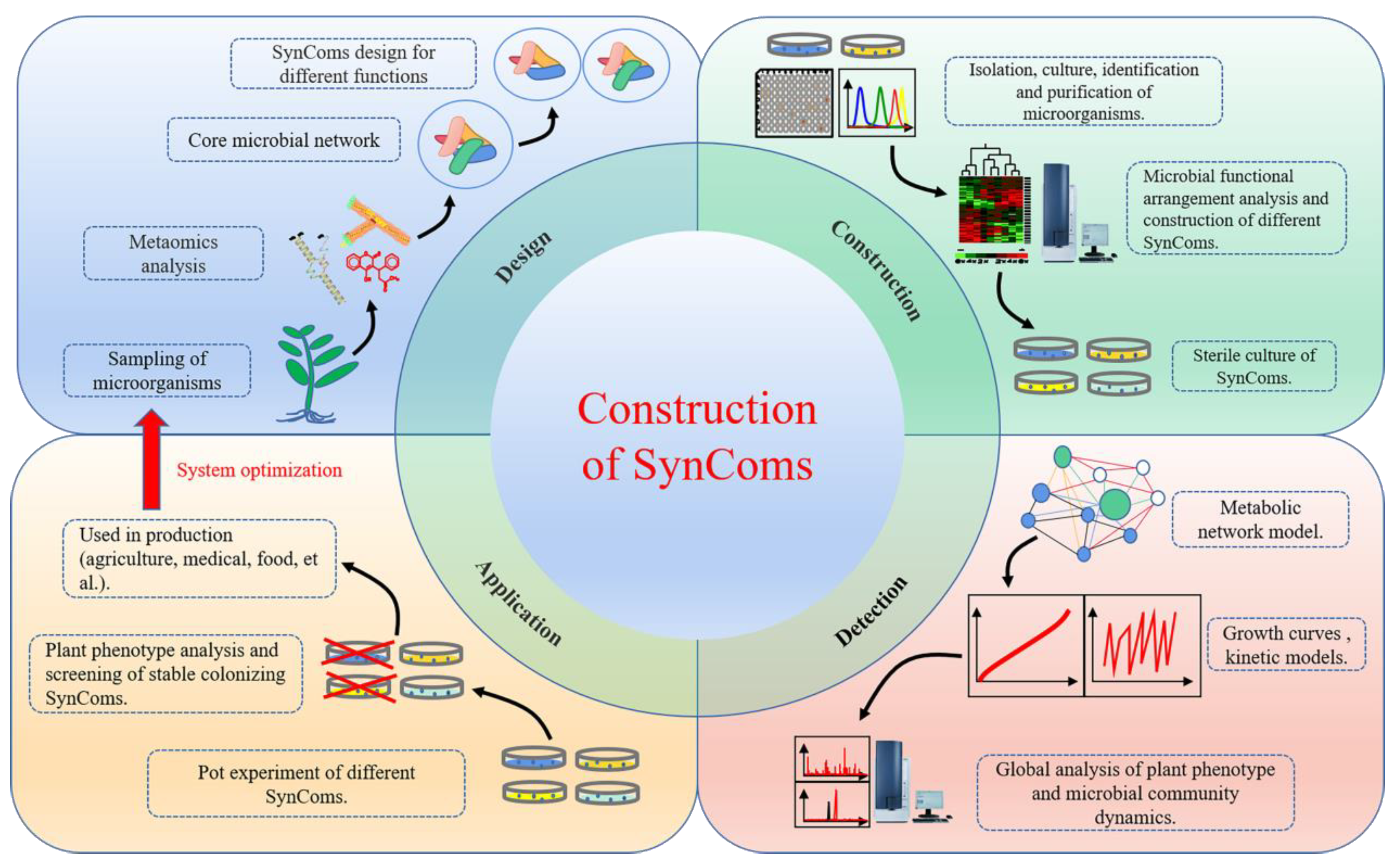
1. Construction of the SynComs Model for Endophyte–Plant Interactions
2. Application of Omics to Elucidate the Mechanisms by Which Endophytes Promote the Occurrence and Accumulation of Plant Secondary Metabolites
3. Endophytes Advance the Application of Plant SMs
References
- Karkaria, B.D.; Fedorec, A.J.H.; Barnes, C.P. Automated design of synthetic microbial communities. Nat. Commun. 2021, 12, 672.
- Shou, W.; Ram, S.; Vilar, J.M. Synthetic cooperation in engineered yeast populations. Proc. Natl. Acad. Sci. USA 2007, 104, 1877–1882.
- Balagaddé, F.K.; Song, H.; Ozaki, J.; Collins, C.H.; Barnet, M.; Arnold, F.H.; Quake, S.R.; You, L. A synthetic Escherichia coli predator-prey ecosystem. Mol. Syst. Biol. 2008, 4, 187.
- Kim, H.J.; Boedicker, J.Q.; Choi, J.; Ismagilov, R.F. Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc. Natl. Acad. Sci. USA 2008, 105, 18188–18193.
- Klitgord, N.; Segrè, D. Environments that induce synthetic microbial ecosystems. PLoS Comput. Biol. 2010, 6, e1001002.
- Faith, J.J.; McNulty, N.P.; Rey, F.E.; Gordon, J.I. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science 2011, 333, 101–104.
- Grosskopf, T.; Soyer, O.S. Synthetic microbial communities. Curr. Opin. Microbiol. 2014, 18, 72–77.
- Bodenhausen, N.; Bortfeld-Miller, M.; Ackermann, M.; Vorholt, J.A. A synthetic community approach reveals plant genotypes affecting the phyllosphere microbiota. PLoS Genet. 2014, 10, e1004283.
- Zhang, J.; Liu, Y.X.; Zhang, N.; Hu, B.; Jin, T.; Xu, H.; Qin, Y.; Yan, P.; Zhang, X.; Guo, X.; et al. NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat. Biotechnol. 2019, 37, 676–684.
- Niu, B.; Paulson, J.N.; Zheng, X.; Kolter, R. Simplified and representative bacterial community of maize roots. Proc. Natl. Acad. Sci. USA 2017, 114, E2450–E2459.
- Widder, S.; Allen, R.J.; Pfeiffer, T.; Curtis, T.P.; Wiuf, C.; Sloan, W.T.; Cordero, O.X.; Brown, S.P.; Momeni, B.; Shou, W.; et al. Challenges in microbial ecology: Building predictive understanding of community function and dynamics. ISME J. 2016, 10, 2557–2568.
- Wang, S.; Yang, C.; Tu, H.; Zhou, J.; Liu, X.; Cheng, Y.; Luo, J.; Deng, X.; Zhang, H.; Xu, J. Characterization and Metabolic Diversity of Flavonoids in Citrus Species. Sci. Rep. 2017, 7, 10549.
- Danko, D.; Bezdan, D.; Afshin, E.E.; Ahsanuddin, S.; Bhattacharya, C.; Butler, D.J.; Chng, K.R.; Donnellan, D.; Hecht, J.; Jackson, K.; et al. A global metagenomic map of urban microbiomes and antimicrobial resistance. Cell 2021, 184, 3376–3393.e17.
- Roesch, L.F.; Fulthorpe, R.R.; Riva, A.; Casella, G.; Hadwin, A.K.; Kent, A.D.; Daroub, S.H.; Camargo, F.A.; Farmerie, W.G.; Triplett, E.W. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007, 1, 283–290.
- Huber, J.A.; Mark Welch, D.B.; Morrison, H.G.; Huse, S.M.; Neal, P.R.; Butterfield, D.A.; Sogin, M.L. Microbial population structures in the deep marine biosphere. Science 2007, 318, 97–100.
- Wheeler, D.A.; Srinivasan, M.; Egholm, M.; Shen, Y.; Chen, L.; McGuire, A.; He, W.; Chen, Y.J.; Makhijani, V.; Roth, G.T.; et al. The complete genome of an individual by massively parallel DNA sequencing. Nature 2008, 452, 872–876.
- Merritt, C.R.; Ong, G.T.; Church, S.E.; Barker, K.; Danaher, P.; Geiss, G.; Hoang, M.; Jung, J.; Liang, Y.; McKay-Fleisch, J. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat. Biotechnol. 2020, 38, 586–599.
- Eveland, A.L.; Satoh-Nagasawa, N.; Goldshmidt, A.; Meyer, S.; Beatty, M.; Sakai, H.; Ware, D.; Jackson, D. Digital gene expression signatures for maize development. Plant Physiol. 2010, 154, 1024–1039.
- Liu, Q.; Ding, C.; Lang, X.; Guo, G.; Chen, J.; Su, X. Small noncoding RNA discovery and profiling with sRNAtools based on high-throughput sequencing. Brief. Bioinform. 2021, 22, 463–473.
- Lu, C.; Tej, S.S.; Luo, S.; Haudenschild, C.D.; Meyers, B.C.; Green, P.J. Elucidation of the small RNA component of the transcriptome. Science 2005, 309, 1567–1569.
- Jiang, M.; Jia, K.; Wang, L.; Li, W.; Chen, B.; Liu, Y.; Wang, H.; Zhao, S.; He, Y.; Zhou, C. Alterations of DNA damage response pathway: Biomarker and therapeutic strategy for cancer immunotherapy. Acta Pharm. Sin. B 2021, 11, 2983–2994.
- Wang, X.; Niu, Q.W.; Teng, C.; Li, C.; Mu, J.; Chua, N.H.; Zuo, J. Overexpression of PGA37/MYB118 and MYB115 promotes vegetative-to-embryonic transition in Arabidopsis. Cell Res. 2009, 19, 224–235.
- Battu, L.; Reddy, M.M.; Goud, B.S.; Ulaganathan, K.; Kandasamy, U. Genome inside genome: NGS based identification and assembly of endophytic Sphingopyxis granuli and Pseudomonas aeruginosa genomes from rice genomic reads. Genomics 2017, 109, 141–146.
- Cokus, S.J.; Feng, S.; Zhang, X.; Chen, Z.; Merriman, B.; Haudenschild, C.D.; Pradhan, S.; Nelson, S.F.; Pellegrini, M.; Jacobsen, S.E. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 2008, 452, 215–219.
- Li, L.; Liu, C.; Wen, W.; Li, Q.; Pan, T.; Li, Z.; Qian, G.; He, Y.; Xu, D. Dendrobine biosynthesis in Dendrobium nobile in four different habitats is affected by the variations in the endophytic fungal community. Front. Microbiol. 2022, 13, 981070.
- Zhang, C.; He, J.; Dai, H.; Wang, G.; Zhang, X.; Wang, C.; Shi, J.; Chen, X.; Wang, D.; Wang, E. Discriminating symbiosis and immunity signals by receptor competition in rice. Proc. Natl. Acad. Sci. USA 2021, 118, e2023738118.
- Franzosa, E.A.; Hsu, T.; Sirota-Madi, A.; Shafquat, A.; Abu-Ali, G.; Morgan, X.C.; Huttenhower, C. Sequencing and beyond: Integrating molecular ‘omics’ for microbial community profiling. Nat. Rev. Microbiol. 2015, 13, 360–372.
- Hou, S.; Thiergart, T.; Vannier, N.; Mesny, F.; Ziegler, J.; Pickel, B.; Hacquard, S. A microbiota-root-shoot circuit favours Arabidopsis growth over defence under suboptimal light. Nat. Plants 2021, 7, 1078–1092.
- Dai, X.; Zhu, Y.; Luo, Y.; Song, L.; Liu, D.; Liu, L.; Chen, F.; Wang, M.; Li, J.; Zeng, X.; et al. Metagenomic insights into the fibrolytic microbiome in yak rumen. PLoS ONE 2012, 7, e40430.
- Chen, W.; Guillaume-Gentil, O.; Rainer, P.Y.; Gäbelein, C.G.; Saelens, W.; Gardeux, V.; Klaeger, A.; Dainese, R.; Zachara, M.; Zambelli, T.; et al. Live-seq enables temporal transcriptomic recording of single cells. Nature 2022, 608, 733–740.
- Prosser, J.I. Dispersing misconceptions and identifying opportunities for the use of ‘omics’ in soil microbial ecology. Nat. Rev. Microbiol. 2015, 13, 439–446.
- OuYang, X.L.; Liu, J.L.; Gong, G.L.; Shen, G.N.; Yuan, Y.; Gao, Y.M.; Yan, A.; Wang, W.D. Advances in Endophytes and Secondary Metabolites of Ginkgo biloba. J. Heilongjiang Bayi Agric. Univ. 2021, 33, 72–78.
- Torres-Mendoza, D.; Ortega, H.E.; Cubilla-Rios, L. Patents on Endophytic Fungi Related to Secondary Metabolites and Biotransformation Applications. J. Fungi. 2020, 6, 58.
- Singh, D.; Thapa, S.; Mahawar, H.; Kumar, D.; Geat, N.; Singh, S.K. Prospecting potential of endophytes for modulation of biosynthesis of therapeutic bioactive secondary metabolites and plant growth promotion of medicinal and aromatic plants. Antonie Leeuwenhoek 2022, 115, 699–730.
- Geng, X. Endophytic Fungi and Secondary Metabolites of Rehmannia Glutinosa Based on Traditional Chinese Medicine Fingerprints. Contrast Media Mol. Imaging 2022, 202, 7701198.
- Sankari, S.; Babu, V.M.P.; Bian, K.; Alhhazmi, A.; Andorfer, M.C.; Avalos, D.M.; Smith, T.A.; Yoon, K.; Drennan, C.L.; Yaffe, M.B.; et al. A haem-sequestering plant peptide promotes iron uptake in symbiotic bacteria. Nat. Microbiol. 2022, 7, 1453–1465.




