Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Wei Li | -- | 2952 | 2023-02-22 16:15:47 | | | |
| 2 | Sirius Huang | Meta information modification | 2952 | 2023-02-23 06:31:35 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Albadari, N.; Li, W. Survivin Small Molecules Inhibitors. Encyclopedia. Available online: https://encyclopedia.pub/entry/41551 (accessed on 12 January 2026).
Albadari N, Li W. Survivin Small Molecules Inhibitors. Encyclopedia. Available at: https://encyclopedia.pub/entry/41551. Accessed January 12, 2026.
Albadari, Najah, Wei Li. "Survivin Small Molecules Inhibitors" Encyclopedia, https://encyclopedia.pub/entry/41551 (accessed January 12, 2026).
Albadari, N., & Li, W. (2023, February 22). Survivin Small Molecules Inhibitors. In Encyclopedia. https://encyclopedia.pub/entry/41551
Albadari, Najah and Wei Li. "Survivin Small Molecules Inhibitors." Encyclopedia. Web. 22 February, 2023.
Copy Citation
Survivin, as a member of the inhibitor of apoptosis proteins (IAPs) family, acts as a suppressor of apoptosis and plays a central role in cell division. Survivin has been considered as an important cancer drug target because it is highly expressed in many types of human cancers, while it is effectively absent from terminally differentiated normal tissues. Moreover, survivin is involved in tumor cell resistance to chemotherapy and radiation.
survivin
apoptosis
mitosis
chemoresistance
survivin small molecules inhibitors
1. Introduction
Programmed cell death serves fundamental functions during mammalian tissue development. Apoptosis is a highly regulated and controlled process and represents one form of programmed cell death. Defective and insufficient apoptosis processes can result in uncontrolled cell proliferation and cancer. Apoptosis is initiated by activating either the intrinsic or the extrinsic pathway, and it is executed by caspases. The intrinsic pathway is activated by endogenous stress signals or irradiation, which depends on the release of Cytochrome c (Cyt-c) from the mitochondria. In contrast, the extrinsic pathway (also known as the death receptor pathway) is mitochondrion-independent and is activated by extracellular ligands binding to cell-surface death receptors [1]. Apoptosis can be blocked by endogenous proteins such as the inhibitors of apoptosis proteins (IAPs). IAP proteins were first identified in baculoviruses, where they could inhibit the host’s defensive apoptotic response to infected insect cells and enhance viral replication [2]. Subsequently, several cellular IAP homologs were found in diverse organisms, including vertebrates, insects, and yeasts [3][4]. The human IAPs are a family of eight structurally and functionally related proteins. All IAP family members have one to three copies of a baculovirus IAP repeat (BIR), a domain with ~70 amino acids, which is the main mediator of the antiapoptotic function. Survivin is the smallest member of the IAPs and has only one BIR domain [5]. Survivin is a multi-tasking protein that is both essential for mitosis and can inhibit apoptosis. It has gained significant attention as a potential therapeutic target for cancer partially because it is expressed only in most rapidly dividing cells, such as cancer cells, while its expression is very low in differentiated normal cells. In addition, survivin expression correlates positively with chemoresistance, radiation insensitivity, and poor patient prognosis. Moreover, survivin plays a role in promoting tumor cell survival and cancer metastasis.
2. Structure and Cellular Functions of Survivin
Human survivin is a small protein with a molecular weight of 16.5 kDa and contains 142 amino acid residues. Structurally, human survivin closely resembles the BIR-containing proteins from yeasts and C. elegans [3][6]. Human survivin has a signal BIR domain (aa 18–88) in the N-terminal, followed by a linker region (aa 89–102) and an extended α-helix (aa 98–142) in the C-terminal (Figure 1) [6]. The BIR domain is stabilized by a zinc finger created by four amino acids: Cys57, Cys60, His77, and Cys84 [6]. Survivin exists in the body as both a monomer and a homodimer. Survivin monomers form a homodimer through interactions located mainly in the linker region and residues 6–10 in the N-terminal region of the BIR domain [5][7]. Survivin homodimerization for function is not always needed, since both the survivin monomer and dimer are functional. While most IAP proteins are predominantly cytosolic, survivin has been found in the cytoplasm, nucleus, mitochondria [8], exosomes [9], outer surface of the cell membrane, and extracellular matrix. Survivin possesses a dual function in the body: protection from apoptosis and regulation of cell division [10]. Recent reports also suggested that survivin is involved in autophagy [11], angiogenesis [12], and stemness [13].
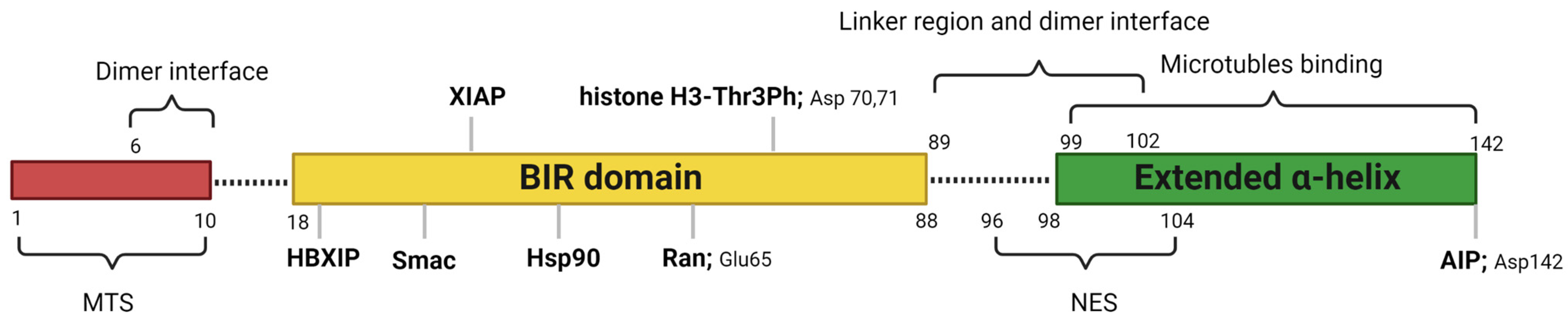
Figure 1. Critical features of survivin structure and its interaction sites with its key partners. Survivin has a single BIR domain (18–88 aa), a linker region (89–102 aa), and an extended α helix (99–142 aa) in its C-terminal. The first 10 amino acids in its N-terminus are proline-rich and represent the MTS for mitochondrial importation of survivin. The BAR domain is functionally significant for survivin interactions with Smac, Ran, XIAP, HBXIP, and histone 3 of the centromeric chromatin. The linker region mediates the dimerization along with residues 6–10 in the N-terminal region of the survivin. Borealin replaces one survivin monomer through the dimer interface to form a survivin–borealin heterodimer. The C-terminal of survivin, the N-terminal of borealin, and INCENP form the triple helix unit as part of CPC. The survivin helical region has tubulin- and AIP-binding sites. Survivin has an NES between the BIR domain and the C-terminal helix, which is masked by the homodimerization interface. Moreover, survivin has a non-classical bipartite NES in the C-terminal. This figure was created with BioRender.com.
2.1. Cytoplasmic and Mitochondrial Survivin
A predominant cytoplasmic and mitochondrial localization of survivin is essential for its anti-apoptotic activity [14]. Many reports showed its upregulation in the cytoplasmic expression in cancer cells [15]. Survivin is exported from the nucleus to the cytoplasm by chromosome region maintenance 1 (Crm1, also known as exportin-1) [16]. Survivin–Crm1 interaction is mediated by a centrally located leucine-rich nuclear export sequence (NES) that is placed between the BIR domain and the C-terminal helix of survivin and a non-classical bipartite NES in the C-terminus [17][18]. The central NES is primarily active in survivin monomers because it is partially masked by the homodimerization interface [19]. Survivin physically associates with the X-linked inhibitor of apoptosis protein (XIAP), another member of the IAPs, through its BIR domain, protects it from ubiquitination, and enhances its stability. A stabilized survivin–XIAP complex suppresses caspase-9 activity and blocks apoptosis in vivo (Figure 2) [20]. However, XIAP-associated factor 1 (XAF1), a nuclear protein that binds to XIAP and suppresses its anti-caspase activity, reverses the inhibition of the ubiquitination of XIAP by survivin and activates the XIAP E3 ligase to target and promote survivin degradation [21][22]. Survivin also augments the anti-apoptotic function of XIAP via another mechanism, where survivin binds to the second mitochondria-derived activator of caspases (Smac). Smac, also known as DIABLO, is a mitochondrial protein that is released into the cytosol following the increase in the mitochondrial outer membrane permeability (MOMP) during apoptosis. Smac antagonizes IAPs, including XIAP, cellular inhibitor of apoptosis protein 1 (CIAP-1), and cellular inhibitor of apoptosis protein 2 (CIAP-2), and promotes Cyt-c-dependent caspase activation [23]. Survivin sequesters Smac away from other IAPs and protects the cell from mitochondria-regulated apoptosis [24]. Survivin interacts with Smac via its BIR domain, thereby freeing XIAP and allowing it to block caspases without being antagonized [24]. The BIR domain of survivin is also necessary for binding with the hepatitis B virus X-interacting protein (HBXIP, also known as LAMTOR5). HBXIP operates as a cofactor for survivin and allows the HBXIP–survivin complex to bind pro-caspase-9 and preclude pro-caspase-9 recruitment to activated apoptotic protease activating factor-1 (Apaf-1) and, thus, suppress activation of caspase-9 [25]. In addition to caspase inhibition, survivin has been reported to inhibit caspase-independent apoptosis. Survivin blocks the release of apoptosis-inducing factor (AIF), the primary mediator of caspase-independent apoptosis, from the mitochondrial intermembrane space (IMS) and its nuclear translocation [26]. In response to different apoptosis stimuli, AIF translocates from IMS to the nucleus, causing DNA fragmentation and chromatin condensation [26][27]. Survivin importation into mitochondria is directed through the mitochondrial targeting sequence (MTS) (1–10 aa) located in its N-terminus [28]. In addition, survivin directly associates with aryl hydrocarbon receptor-interacting protein (AIP) [29]. This interaction is mediated by the survivin carboxyl terminus coiled coil and three tetratricopeptide motifs located in the carboxyl-terminal end of AIP. Aspartic acid 142 (Asp142), the last amino acid in survivin, plays a critical role in AIP recognition [29]. The survivin–AIP complex stabilizes survivin levels and enhances its anti-apoptosis function in the mitochondria [29]. Survivin also interacts with heat shock protein 90 (Hsp90), where Hsp90 preserves survivin stability in vivo [30]. Hsp90 association with survivin involves the ATPase domain of Hsp90 and the survivin BIR domain. Disruption of the survivin–Hsp90 interaction results in the proteasomal degradation of survivin and mitochondrial-dependent apoptosis [30].
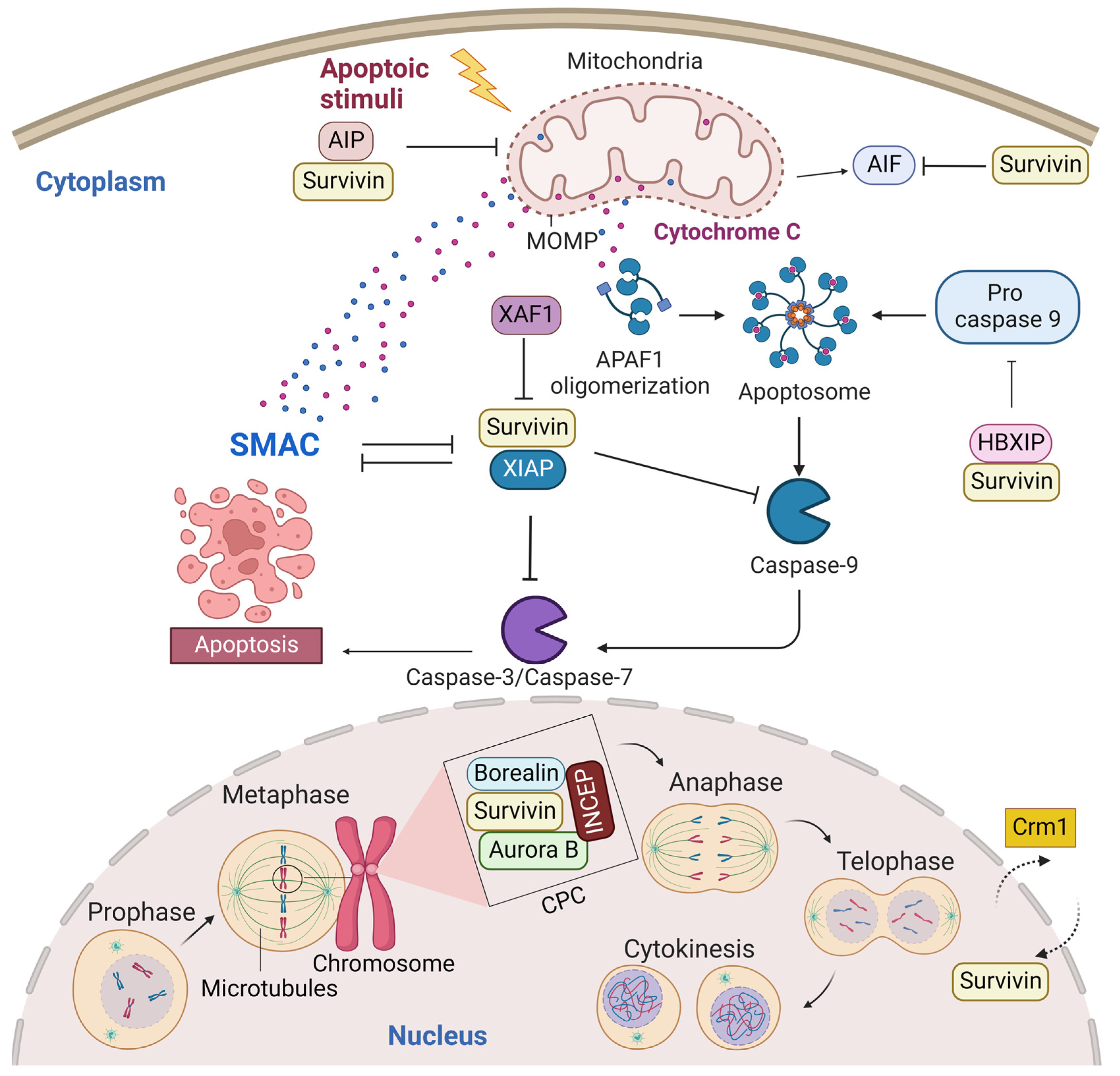
Figure 2. Survivin is an antiapoptotic protein and essential mitotic protein. Cytoplasmic survivin and mitochondrial survivin inhibit apoptosis, while nuclear survivin regulates cell division. Crm1 mediates the exportation of survivin from the nucleus to the cytoplasm. Survivin plays a significant role in inhibiting the intrinsic apoptotic pathway. The intrinsic apoptotic pathway is activated by endogenous stress signals or irradiation and depends on the release of Cyt-c from the mitochondria. Cyt-c binds with Apaf-1 and ATP, which bind to pro-caspase-9 and create a protein complex known as an apoptosome. Pro-caspase-9 is cleaved to its active form of caspase-9 by the apoptosome, which in turn activates the effector caspase-3/6/7, resulting in cell apoptosis. Survivin stabilizes XIAP and blocks Smac from antagonizing XIAP. Survivin–XIAP complex blocks caspase-9 and inhibits apoptosis. Survivin-HBXIP also suppresses pro-caspase-9 and blocks the activation of caspase-9. The survivin–AIP complex stabilizes survivin and promotes its anti-apoptotic function in the mitochondria. Mitotic survivin is part of the CPC and directs Aurora-B kinase to centromere during mitosis. This figure was created with BioRender.com.
2.2. Nuclear Survivin
Nuclear survivin plays a pivotal role in the coordination of mitosis and cytokinesis. Survivin localization in the nucleus, thus far, is thought to occur by passive diffusion, as no classical nuclear localization signal (NLS) exists within the protein. Some reports showed that the nuclear localization of survivin is proapoptotic and increases the susceptibility of cancer cells to conventional chemotherapy and radiation treatment. For example, Colnaghi et al. showed that mutant survivin accumulating in the nucleus could no longer protect cells against the ionizing radiation or apoptosis induced by the TNF-related apoptosis-inducing ligand [14][31]. Moreover, when Connell et al. artificially forced wild-type human survivin expression in the nucleus, they observed that the nuclear localization of survivin prevented it from acting as an inhibitor of apoptosis [32]. However, other reports showed that nuclear survivin is antiapoptotic and associated with poor prognosis in various tumor types [15][33][34]. This contradiction in data regarding the role of nuclear survivin as a predictor for prognosis may be tumor-type-specific and/or due to the variable criteria used to classify a tumor as nuclear survivin or cytoplasmic survivin. Nuclear survivin has a highly dynamic and characteristic localization pattern during mitosis. The localization pattern of survivin during mitosis is typical for chromosomal passenger proteins. In fact, the survivin monomer can interact with the chromosomal passage protein, Borealin, during mitosis [35]. Borealin uses the same survivin–survivin dimerization interface to interact and replace one survivin monomer to form a survivin–borealin heterodimer [35]. Survivin, together with borealin and the inner centromere protein (INCENP) form the non-enzymatic regulatory components of the chromosomal passenger complex (CPC), an essential mitotic complex (Figure 2). The N-termini of borealin and survivin associate with INCENP to form a tight three-helical bundle and create a single structural unit [35]. The association of the core “passenger” proteins controls the activity and localization of the CPC enzymatic component, the Aurora B kinase, both temporally and spatially [36]. Survivin targets the CPC to the centromeres during prometaphase, ensuring that chromosomes are correctly aligned before they are segregated at anaphase. The BIR domain of survivin (residues Asp70 and Asp71) recognizes histone 3 in centromeric chromatin that has been phosphorylated at Thr3 by the haspin kinase [37][38]. Survivin binds to the N-terminal tail of the histone H3 carrying the Thr3 phosphorylation mark (Thr3ph) [37][38]. During prometaphase, survivin is phosphorylated at Thr117 by the Aurora B kinase to ensure that its association with the centromeres remains dynamic until all chromosomes have oriented [39][40][41][42]. During the metaphase–anaphase transition, survivin dissociates from the centromere and tethers at the spindle midzone, while the sister chromatids migrate to the poles. During telophase, survivin is located in the midbodies at the intercellular bridge and is degraded after cytokinesis. During cytokinesis, survivin delineates the cleavage plane prior to actomyosin recruitment [43].
Most studies indicate a close association between survivin and microtubules. Survivin was shown to bind to the polymerized microtubules of the mitotic spindle and the midzone [44], to centrosomes [39][43], or to kinetochores [45]. The binding site was hypothesized to be in the survivin helical region [7]. Survivin also regulates the microtubule dynamics and nucleation, as Rosa et al. demonstrated: depletion of survivin increased both the amount of microtubules nucleation and the incidence of microtubule catastrophe. In contrast, its overexpression reduced the microtubules’ nucleated by centrosomes and suppressed the microtubule dynamics in mitotic spindles and bidirectional growth of microtubules in midbodies during cytokinesis [46]. It is worth mentioning that these survivin effects on microtubules are independent of Aurora B expression or activity [46]. Survivin suppresses microtubule dynamics through its interaction with Ran, which promotes the delivery of the Ran effector molecule, the targeting protein for Xenopus kinesin-like protein 2 (TPX2), to microtubules for proper spindle formation [47]. The mitotic spindle is a bipolar-microtubule-based structure that segregates chromosomes during the cell cycle. Spindle assembly and chromosome segregation depend on the function of various microtubule motor proteins, such as microtubule-associated proteins (MAPs), which are required to regulate microtubule dynamics and the other molecules, such as the small GTPase Ran, involved in microtubule polymerization during mitosis [47]. TPX2 is a microtubule-associated protein that acts as a spindle assembly factor (SAF), mediates the binding of the COOH-terminal domain of Xenopus kinesin-like protein 2 to microtubules, and colocalizes with spindle microtubules in the M-phase in vivo and in vitro [48][49]. Survivin physically interacts with Ran via a discrete binding interface centered on Glutamic acid 65 (Glu65) in survivin [47]. Ran also regulates the Crm1–survivin/cargoes interaction, where cargoes’ binding and the release of Crm1 are controlled by the asymmetric distribution of the two nucleotide states of Ran, the so-called RanGTP gradient. The Ran guanine nucleotide exchange factor (RanGEF), RCC1, facilitates Ran binding to Crm1. Upon export to the cytoplasm, the trimeric exportin/RanGTP/substrate complex is disassembled by the RanGTP hydrolysis induced by the RanGTPase-activating protein (RanGAP) and RanGTP-binding protein 1 (RanBP1) [50].
3. Expression and Isoforms of Survivin
Survivin is scarcely expressed in resting adult tissue yet is found in adult myeloid stem cells, adult marrow, umbilical cord blood CD34+ cells, peripheral blood mononuclear cells, and T lymphocytes; during embryogenesis; and in most human cancers [6][51][52]. Survivin is required for mitosis during development, since embryos with homozygous general deletion of survivin are embryonically lethal 4.5 days before post-coitum [43][53]. Survivin null mouse embryos showed degenerated blastomeres, micronuclei formation, variable nuclear sizes, irregular nuclear morphology, multinucleation, the absence of normal mitotic spindle structures and intercellular midbodies, reduced microtubule networks around the cells, and bundling of microtubules [43]. Consistent with its role in controlling mitotic progression, survivin levels are regulated in a cell-cycle-dependent fashion. Normally, survivin is maximally expressed during the G2/M phase of the cell cycle, where it associates with the mitotic spindle microtubules and performs functions essential for chromosome segregation and cytokinesis [44]. Survivin presents at very low levels in G1 and S phases, since it is ubiquitylated and degraded after mitosis by the 26S proteasome [54]. Survivin is preferentially degraded in the nucleus in a cdh1/APC-dependent manner [32]. The anaphase-promoting complex or cyclosome (APC/C), an E3 ubiquitin ligase, marks survivin for degradation by the 26S proteasome, to trigger the transition from metaphase to anaphase. However, when overexpressed, as it is in cancer cells, survivin is present in interphase and shuttles between the cytoplasm and nucleus. The BIRC5 gene encodes human survivin, so it is also known as a baculoviral inhibitor of apoptosis repeat-containing 5 (BIRC5). BIRC5 represents one of 40 genes to be expressed at elevated levels in all cancer tissues but not in normal cells [55]. Human BIRC5 is mapped to chromosome 17q25, and its promoter possesses a canonical CpG island and numerous Sp1 sites but no TATA box with cell cycle-regulatory sequences (cell-cycle-dependent element (CDE)/cell-cycle gene homology region (CHR)), which ensures its cell-cycle-dependent expression [56]. Moreover, the survivin promoter has multiple sites for binding prooncogenic transcription factors, including those that may be responsible for its differential expression in normal and cancer tissues [57][58]. BIRC5 has four exons and three introns, and its alternative transcription gives rise to six different transcripts, survivin, survivin-ΔEx3, survivin-2B, survivin-3B, survivin 2α, and survivin-3α, with additional variants found in the Est database (Figure 3) [6][59]. Survivin is the predominant (mature) wild-type form, and it is derived from exons 1–4 [60][61][62]. Survivin-ΔEx3 (137 amino acids) lacks exon 3; thus, its BIR domain is truncated at amino acid position 74, and it has a characteristic frameshift in the translation in its carboxyl terminus [61]. However, Survivin 2α (74 amino acids) contains the coding sequences from exon 1,2 and one additional amino acid before termination, and it lacks the entire carboxy-terminal coiled-coil domain [63]. Survivin-3α is also a truncated variant, similar to survivin-2α, and is not widely studied [64]. Survivin-2B (165 amino acids) retains a part of intron 2 as a cryptic exon that creates an additional and alternative exon named 2B that encodes the insertion of 23 additional amino acids into the BIR domain at essentially the same position (amino acid 74), where the BIR domain of survivin-ΔEx3 is truncated [61]. Survivin-3B (120 amino acids) is coded by exons 1,2, and 3. It consists of the N-terminal 113 amino acids of survivin and seven new amino acid sequences at the C-terminal tail encoded by exon 3B from intron 3 of survivin [62]. It also contains the BIR domain but lacks the carboxyl-terminal coiled-coil domain [62]. However, not all variants have been unambiguously shown to be expressed in vivo [65]. In addition, there are conflicting reports regarding the biological functions of survivin splice variants, as they may undergo homo-/heterodimerization, particularly with wild-type survivin. Recent studies looked at their potential contributions to disease and their significance as biomarkers and diagnostic tools in cancer [60]. While the expression levels of survivin and survivin-ΔEx3 correlate with tumor aggressiveness and resistance to therapy, survivin-2B and survivin-2α were reported to be cytoprotective and proapoptotic [61][63][66]. However, Knauer and colleagues reported that only survivin-3B, among the survivin isoforms, protected cells against cisplatin- or irradiation-induced apoptosis [65].
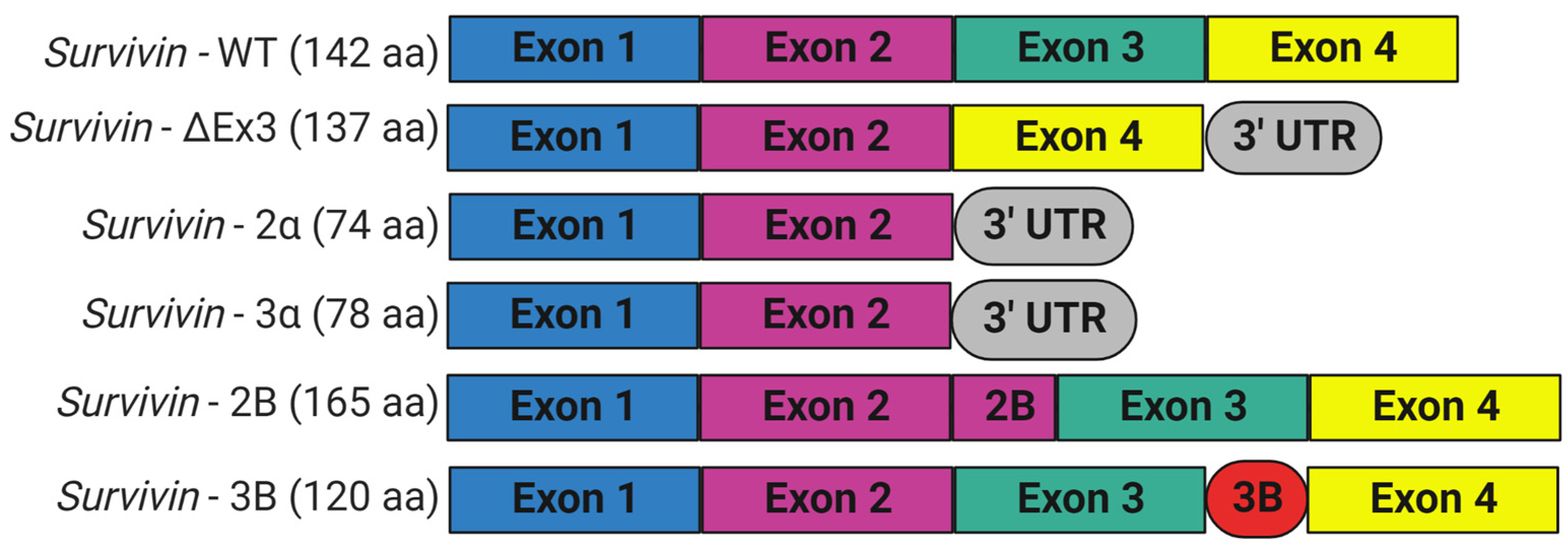
Figure 3. Schematic representation of alternative splice variants of survivin encoded by BIRC5 gene. Six different variants were identified. The wild-type survivin with four exons: survivin-ΔEx3 lacks exon 3 and shows a frameshift with the extension of the reading frame into the open reading frame of the 3′ untranslated region; survivin-2α and survivin-3α have exon 1 and 2; survivin-2B has an additional exon (exon 2B) inserted between exon 2 and 3; and survivin-3B has a novel exon (exon 3B) flanked by exon 3 and 4. This figure was created with BioRender.com.
References
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541.
- Crook, N.E.; Clem, R.J.; Miller, L.K. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J. Virol. 1993, 67, 2168–2174.
- Uren, A.G.; Coulson, E.J.; Vaux, D.L. Conservation of baculovirus inhibitor of apoptosis repeat proteins (BIRPs) in viruses, nematodes, vertebrates and yeasts. Trends Biochem. Sci. 1998, 23, 159–162.
- Uren, A.G.; Beilharz, T.; O’Connell, M.J.; Bugg, S.J.; van Driel, R.; Vaux, D.L.; Lithgow, T. Role for yeast inhibitor of apoptosis (IAP)-like proteins in cell division. Proc. Natl. Acad. Sci. USA 1999, 96, 10170–10175.
- Muchmore, S.W.; Chen, J.; Jakob, C.; Zakula, D.; Matayoshi, E.D.; Wu, W.; Zhang, H.; Li, F.; Ng, S.C.; Altieri, D.C. Crystal structure and mutagenic analysis of the inhibitor-of-apoptosis protein survivin. Mol. Cell 2000, 6, 173–182.
- Ambrosini, G.; Adida, C.; Altieri, D.C. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat. Med. 1997, 3, 917–921.
- Verdecia, M.A.; Huang, H.; Dutil, E.; Kaiser, D.A.; Hunter, T.; Noel, J.P. Structure of the human anti-apoptotic protein survivin reveals a dimeric arrangement. Nat. Struct. Biol. 2000, 7, 602–608.
- Fortugno, P.; Wall, N.R.; Giodini, A.; O’Connor, D.S.; Plescia, J.; Padgett, K.M.; Tognin, S.; Marchisio, P.C.; Altieri, D.C. Survivin exists in immunochemically distinct subcellular pools and is involved in spindle microtubule function. J. Cell Sci. 2002, 115, 575–585.
- Khan, S.; Aspe, J.R.; Asumen, M.G.; Almaguel, F.; Odumosu, O.; Acevedo-Martinez, S.; De Leon, M.; Langridge, W.H.; Wall, N.R. Extracellular, cell-permeable survivin inhibits apoptosis while promoting proliferative and metastatic potential. Br. J. Cancer 2009, 100, 1073–1086.
- Altieri, D.C. Validating survivin as a cancer therapeutic target. Nat. Rev. Cancer 2003, 3, 46–54.
- Humphry, N.J.; Wheatley, S.P. Survivin inhibits excessive autophagy in cancer cells but does so independently of its interaction with LC3. Biol. Open 2018, 7, bio037374.
- Sanhueza, C.; Wehinger, S.; Castillo Bennett, J.; Valenzuela, M.; Owen, G.I.; Quest, A.F. The twisted survivin connection to angiogenesis. Mol. Cancer 2015, 14, 198.
- Mull, A.N.; Klar, A.; Navara, C.S. Differential localization and high expression of SURVIVIN splice variants in human embryonic stem cells but not in differentiated cells implicate a role for SURVIVIN in pluripotency. Stem Cell Res. 2014, 12, 539–549.
- Knauer, S.K.; Kramer, O.H.; Knosel, T.; Engels, K.; Rodel, F.; Kovacs, A.F.; Dietmaier, W.; Klein-Hitpass, L.; Habtemichael, N.; Schweitzer, A.; et al. Nuclear export is essential for the tumor-promoting activity of survivin. FASEB J. 2007, 21, 207–216.
- Li, F.; Yang, J.; Ramnath, N.; Javle, M.M.; Tan, D. Nuclear or cytoplasmic expression of survivin: What is the significance? Int. J. Cancer 2005, 114, 509–512.
- Rodriguez, J.A.; Span, S.W.; Ferreira, C.G.; Kruyt, F.A.; Giaccone, G. CRM1-mediated nuclear export determines the cytoplasmic localization of the antiapoptotic protein Survivin. Exp. Cell Res. 2002, 275, 44–53.
- Knauer, S.K.; Bier, C.; Habtemichael, N.; Stauber, R.H. The Survivin-Crm1 interaction is essential for chromosomal passenger complex localization and function. EMBO Rep. 2006, 7, 1259–1265.
- Stauber, R.H.; Rabenhorst, U.; Rekik, A.; Engels, K.; Bier, C.; Knauer, S.K. Nucleocytoplasmic shuttling and the biological activity of mouse survivin are regulated by an active nuclear export signal. Traffic 2006, 7, 1461–1472.
- Engelsma, D.; Rodriguez, J.A.; Fish, A.; Giaccone, G.; Fornerod, M. Homodimerization antagonizes nuclear export of survivin. Traffic 2007, 8, 1495–1502.
- Dohi, T.; Okada, K.; Xia, F.; Wilford, C.E.; Samuel, T.; Welsh, K.; Marusawa, H.; Zou, H.; Armstrong, R.; Matsuzawa, S.; et al. An IAP-IAP complex inhibits apoptosis. J. Biol. Chem. 2004, 279, 34087–34090.
- Liston, P.; Fong, W.G.; Kelly, N.L.; Toji, S.; Miyazaki, T.; Conte, D.; Tamai, K.; Craig, C.G.; McBurney, M.W.; Korneluk, R.G. Identification of XAF1 as an antagonist of XIAP anti-Caspase activity. Nat. Cell. Biol. 2001, 3, 128–133.
- Arora, V.; Cheung, H.H.; Plenchette, S.; Micali, O.C.; Liston, P.; Korneluk, R.G. Degradation of survivin by the X-linked inhibitor of apoptosis (XIAP)-XAF1 complex. J. Biol. Chem. 2007, 282, 26202–26209.
- Du, C.; Fang, M.; Li, Y.; Li, L.; Wang, X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell 2000, 102, 33–42.
- Song, Z.; Yao, X.; Wu, M. Direct interaction between survivin and Smac/DIABLO is essential for the anti-apoptotic activity of survivin during taxol-induced apoptosis. J. Biol. Chem. 2003, 278, 23130–23140.
- Marusawa, H.; Matsuzawa, S.; Welsh, K.; Zou, H.; Armstrong, R.; Tamm, I.; Reed, J.C. HBXIP functions as a cofactor of survivin in apoptosis suppression. EMBO J. 2003, 22, 2729–2740.
- Liu, T.; Brouha, B.; Grossman, D. Rapid induction of mitochondrial events and caspase-independent apoptosis in Survivin-targeted melanoma cells. Oncogene 2004, 23, 39–48.
- Liu, T.; Biddle, D.; Hanks, A.N.; Brouha, B.; Yan, H.; Lee, R.M.; Leachman, S.A.; Grossman, D. Activation of dual apoptotic pathways in human melanocytes and protection by survivin. J. Investig. Dermatol. 2006, 126, 2247–2256.
- Dunajova, L.; Cash, E.; Markus, R.; Rochette, S.; Townley, A.R.; Wheatley, S.P. The N-terminus of survivin is a mitochondrial-targeting sequence and Src regulator. J. Cell Sci 2016, 129, 2707–2712.
- Kang, B.H.; Altieri, D.C. Regulation of survivin stability by the aryl hydrocarbon receptor-interacting protein. J. Biol. Chem. 2006, 281, 24721–24727.
- Fortugno, P.; Beltrami, E.; Plescia, J.; Fontana, J.; Pradhan, D.; Marchisio, P.C.; Sessa, W.C.; Altieri, D.C. Regulation of survivin function by Hsp90. Proc. Natl. Acad. Sci. USA 2003, 100, 13791–13796.
- Colnaghi, R.; Connell, C.M.; Barrett, R.M.; Wheatley, S.P. Separating the anti-apoptotic and mitotic roles of survivin. J. Biol. Chem. 2006, 281, 33450–33456.
- Connell, C.M.; Colnaghi, R.; Wheatley, S.P. Nuclear survivin has reduced stability and is not cytoprotective. J. Biol. Chem. 2008, 283, 3289–3296.
- Krieg, S.; Roderburg, C.; Fung, S.; Luedde, T.; Knoefel, W.T.; Krieg, A. Nuclear survivin is a prognosticator in gastroenteropancreatic neuroendocrine neoplasms: A meta-analysis. J. Cancer Res. Clin. Oncol. 2022, 148, 2235–2246.
- Vay, C.; Babaei, S.; Safi, S.A.; Dizdar, L.; Rehders, A.; Haeberle, L.; Roderburg, C.; Loosen, S.H.; Esposito, I.; Knoefel, W.T.; et al. Clinicopathological and Prognostic Value of Survivin Expression in Surgically Resected Pancreatic Ductal Adenocarcinoma. Cancers 2022, 14, 3494.
- Jeyaprakash, A.A.; Klein, U.R.; Lindner, D.; Ebert, J.; Nigg, E.A.; Conti, E. Structure of a Survivin-Borealin-INCENP core complex reveals how chromosomal passengers travel together. Cell 2007, 131, 271–285.
- Carmena, M.; Ruchaud, S.; Earnshaw, W.C. Making the Auroras glow: Regulation of Aurora A and B kinase function by interacting proteins. Curr. Opin. Cell. Biol. 2009, 21, 796–805.
- Jeyaprakash, A.A.; Basquin, C.; Jayachandran, U.; Conti, E. Structural basis for the recognition of phosphorylated histone h3 by the survivin subunit of the chromosomal passenger complex. Structure 2011, 19, 1625–1634.
- Kelly, A.E.; Ghenoiu, C.; Xue, J.Z.; Zierhut, C.; Kimura, H.; Funabiki, H. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science 2010, 330, 235–239.
- Wheatley, S.P.; Carvalho, A.; Vagnarelli, P.; Earnshaw, W.C. INCENP is required for proper targeting of Survivin to the centromeres and the anaphase spindle during mitosis. Curr. Biol. 2001, 11, 886–890.
- Wheatley, S.P.; Barrett, R.M.; Andrews, P.D.; Medema, R.H.; Morley, S.J.; Swedlow, J.R.; Lens, S.M. Phosphorylation by aurora-B negatively regulates survivin function during mitosis. Cell Cycle 2007, 6, 1220–1230.
- Lens, S.M.; Wolthuis, R.M.; Klompmaker, R.; Kauw, J.; Agami, R.; Brummelkamp, T.; Kops, G.; Medema, R.H. Survivin is required for a sustained spindle checkpoint arrest in response to lack of tension. EMBO J. 2003, 22, 2934–2947.
- Carvalho, A.; Carmena, M.; Sambade, C.; Earnshaw, W.C.; Wheatley, S.P. Survivin is required for stable checkpoint activation in taxol-treated HeLa cells. J. Cell Sci. 2003, 116, 2987–2998.
- Uren, A.G.; Wong, L.; Pakusch, M.; Fowler, K.J.; Burrows, F.J.; Vaux, D.L.; Choo, K.H. Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr. Biol. 2000, 10, 1319–1328.
- Li, F.; Ambrosini, G.; Chu, E.Y.; Plescia, J.; Tognin, S.; Marchisio, P.C.; Altieri, D.C. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature 1998, 396, 580–584.
- Skoufias, D.A.; Mollinari, C.; Lacroix, F.B.; Margolis, R.L. Human survivin is a kinetochore-associated passenger protein. J. Cell Biol. 2000, 151, 1575–1582.
- Rosa, J.; Canovas, P.; Islam, A.; Altieri, D.C.; Doxsey, S.J. Survivin modulates microtubule dynamics and nucleation throughout the cell cycle. Mol. Biol. Cell 2006, 17, 1483–1493.
- Xia, F.; Canovas, P.M.; Guadagno, T.M.; Altieri, D.C. A survivin-ran complex regulates spindle formation in tumor cells. Mol. Cell. Biol. 2008, 28, 5299–5311.
- Garrett, S.; Auer, K.; Compton, D.A.; Kapoor, T.M. hTPX2 is required for normal spindle morphology and centrosome integrity during vertebrate cell division. Curr. Biol. 2002, 12, 2055–2059.
- Gruss, O.J.; Wittmann, M.; Yokoyama, H.; Pepperkok, R.; Kufer, T.; Sillje, H.; Karsenti, E.; Mattaj, I.W.; Vernos, I. Chromosome-induced microtubule assembly mediated by TPX2 is required for spindle formation in HeLa cells. Nat. Cell. Biol. 2002, 4, 871–879.
- Weis, K. Regulating access to the genome: Nucleocytoplasmic transport throughout the cell cycle. Cell 2003, 112, 441–451.
- Fukuda, S.; Pelus, L.M. Regulation of the inhibitor-of-apoptosis family member survivin in normal cord blood and bone marrow CD34(+) cells by hematopoietic growth factors: Implication of survivin expression in normal hematopoiesis. Blood 2001, 98, 2091–2100.
- Fukuda, S.; Foster, R.G.; Porter, S.B.; Pelus, L.M. The antiapoptosis protein survivin is associated with cell cycle entry of normal cord blood CD34(+) cells and modulates cell cycle and proliferation of mouse hematopoietic progenitor cells. Blood 2002, 100, 2463–2471.
- Martini, E.; Schneider, E.; Neufert, C.; Neurath, M.F.; Becker, C. Survivin is a guardian of the intestinal stem cell niche and its expression is regulated by TGF-beta. Cell Cycle 2016, 15, 2875–2881.
- Zhao, J.; Tenev, T.; Martins, L.M.; Downward, J.; Lemoine, N.R. The ubiquitin-proteasome pathway regulates survivin degradation in a cell cycle-dependent manner. J. Cell Sci. 2000, 113 Pt 23, 4363–4371.
- Velculescu, V.E.; Madden, S.L.; Zhang, L.; Lash, A.E.; Yu, J.; Rago, C.; Lal, A.; Wang, C.J.; Beaudry, G.A.; Ciriello, K.M.; et al. Analysis of human transcriptomes. Nat. Genet. 1999, 23, 387–388.
- Li, F.; Altieri, D.C. Transcriptional analysis of human survivin gene expression. Biochem. J. 1999, 344 Pt 2, 305–311.
- Warrier, N.M.; Agarwal, P.; Kumar, P. Emerging Importance of Survivin in Stem Cells and Cancer: The Development of New Cancer Therapeutics. Stem Cell. Rev. Rep. 2020, 16, 828–852.
- Zhang, M.; Yang, J.; Li, F. Transcriptional and post-transcriptional controls of survivin in cancer cells: Novel approaches for cancer treatment. J. Exp. Clin. Cancer Res. 2006, 25, 391–402.
- Adamopoulos, P.G.; Tsiakanikas, P.; Adam, E.E.; Scorilas, A. Unraveling novel survivin mRNA transcripts in cancer cells using an in-house developed targeted high-throughput sequencing approach. Genomics 2021, 113, 573–581.
- Sah, N.K.; Seniya, C. Survivin splice variants and their diagnostic significance. Tumour Biol. 2015, 36, 6623–6631.
- Mahotka, C.; Wenzel, M.; Springer, E.; Gabbert, H.E.; Gerharz, C.D. Survivin-deltaEx3 and survivin-2B: Two novel splice variants of the apoptosis inhibitor survivin with different antiapoptotic properties. Cancer Res. 1999, 59, 6097–6102.
- Badran, A.; Yoshida, A.; Ishikawa, K.; Goi, T.; Yamaguchi, A.; Ueda, T.; Inuzuka, M. Identification of a novel splice variant of the human anti-apoptopsis gene survivin. Biochem. Biophys. Res. Commun. 2004, 314, 902–907.
- Caldas, H.; Honsey, L.E.; Altura, R.A. Survivin 2alpha: A novel Survivin splice variant expressed in human malignancies. Mol. Cancer 2005, 4, 11.
- Necochea-Campion, R.; Chen, C.S.; Mirshahidi, S.; Howard, F.D.; Wall, N.R. Clinico-pathologic relevance of Survivin splice variant expression in cancer. Cancer Lett. 2013, 339, 167–174.
- Knauer, S.K.; Bier, C.; Schlag, P.; Fritzmann, J.; Dietmaier, W.; Rodel, F.; Klein-Hitpass, L.; Kovacs, A.F.; Doring, C.; Hansmann, M.L.; et al. The survivin isoform survivin-3B is cytoprotective and can function as a chromosomal passenger complex protein. Cell Cycle 2007, 6, 1502–1509.
- Caldas, H.; Fangusaro, J.R.; Boue, D.R.; Holloway, M.P.; Altura, R.A. Dissecting the role of endothelial SURVIVIN DeltaEx3 in angiogenesis. Blood 2007, 109, 1479–1489.
More
Information
Subjects:
Cell Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
990
Revisions:
2 times
(View History)
Update Date:
23 Feb 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




