
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | ERICA GIANAZZA | -- | 6375 | 2023-02-20 10:21:33 | | | |
| 2 | Lindsay Dong | Meta information modification | 6375 | 2023-02-22 01:44:14 | | |
Video Upload Options
Lipid-lowering therapies are widely used to prevent the development of atherosclerotic cardiovascular disease (ASCVD) and related mortality worldwide. “Omics” technologies have been successfully applied to investigate the mechanisms of action of these drugs, their pleiotropic effects, and their side effects, aiming to identify novel targets for future personalized medicine with an improvement of the efficacy and safety associated with the treatment. Pharmacometabolomics is a branch of metabolomics that is focused on the study of drug effects on metabolic pathways that are implicated in the variation of response to the treatment considering also the influences from a specific disease, environment, and concomitant pharmacological therapies. The integration of pharmacometabolomics data with the information obtained from the other “omics” approaches could help in the comprehension of the biological mechanisms underlying the use of lipid-lowering drugs in view of defining a precision medicine to improve the efficacy and reduce the side effects associated with the treatment.
1. Introduction
2. Methodological Approaches for Pharmacometabolomics and Pharmacolipidomics
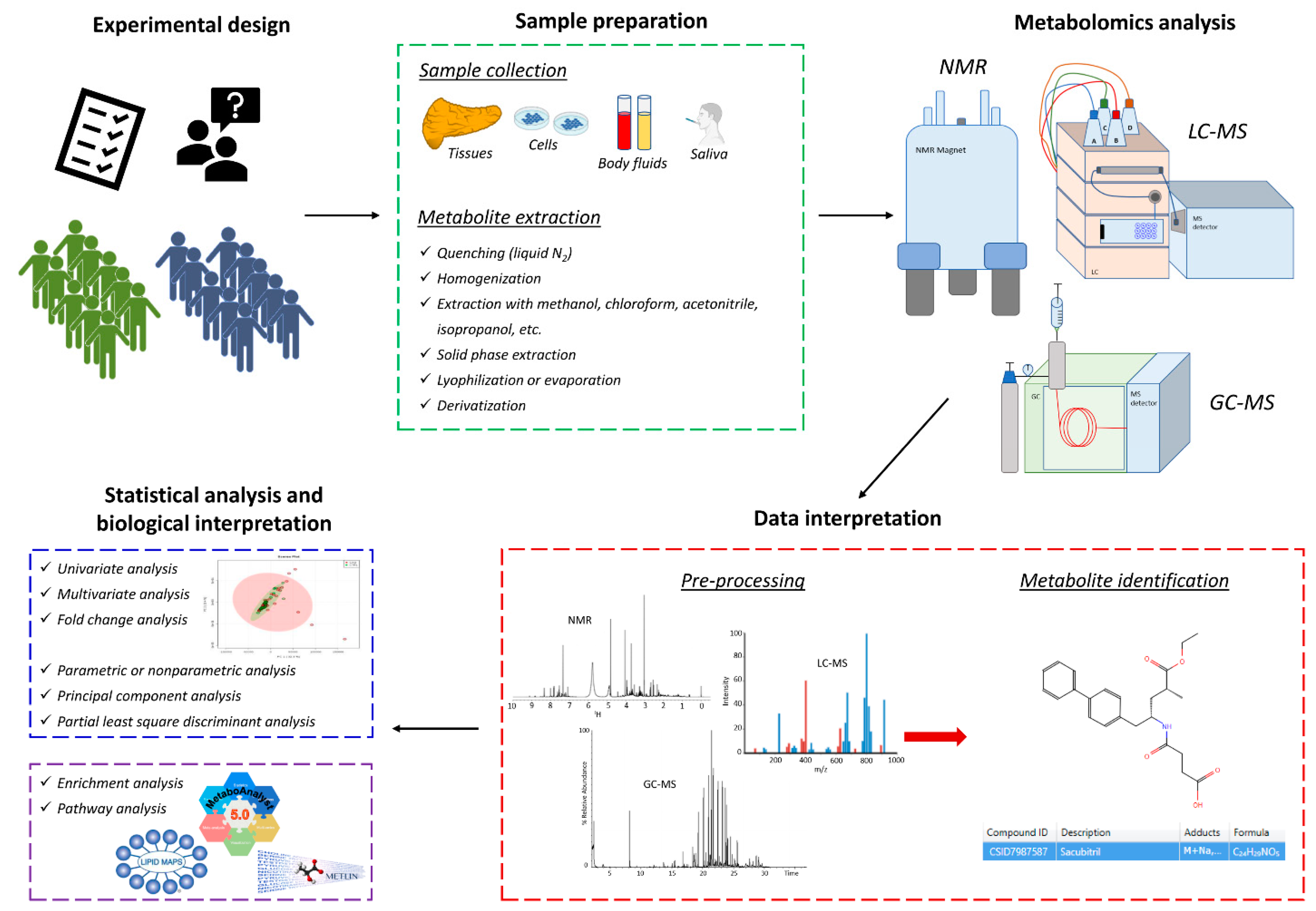
2.1. Nuclear Magnetic Resonance (NMR)
2.2. Gas Chromatography-Mass Spectrometry (GC-MS)
2.3. Liquid Chromatography-Mass Spectrometry (LC-MS)
3. Lipid-Lowering Therapies and Metabolomics
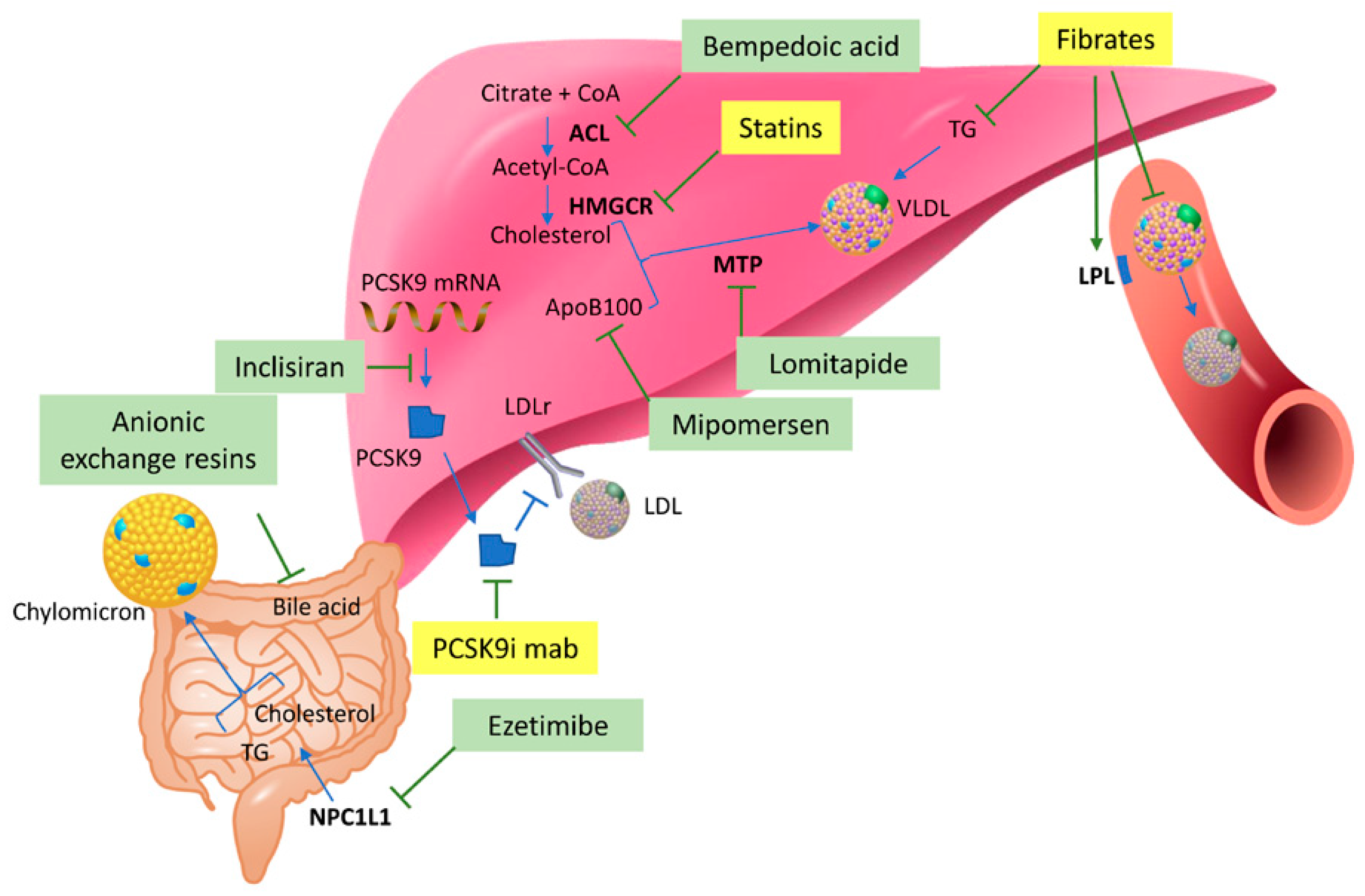
3.1. Statins
3.1.1. Statin Response Variability
3.1.2. Alterations in Gut Microbiota by Statin Therapy
3.1.3. Adverse Effects of Statins
Statins are highly effective and safe for most people, but they can cause minor or severe side effects that should never be neglected. Statin-related myotoxicity, for example, can range from mild muscle pain up to rhabdomyolysis, which is a serious and fatal disorder that sometimes occurs in patients following pharmacological treatment [45]. Statin-associated rhabdomyolysis risk has been reported as dose-dependent and concentration-dependent [46].
Metabolomics provides an accurate signature of all metabolite changes in biological fluids, cells, and tissues that can be a source for biomarker discovery. A metabolomic analysis of skeletal muscle and plasma using LC-MS and GC-MS was performed on a rat model treated with two myotoxicants, cerivastatin and tetramethyl-p-phenylenediamine, to induce a skeletal injury and identify candidate biomarkers for skeletal muscle toxicity [47]. They observed in skeletal muscle a significant increase in 2-hydroxyglutarate in cerivastatin-treated rats and hexanoylcarnitine in both types of treated rats. These increases were also measured in plasma samples at different times after dosing, demonstrating the possibility to use plasma 2-hydroxyglutarate and hexanoylcarnitine as valid and easily detectable biomarkers for the early detection of skeletal muscle toxicity in rats, with better sensitivity than the conventional markers creatine kinase and aspartate aminotransferase whose utility in clinics is limited due to their low diagnostic power [48]. Moreover, this study confirmed the importance and benefit of metabolomics for biomarker discovery in toxicological studies.
Among the potential statin-related adverse events, there is also an increased incidence of type II diabetes mellitus that can lead to premature discontinuation of treatment. Therefore, it is important to evaluate a correlation between statin-induced metabolic changes and statin-induced hyperglycemia and insulin resistance, to identify pre-drug treatment metabolites predictive of increased diabetic risk [49]. In this regard, a pharmacometabolomic study was performed by GC-TOF-MS on plasma pre- and post-treatment with simvastatin for 6 weeks from patients enrolled for the CAP study [49] to measure changes in intermediary metabolism and the associated high plasma glucose levels as a potentially adverse response to simvastatin. Some patients developed hyperglycemia and pre-diabetes, as well as a dysfunction of beta cells and insulin resistance in more than 50% of patients following statin therapy. An initial metabolic profile of simvastatin-induced insulin resistance was identified, including ethanolamine, hydroxylamine, hydroxycarbamate, and isoleucine, which can be predictive biomarkers of individuals at risk of developing a statin-induced new onset pre-type II diabetes mellitus [49]. In particular, the metabolite ethanolamine was identified as the most likely to predict simvastatin-induced diabetic risk, indicating that decarboxylation and oxidation were significantly associated with statin-induced hyperglycemia and insulin resistance. Pharmacometabolomics allows having a baseline metabolic signature before starting the drug therapy that can be then used to find predictive biomarkers able to stratify patients and to identify subjects who are at higher risk of adverse side effects, enabling personalized selection of the most appropriate medication for each patient and personalized monitoring of their prognosis.
3.1.4. Beneficial Effects of Statins
3.2. PCSK9 Inhibitors
3.3. Fibrates
Fibrates are frequently used in combination with statins, working in synergy to reduce plasma lipids, even if this type of treatment is associated with a higher incidence of fatal side effects, such as acute tubular necrosis and rhabdomyolysis. Several hypotheses have been formulated including pharmacokinetic interference, displacement of statins from their binding sites, synergistic action on skeletal muscle, or inhibition of statin glucuronidation by fibrates.
3.4. Nutraceutical and Dietary Habits
4. Conclusions
References
- Larsen, L.E.; Stoekenbroek, R.M.; Kastelein, J.J.P.; Holleboom, A.G. Moving Targets: Recent Advances in Lipid-Lowering Therapies. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 349–359.
- Gowda, G.A.; Zhang, S.; Gu, H.; Asiago, V.; Shanaiah, N.; Raftery, D. Metabolomics-based methods for early disease diagnostics. Expert Rev. Mol. Diagn. 2008, 8, 617–633.
- Nicholson, J.K.; Lindon, J.C. Systems biology: Metabonomics. Nature 2008, 455, 1054–1056.
- Holmes, E.; Wilson, I.D.; Nicholson, J.K. Metabolic phenotyping in health and disease. Cell 2008, 134, 714–717.
- Burt, T.; Nandal, S. Pharmacometabolomics in Early-Phase Clinical Development. Clin. Transl. Sci. 2016, 9, 128–138.
- Kim, H.; Yoon, Y. Pharmacometabolomics: Current Applications and Future Perspectives. Transl. Clin. Pharmacol. 2014, 22, 8–10.
- Rattray, N.; Daouk, R. Pharmacometabolomics and Precision Medicine Special Issue Editorial. Metabolomics 2017, 13, 59.
- Huang, Q.; Aa, J.; Jia, H.; Xin, X.; Tao, C.; Liu, L.; Zou, B.; Song, Q.; Shi, J.; Cao, B.; et al. A Pharmacometabonomic Approach To Predicting Metabolic Phenotypes and Pharmacokinetic Parameters of Atorvastatin in Healthy Volunteers. J. Proteome Res. 2015, 14, 3970–3981.
- Clayton, T.A.; Lindon, J.C.; Cloarec, O.; Antti, H.; Charuel, C.; Hanton, G.; Provost, J.P.; Le Net, J.L.; Baker, D.; Walley, R.J.; et al. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature 2006, 440, 1073–1077.
- Wilson, I.D. Drugs, bugs, and personalized medicine: Pharmacometabonomics enters the ring. Proc. Natl. Acad. Sci. USA 2009, 106, 14187–14188.
- Winnike, J.H.; Li, Z.; Wright, F.A.; Macdonald, J.M.; O’Connell, T.M.; Watkins, P.B. Use of pharmaco-metabonomics for early prediction of acetaminophen-induced hepatotoxicity in humans. Clin. Pharmacol. Ther. 2010, 88, 45–51.
- Alarcon-Barrera, J.C.; Kostidis, S.; Ondo-Mendez, A.; Giera, M. Recent advances in metabolomics analysis for early drug development. Drug Discov. Today 2022, 27, 1763–1773.
- Harrieder, E.M.; Kretschmer, F.; Bocker, S.; Witting, M. Current state-of-the-art of separation methods used in LC-MS based metabolomics and lipidomics. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2022, 1188, 123069.
- Macedo, A.N.; Faccio, A.T.; Fukuji, T.S.; Canuto, G.A.B.; Tavares, M.F.M. Analytical Platforms for Mass Spectrometry-Based Metabolomics of Polar and Ionizable Metabolites. Adv. Exp. Med. Biol. 2021, 1336, 215–242.
- Banoei, M.M.; Donnelly, S.J.; Mickiewicz, B.; Weljie, A.; Vogel, H.J.; Winston, B.W. Metabolomics in critical care medicine: A new approach to biomarker discovery. Clin. Investig. Med. 2014, 37, E363–E376.
- Paglia, G.; Del Greco, F.M.; Sigurdsson, B.B.; Rainer, J.; Volani, C.; Hicks, A.A.; Pramstaller, P.P.; Smarason, S.V. Influence of collection tubes during quantitative targeted metabolomics studies in human blood samples. Clin. Chim. Acta 2018, 486, 320–328.
- Volani, C.; Caprioli, G.; Calderisi, G.; Sigurdsson, B.B.; Rainer, J.; Gentilini, I.; Hicks, A.A.; Pramstaller, P.P.; Weiss, G.; Smarason, S.V.; et al. Pre-analytic evaluation of volumetric absorptive microsampling and integration in a mass spectrometry-based metabolomics workflow. Anal. Bioanal. Chem. 2017, 409, 6263–6276.
- Gómez-Cebrián, N.; Ferreiro, P.V.; Hueso, F.J.C.; Andrés, J.L.P.; Puchades-Carrasco, L.; Pineda-Lucena, A. Pharmacometabolomics by NMR in Oncology: A Systematic Review. Pharmaceuticals 2021, 14, 1015.
- Smith, L.; Villaret-Cazadamont, J.; Claus, S.P.; Canlet, C.; Guillou, H.; Cabaton, N.J.; Ellero-Simatos, S. Important Considerations for Sample Collection in Metabolomics Studies with a Special Focus on Applications to Liver Functions. Metabolites 2020, 10, 104.
- Zierer, J.; Jackson, M.A.; Kastenmuller, G.; Mangino, M.; Long, T.; Telenti, A.; Mohney, R.P.; Small, K.S.; Bell, J.T.; Steves, C.J.; et al. The fecal metabolome as a functional readout of the gut microbiome. Nat. Genet. 2018, 50, 790–795.
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted Metabolomics Strategies—Challenges and Emerging Directions. J. Am. Soc. Mass Spectrom. 2016, 27, 1897–1905.
- Marshall, D.D.; Powers, R. Beyond the paradigm: Combining mass spectrometry and nuclear magnetic resonance for metabolomics. Prog. Nucl. Magn. Reson. Spectrosc. 2017, 100, 1–16.
- Au, A. Metabolomics and Lipidomics of Ischemic Stroke. Adv. Clin. Chem. 2018, 85, 31–69.
- Han, X.; Yang, K.; Gross, R.W. Multi-dimensional mass spectrometry-based shotgun lipidomics and novel strategies for lipidomic analyses. Mass Spectrom. Rev. 2012, 31, 134–178.
- Belhaj, M.R.; Lawler, N.G.; Hoffman, N.J. Metabolomics and Lipidomics: Expanding the Molecular Landscape of Exercise Biology. Metabolites 2021, 11, 151.
- Haslauer, K.E.; Hemmler, D.; Schmitt-Kopplin, P.; Heinzmann, S.S. Guidelines for the Use of Deuterium Oxide (D2O) in (1)H NMR Metabolomics. Anal. Chem. 2019, 91, 11063–11069.
- Emwas, A.H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.A.N.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. NMR Spectroscopy for Metabolomics Research. Metabolites 2019, 9, 123.
- Bodi, V.; Marrachelli, V.G.; Husser, O.; Chorro, F.J.; Vina, J.R.; Monleon, D. Metabolomics in the diagnosis of acute myocardial ischemia. J. Cardiovasc. Transl. Res. 2013, 6, 808–815.
- Amor, A.J.; Vinagre, I.; Valverde, M.; Urquizu, X.; Meler, E.; Lopez, E.; Alonso, N.; Pane, A.; Gimenez, M.; Codina, L.; et al. Nuclear magnetic resonance-based metabolomic analysis in the assessment of preclinical atherosclerosis in type 1 diabetes and preeclampsia. Diabetes Res. Clin. Pract. 2021, 171, 108548.
- Fiehn, O. Metabolomics by Gas Chromatography-Mass Spectrometry: Combined Targeted and Untargeted Profiling. Curr. Protoc. Mol. Biol. 2016, 114, 30.4.1–30.4.32.
- Paglia, G.; Smith, A.J.; Astarita, G. Ion mobility mass spectrometry in the omics era: Challenges and opportunities for metabolomics and lipidomics. Mass Spectrom. Rev. 2021, 41, 722–765.
- Alseekh, S.; Aharoni, A.; Brotman, Y.; Contrepois, K.; D’Auria, J.; Ewald, J.; Ewald, J.C.; Fraser, P.D.; Giavalisco, P.; Hall, R.D.; et al. Mass spectrometry-based metabolomics: A guide for annotation, quantification and best reporting practices. Nat. Methods 2021, 18, 747–756.
- Gowda, S.G.B.; Gowda, D.; Hou, F.; Chiba, H.; Parcha, V.; Arora, P.; Halade, G.V.; Hui, S.P. Temporal lipid profiling in the progression from acute to chronic heart failure in mice and ischemic human hearts. Atherosclerosis 2022, 363, 30–41.
- Ruscica, M.; Banach, M.; Sahebkar, A.; Corsini, A.; Sirtori, C.R. ETC-1002 (Bempedoic acid) for the management of hyperlipidemia: From preclinical studies to phase 3 trials. Expert Opin. Pharmacother. 2019, 20, 791–803.
- Mach, F.; Baigent, C.; Catapano, A.L.; Koskinas, K.C.; Casula, M.; Badimon, L.; Chapman, M.J.; De Backer, G.G.; Delgado, V.; Ference, B.A.; et al. 2019 ESC/EAS Guidelines for the management of dyslipidaemias: Lipid modification to reduce cardiovascular risk. Eur. Heart J. 2020, 41, 111–188.
- Bilen, O.; Ballantyne, C.M. Bempedoic Acid (ETC-1002): An Investigational Inhibitor of ATP Citrate Lyase. Curr. Atheroscler. Rep. 2016, 18, 61.
- Xu, Q.Y.; Liu, Y.H.; Zhang, Q.; Ma, B.; Yang, Z.D.; Liu, L.; Yao, D.; Cui, G.B.; Sun, J.J.; Wu, Z.M. Metabolomic analysis of simvastatin and fenofibrate intervention in high-lipid diet-induced hyperlipidemia rats. Acta Pharmacol. Sin. 2014, 35, 1265–1273.
- Muller, A.L.; Freed, D.H. Basic and Clinical Observations of Mevalonate Depletion on the Mevalonate Signaling Pathway. Curr. Mol. Pharmacol. 2017, 10, 6–12.
- Liao, J.K.; Laufs, U. Pleiotropic effects of statins. Annu. Rev. Pharmacol. Toxicol. 2005, 45, 89–118.
- Silva, L.F.; Ravi, R.; Vangipurapu, J.; Laakso, M. Metabolite Signature of Simvastatin Treatment Involves Multiple Metabolic Pathways. Metabolites 2022, 12, 753.
- Krauss, R.M.; Zhu, H.; Kaddurah-Daouk, R. Pharmacometabolomics of statin response. Clin. Pharmacol. Ther. 2013, 94, 562–565.
- Trupp, M.; Zhu, H.; Wikoff, W.R.; Baillie, R.A.; Zeng, Z.B.; Karp, P.D.; Fiehn, O.; Krauss, R.M.; Kaddurah-Daouk, R. Metabolomics reveals amino acids contribute to variation in response to simvastatin treatment. PLoS ONE 2012, 7, e38386.
- Hu, T.; Zhang, J.L. Mass-spectrometry-based lipidomics. J. Sep. Sci. 2018, 41, 351–372.
- Zhang, S.; Yuan, L.; Li, H.; Han, L.; Jing, W.; Wu, X.; Ullah, S.; Liu, R.; Wu, Y.; Xu, J. The Novel Interplay between Commensal Gut Bacteria and Metabolites in Diet-Induced Hyperlipidemic Rats Treated with Simvastatin. J. Proteome Res. 2022, 21, 808–821.
- Hussain, K.; Xavier, A. Rosuvastatin-related rhabdomyolysis causing severe proximal paraparesis and acute kidney injury. BMJ Case Rep. 2019, 12, e229244.
- Graham, D.J.; Staffa, J.A.; Shatin, D.; Andrade, S.E.; Schech, S.D.; La Grenade, L.; Gurwitz, J.H.; Chan, K.A.; Goodman, M.J.; Platt, R. Incidence of hospitalized rhabdomyolysis in patients treated with lipid-lowering drugs. JAMA 2004, 292, 2585–2590.
- Obayashi, H.; Kobayashi, N.; Nezu, Y.; Yamoto, T.; Shirai, M.; Asai, F. Plasma 2-hydroxyglutarate and hexanoylcarnitine levels are potential biomarkers for skeletal muscle toxicity in male Fischer 344 rats. J. Toxicol. Sci. 2017, 42, 385–396.
- Laaksonen, R. STOMPing forward: Statins, muscle complaints and CK. Atherosclerosis 2013, 230, 256–257.
- Elbadawi-Sidhu, M.; Baillie, R.A.; Zhu, H.; Chen, Y.I.; Goodarzi, M.O.; Rotter, J.I.; Krauss, R.M.; Fiehn, O.; Kaddurah-Daouk, R. Pharmacometabolomic signature links simvastatin therapy and insulin resistance. Metabolomics 2017, 13, 11.
- Pallares-Mendez, R.; Aguilar-Salinas, C.A.; Cruz-Bautista, I.; Del Bosque-Plata, L. Metabolomics in diabetes, a review. Ann. Med. 2016, 48, 89–102.
- Ooga, T.; Sato, H.; Nagashima, A.; Sasaki, K.; Tomita, M.; Soga, T.; Ohashi, Y. Metabolomic anatomy of an animal model revealing homeostatic imbalances in dyslipidaemia. Mol. Biosyst. 2011, 7, 1217–1223.
- Christensen, J.J.; Ulven, S.M.; Retterstol, K.; Narverud, I.; Bogsrud, M.P.; Henriksen, T.; Bollerslev, J.; Halvorsen, B.; Aukrust, P.; Holven, K.B. Comprehensive lipid and metabolite profiling of children with and without familial hypercholesterolemia: A cross-sectional study. Atherosclerosis 2017, 266, 48–57.
- Zhao, Z.; Du, S.; Shen, S.; Luo, P.; Ding, S.; Wang, G.; Wang, L. Comparative efficacy and safety of lipid-lowering agents in patients with hypercholesterolemia: A frequentist network meta-analysis. Medicine 2019, 98, e14400.
- Artenstein, A.W.; Opal, S.M. Proprotein convertases in health and disease. N. Engl. J. Med. 2011, 365, 2507–2518.
- Pecin, I.; Hartgers, M.L.; Hovingh, G.K.; Dent, R.; Reiner, Z. Prevention of cardiovascular disease in patients with familial hypercholesterolaemia: The role of PCSK9 inhibitors. Eur. J. Prev. Cardiol. 2017, 24, 1383–1401.
- Ference, B.A.; Robinson, J.G.; Brook, R.D.; Catapano, A.L.; Chapman, M.J.; Neff, D.R.; Voros, S.; Giugliano, R.P.; Smith, G.D.; Fazio, S.; et al. Variation in PCSK9 and HMGCR and Risk of Cardiovascular Disease and Diabetes. N. Engl. J. Med. 2016, 375, 2144–2153.
- Sabatine, M.S.; Giugliano, R.P.; Keech, A.C.; Honarpour, N.; Wiviott, S.D.; Murphy, S.A.; Kuder, J.F.; Wang, H.; Liu, T.; Wasserman, S.M.; et al. Evolocumab and Clinical Outcomes in Patients with Cardiovascular Disease. N. Engl. J. Med. 2017, 376, 1713–1722.
- Schwartz, G.G.; Steg, P.G.; Szarek, M.; Bhatt, D.L.; Bittner, V.A.; Diaz, R.; Edelberg, J.M.; Goodman, S.G.; Hanotin, C.; Harrington, R.A.; et al. Alirocumab and Cardiovascular Outcomes after Acute Coronary Syndrome. N. Engl. J. Med. 2018, 379, 2097–2107.
- Gallego-Colon, E.; Daum, A.; Yosefy, C. Statins and PCSK9 inhibitors: A new lipid-lowering therapy. Eur. J. Pharmacol. 2020, 878, 173114.
- Di Minno, A.; Orsini, R.C.; Chiesa, M.; Cavalca, V.; Calcaterra, I.; Tripaldella, M.; Anesi, A.; Fiorelli, S.; Eligini, S.; Colombo, G.I.; et al. Treatment with PCSK9 Inhibitors in Patients with Familial Hypercholesterolemia Lowers Plasma Levels of Platelet-Activating Factor and Its Precursors: A Combined Metabolomic and Lipidomic Approach. Biomedicines 2021, 9, 1073.
- Ohta, T.; Masutomi, N.; Tsutsui, N.; Sakairi, T.; Mitchell, M.; Milburn, M.V.; Ryals, J.A.; Beebe, K.D.; Guo, L. Untargeted metabolomic profiling as an evaluative tool of fenofibrate-induced toxicology in Fischer 344 male rats. Toxicol. Pathol. 2009, 37, 521–535.
- Patterson, A.D.; Slanar, O.; Krausz, K.W.; Li, F.; Hofer, C.C.; Perlik, F.; Gonzalez, F.J.; Idle, J.R. Human urinary metabolomic profile of PPARalpha induced fatty acid beta-oxidation. J. Proteome Res. 2009, 8, 4293–4300.
- Lu, Y.; Boekschoten, M.V.; Wopereis, S.; Muller, M.; Kersten, S. Comparative transcriptomic and metabolomic analysis of fenofibrate and fish oil treatments in mice. Physiol. Genom. 2011, 43, 1307–1318.
- Hertog, M.G.; Feskens, E.J.; Hollman, P.C.; Katan, M.B.; Kromhout, D. Dietary antioxidant flavonoids and risk of coronary heart disease: The Zutphen Elderly Study. Lancet 1993, 342, 1007–1011.
- Mink, P.J.; Scrafford, C.G.; Barraj, L.M.; Harnack, L.; Hong, C.P.; Nettleton, J.A.; Jacobs, D.R., Jr. Flavonoid intake and cardiovascular disease mortality: A prospective study in postmenopausal women. Am. J. Clin. Nutr. 2007, 85, 895–909.
- Grosso, G. Effects of Polyphenol-Rich Foods on Human Health. Nutrients 2018, 10, 1089.
- Sommella, E.; Badolati, N.; Riccio, G.; Salviati, E.; Bottone, S.; Dentice, M.; Campiglia, P.; Tenore, G.C.; Stornaiuolo, M.; Novellino, E. A Boost in Mitochondrial Activity Underpins the Cholesterol-Lowering Effect of Annurca Apple Polyphenols on Hepatic Cells. Nutrients 2019, 11, 163.
- Tenore, G.C.; Caruso, D.; Buonomo, G.; D’Avino, M.; Campiglia, P.; Marinelli, L.; Novellino, E. A Healthy Balance of Plasma Cholesterol by a Novel Annurca Apple-Based Nutraceutical Formulation: Results of a Randomized Trial. J. Med. Food 2017, 20, 288–300.
- Ding, Z.; Hani, A.; Li, W.; Gao, L.; Ke, W.; Guo, X. Influence of a cholesterol-lowering strain Lactobacillus plantarum LP3 isolated from traditional fermented yak milk on gut bacterial microbiota and metabolome of rats fed with a high-fat diet. Food Funct. 2020, 11, 8342–8353.
- Hughes, D.A. Plant polyphenols: Modifiers of immune function and risk of cardiovascular disease. Nutrition 2005, 21, 422–423.
- Zhou, M.; Wang, S.; Zhao, A.; Wang, K.; Fan, Z.; Yang, H.; Liao, W.; Bao, S.; Zhao, L.; Zhang, Y.; et al. Transcriptomic and metabonomic profiling reveal synergistic effects of quercetin and resveratrol supplementation in high fat diet fed mice. J. Proteome Res. 2012, 11, 4961–4971.




